Abstract
Deregulated expression of HOXB13 in a subset of estrogen receptor-positive breast cancer patients treated with tamoxifen monotherapy is associated with an aggressive clinical course and poor outcome. Because the ovary is another hormone-responsive organ, we investigated whether HOXB13 plays a role in ovarian cancer progression. We show that HOXB13 is expressed in multiple human ovarian cancer cell lines and tumors and that knockdown of endogenous HOXB13 by RNA interference in human ovarian cancer cell lines is associated with reduced cell proliferation. Ectopic expression of HOXB13 is capable of transforming p53−/− mouse embryonic fibroblasts and promotes cell proliferation and anchorage-independent growth in mouse ovarian cancer cell lines that contain genetic alterations in p53, myc, and ras. In this genetically defined cell line model of ovarian cancer, we demonstrate that HOXB13 collaborates with activated ras to markedly promote tumor growth in vivo and that HOXB13 confers resistance to tamoxifen-mediated apoptosis. Taken together, our results support a pro-proliferative and pro-survival role for HOXB13 in ovarian cancer.
Keywords: estrogen, Ras, tamoxifen, homeobox, mouse model
The HOX family of homeobox genes is an important group of developmental transcriptional regulators that are critical for various aspects of differentiation and morphogenesis (1). Similar to other genes that regulate normal growth and differentiation, HOX genes have been implicated in different aspects of the oncogenic process, because ectopic expression of HOX genes promotes cellular transformation in vitro and tumorigenesis in vivo (2). Of particular interest to human tumorigenesis is the observation that various human tumors including breast, colon, prostate, and lung carcinomas display altered HOX gene expression (3–7). Several HOX family members have been implicated in ovarian cancer differentiation (8, 9), although it is unknown whether HOX genes play a direct role in ovarian cancer progression.
We recently demonstrated that dysregulated HOXB13 expression in human breast cancer is directly correlated with poor clinical outcome in estrogen receptor (ER)-positive breast cancer patients treated with tamoxifen monotherapy (10). In preliminary functional studies, we demonstrated that ectopic expression of HOXB13 in a nontransformed human mammary epithelial cell confers increased cell migration and invasion, two characteristics associated with tumor aggressiveness (10). Consistent with a possible role in human tumorigenesis, others have recently shown that HOXB13 is overexpressed in human endometrial, ovarian, and cervical carcinomas and that overexpression of HOXB13 is associated with the invasiveness of ovarian and endometrial cancer cells (11–13). Collectively, these observations suggest that HOXB13 may play an important role in tumors arising from endocrine-responsive organs. Herein, we characterized the expression of HOXB13 in ovarian cancer cell lines and tumors. Furthermore, we investigated the potential growth modulatory role of HOXB13 in vitro and in a genetically defined mouse model of ovarian cancer.
Results and Discussion
HOXB13 Is Expressed in Multiple Human Ovarian Cancer Cell Lines and Tumors.
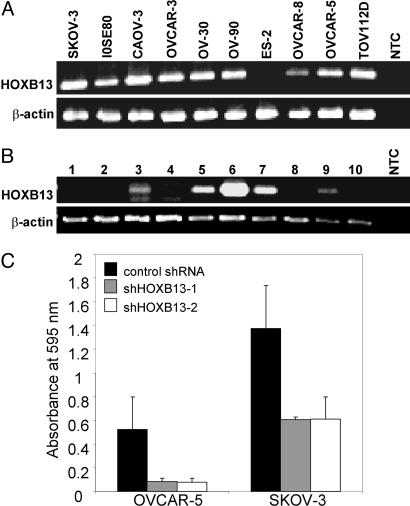
We have recently demonstrated in a cohort of 42 sporadic breast cancer patients that HOXB13 is markedly up-regulated in ≈30% of human breast cancers, whereas little (<15% of all cases) or no expression (>85% of all cases) is observed in patient-matched adult normal breast epithelium (10). To determine the prevalence of HOXB13 gene expression in ovarian cancer, we performed RT-PCR analysis in 10 different human ovarian cell lines and 10 primary ovarian tumors. HOXB13 was expressed in all but one ovarian cancer cell line (Fig. 1A) and in 5 of 9 ovarian carcinomas that were examined (Fig. 1B). These findings are consistent with the previous observations that HOXB13 demonstrates high expression in a subset of ovarian cancers (12) with little to no expression in normal ovarian tissue (12, 14).
Fig. 1.
HOXB13 is expressed in a subset of human ovarian carcinoma cell lines and primary tumors, and its inhibition is associated with decreased proliferation. (A and B) RT-PCR analysis of HOXB13 expression in (A) SKOV-3, IOSE80, CAOV-3, OVCAR-3, OVCAR-5, OVCAR-8, OV-30, OV-90, ES-2, and TOV112D cell lines and in (B) human ovarian carcinoma tissues. Case 1 is a clear cell carcinoma and cases 2–9 are papillary serous carcinoma subtype. Case 10 is a serous papillary carcinoma of the fallopian tube. Amplification of β-actin cDNA was used as a normalizing control. NTC denotes no template control PCR. (C) Endogenous HOXB13 inhibition in OVCAR-5 and SKOV-3 cells by lentiviral RNAi. shHOXB13-1 and shHOXB13-2 represent two different shRNA constructs targeting HOXB13. GFP-targeting shRNA was used as a control. Cell proliferation was assessed by measuring absorbance of the cell-associated dye at 595 nm. Error bars represent the standard deviation of three independent lentiviral infections and cultures.
Knockdown of Endogenous HOXB13 in Human Ovarian Cancer Cell Lines Is Associated with Reduced Cell Proliferation.
To determine the functional role of endogenous HOXB13, we used an RNAi approach in human ovarian cancer cell lines. To efficiently inhibit endogenous HOXB13, we used lentiviral vectors encoding small hairpin RNA (shRNA) constructs targeting HOXB13. Lentiviral infection resulted in a 50–95% knockdown of endogenous HOXB13 as assessed by real-time quantitative RT-PCR (data not shown). To determine the effect of shRNA on cell growth, we performed colony growth assays and assessed proliferation by measuring light absorbance of cell-associated crystal-violet dye. Compared with a nontarget shRNA control, the HOXB13-directed shRNA lentiviral constructs (shHOXB13-1 and shHOXB13-2) resulted in decreased colony formation in SKOV-3 and OVCAR-5 cell lines (Fig. 1C). These results support the hypothesis that expression of HOXB13 modulates growth of ovarian cancer cell lines.
HOXB13 Induces Spindle-Like Morphology, Loss of Contact Inhibition, Anchorage-Independent Cell Proliferation, and Decreased Apoptosis.
The observation that HOXB13 is expressed in a high percentage of human ovarian cancer cell lines and tumors, and that the expression of HOXB13 in human breast cancer is associated with a more aggressive clinical course compared with breast tumors that do not express HOXB13, raises the possibility that HOXB13 may play an important role in tumor progression. The possibility that HOXB13 promotes ovarian tumor growth was tested by ectopic expression of this gene by replication-competent avian leukosis virus long terminal repeat with splice acceptor (RCAS) retroviral delivery in T1 ovarian cancer cells, a cell line generated from mouse ovarian surface epithelial cells that contains genetic alterations in p53, c-myc, and K-ras (15–17) and has undetectable expression of endogenous HOXB13 by RT-PCR analysis (Fig. 2A) and Western blotting (Fig. 2B). These cells display typical epithelial morphology, are sensitive to contact inhibition, and are capable of forming colonies in soft agar. Infection of T1 cells with the RCAS-GFP retrovirus (henceforth designated T1-GFP cells) did not change any of these properties in culture. Ectopic expression of the human HOXB13 in T1 cells resulted in expression of HOXB13 mRNA (Fig. 2A) and protein (Fig. 2B), and, as expected, the human HOXB13 protein exhibited nuclear localization (Fig. 2C). T1 cells infected with RCAS-HOXB13 (T1-HOXB13 cells) displayed a distinct phenotypic change, characterized by a spindle-like morphology (Fig. 2D). When grown at high density, the T1-HOXB13 cells displayed a loss of cell contact inhibition that was accompanied by three-dimensional growth (piling up of cells), diminished cell cohesion (Fig. 2E), a reduced rate of apoptosis (Fig. 2G), and an enhanced rate of proliferation (Fig. 2H) with decreased G1 arrest (Fig. 2I) compared with T1-GFP cells. Furthermore, in an anchorage-independent growth assay, T1-HOXB13 cells displayed rapid proliferation in soft agar compared with the control T1-GFP cells (Fig. 2F).
Fig. 2.
Ectopic expression of HOXB13 in murine T1 ovarian cancer cells. (A) RT-PCR detection of endogenous and ectopic HOXB13 in T1 cells infected with the control RCAS-GFP retrovirus (T1-GFP) and T1 cells infected with the RCAS-HOXB13 retrovirus (T1-HOXB13). (B and C) Western blotting (B) and immunofluorescence (C) detection of the human HOXB13 protein by using a rabbit polyclonal antibody to HOXB13. (D and E) The morphology of T1-GFP and T1-HOXB13 cells in monolayer culture grown at low (D) and high (E) cell densities. (F) Representative images of duplicate independent assays of cell proliferation in soft agar. (G) Representative images of TUNEL staining in T1-GFP and T1-HOXB13 cells grown in monolayer culture at high cell density. (H and I) Growth curve plot of cells (×104) (H) and cell cycle analysis (I) of T1-GFP and T1-HOXB13 cells; data are representative of duplicate independent experiments.
HOXB13 Enhances Tumor Growth.
To determine whether HOXB13 can induce cell transformation and tumor progression in vivo, mouse embryonic fibroblasts (MEFs) were infected in vitro with the retroviral constructs RCAS-GFP and RCAS-HOXB13. HOXB13 was unable to transform wild-type MEFs; however, it induced a tumorigenic transformation of MEFs from p53−/− embryos. Tumor development occurred within 45 days in three of seven mice injected with the p53−/− plus RCAS-HOXB13 MEFs and in zero of seven mice injected with the p53−/− plus RCAS-GFP MEFs (Fig. 3A).
Fig. 3.
HOXB13 induces oncogenic transformation of p53−/− MEFs as well as rapid progression of mouse ovarian epithelial tumors in the presence of activated ras. (A) Tumor formation in mice injected s.c. with p53−/− MEFs that were infected with RCAS-HOXB13. The control p53−/− MEFs were infected with RCAS-GFP. (B–D) Enhanced tumor growth in mice that were injected s.c. (B) or i.p. (C and D) with T1 mouse ovarian cancer cells that were infected with RCAS-HOXB13 in comparison with the T1 cells that were infected with RCAS-GFP. (E–H) Influence of HOXB13 on tumor growth in mice that were injected s.c. with T22 cells (E), vector-transduced T22 cells (F), and T22 cells with activated K-ras (G) or H-ras (H).
To explore the potential collaborative interaction of HOXB13 with known signaling pathways, we ectopically expressed HOXB13 in mouse ovarian epithelial tumor cell lines that contain various combinations of genetic alterations in p53, c-myc, K-ras, and Akt. We have generated four independent mouse ovarian cancer cell lines: T1 and T11 cells that contain genetic alterations in p53, c-myc, and K-ras, and T2 and T22 cell lines with alterations in p53, c-myc, and Akt (Table 1). Two independent T1 and T11 cell lines give rise to ≈0.5 cm3 s.c. tumors 3–4 weeks after injection into nude mice, whereas infection of the T1 and T11 cell lines with the control retrovirus (RCAS-GFP) did not alter the rate of tumor growth. However, infection with RCAS-HOXB13 in T1 and T11 cells resulted in extremely rapid tumor growth and formation of large (≥1.4 cm3) tumors between 9 and 19 days after s.c. injection in 30 of 30 mice (Fig. 3B and Table 1). Similarly, i.p. injection of T1+RCAS-HOXB13 (T1-HOXB13) cells into nude mice resulted in ascites accumulation and tumor formation in four of four mice in 12 days (Fig. 3C), whereas the mice injected with T1 cells infected with RCAS-GFP (T1-GFP) did not show any signs of ascites or tumor formation at that time (Fig. 3D). HOXB13 was more effective in inducing tumor growth in T1 and T11 cells than any other oncogene that we have tested to date, including activated Akt1, Her-2/neu, and middle T-antigen (data not shown). Ectopic expression of HOXB13 without the homeobox domain in T1 cells did not result in enhanced tumor growth (data not shown), implicating DNA binding function in pro-growth activity of HOXB13.
Table 1.
Ability of HOXB13 to enhance growth of tumors with defined genetic alterations
| Mouse ovarian cancer cell line | Genetic alterations | No. of tumors in which V(HOXB13)/V(GFP) > 2 | Days between injection and tumor isolation* |
|---|---|---|---|
| T1 | p53−/−, myc, K-ras | 30/30 | 9–15* |
| T11 | p53−/−, myc, K-ras | 4/4 | 19* |
| T2 | p53−/−, Akt, myc | 0/10 | 20–55 |
| T22 | p53−/−, Akt, myc | 0/4 | 16–27 |
| T22+ pBabe | p53−/−, Akt, myc | 0/10 | 14 |
| T22+K-ras | p53−/−, Akt, myc, K-ras | 10/12 | 14* |
| T22+H-ras | p53−/−, Akt, myc, H-ras | 3/3 | 8* |
V, volume.
*Mice were killed when tumor size on one or both flanks exceeded 1.4 cm3.
In contrast to T1 and T11 cells, when T2 cells infected with RCAS-HOXB13 (T2-HOXB13) were injected into nude mice, the resulting tumors were of comparable size to those generated with uninfected T2 cells and T2 cells infected with RCAS-GFP (T2-GFP). The same results were obtained with an independently derived cell line, T22 (Table 1), indicating that HOXB13 does not enhance tumor proliferation in mouse ovarian cancer cell lines that contain genetic alterations in p53, c-myc, and Akt.
Taken together, these data demonstrate that HOXB13 is capable of enhancing tumorigenesis in the context of genetic alterations in p53, c-myc, and K-ras, but not in the context of genetic alterations in p53, c-myc, and Akt, suggesting that HOXB13 may collaborate with K-ras. To investigate the potential collaborative interaction of HOXB13 with ras, we stably transfected T22 cells with an empty pBabe vector (T22+vector), pBabe containing K-ras (T22+K-ras), or pBabe containing H-ras (T22+H-ras). These cell lines were then infected with RCAS-GFP or RCAS-HOXB13. Unlike T22 and T22+vector cells, which resulted in the same rate of growth whether they were infected with RCAS-GFP or RCAS-HOXB13 (Fig. 3 E and F), the T22+K-ras and T22+H-ras cells resulted in significantly accelerated tumor growth when infected with RCAS-HOXB13 in comparison with identical cells infected with RCAS-GFP (Fig. 3 G and H).
Herein, we have demonstrated through ectopic expression and RNAi-mediated inhibition experiments that HOXB13 is a pro-proliferative modulator of ovarian cancer cell growth. Thus far, studies pertaining to the growth modulatory role of HOXB13 in human tumors have been limited to prostate and renal cell carcinoma cell lines. Interestingly, the results of these studies, in contrast to our findings, suggest that HOXB13 plays a tumor suppressor role in the kidney and prostate (14, 18, 19). These contradictory findings are not necessarily surprising, because it is well known that the activity of HOX proteins depends on the cell in which the protein is acting (20). A significant difference between ovary and organs such as prostate and kidney is that HOXB13 is absent in the normal ovary, whereas it is expressed at high levels in the normal prostate and kidney (14, 18, 19, 21), where it is thought to play a role in epithelial cell differentiation (21, 22). The HOXB13 expression pattern is reversed in cancers derived from these organs; HOXB13 is up-regulated in ovarian cancers (12) and down-regulated in renal and prostate carcinoma cell lines (14, 18, 19). Thus, it is likely that tissue- and cell-specific contextual elements, such as endocrine target organ-specific coactivators and corepressors, may play a vital role in dictating HOXB13 function.
HOXB13 Abrogates the Antagonistic Effect of Tamoxifen.
It is known that HOX genes are tightly regulated by hormones during development and in adult reproductive tissues (reviewed in ref. 23). Interest in the functional role of HOXB13 in tumorigenesis originated from our previous observation that HOXB13 is overexpressed in tamoxifen-resistant human breast cancers. We recently demonstrated that HOXB13 expression is repressed by estradiol and that this repression is abrogated in the presence of tamoxifen (24). These findings suggest a possible functional link between HOXB13 and tamoxifen resistance (i.e., aggressive tumor growth manifested as shortened disease-free survival). T1-GFP and T1-HOXB13 cells express estrogen receptor-α (ER) (Fig. 4A), and in vitro exposure of these cells to estrogen results in increased proliferation, indicating that these cells possess a functional ER (Fig. 4B). To determine whether HOXB13 is capable of modulating ER transcriptional activity, we evaluated whether ectopically expressed HOXB13 could modulate the ability of endogenously expressed ER to activate a promoter containing estrogen response element (ERE) sequences. Introduction of HOXB13 into T1 mouse ovarian cancer cell line results in enhanced estrogen-dependent ERE-luciferase expression (Fig. 4C). To ascertain whether the HOXB13-enhanced transcriptional activation depends on ER, we performed ERE-transcriptional reporter assays in cells in which ER expression was knocked down with Faslodex, a drug that selectively accelerates ER degradation (25). The knockdown of ER protein expression in T1-HOXB13 (Fig. 4C) cells abrogates HOXB13-enhanced transcriptional activation supporting the notion that HOXB13-mediated ERE promoter activity is ER dependent. To determine the possible role of HOXB13 in tamoxifen resistance, we measured and compared tamoxifen-mediated inhibition of ERE-transcriptional activity and tamoxifen-induced apoptosis in T1 cells expressing or lacking the expression of HOXB13. As expected, tamoxifen inhibits ERE-transcriptional activation and induces apoptosis in T1-GFP cells. However, T1 cells expressing HOXB13 are resistant to the inhibitory effects of tamoxifen on ERE-mediated transcriptional activation (Fig. 4D). Furthermore, HOXB13-expressing T1 cells are less sensitive to tamoxifen-induced apoptosis (Fig. 4E).
Fig. 4.
Effect of estradiol and tamoxifen on T1-GFP and T1-HOXB13 cells. (A) Western blot analysis of ER-α and β-actin in T1-GFP and T1-HOXB13 cells. (B) Estradiol responsiveness of T1-GFP and T1-HOXB13 cells. (C) ERE-luc responsiveness requires ER expression. T1-GFP and T1-HOXB13 cells were treated with vehicle or 200 nM Falsodex for 24 h before and 48 h after cells were cotransfected with pERE-luc reporter plasmid and pRL normalization plasmid. Results presented are average of triplicate determinations from a representative assay of three independent experiments. Inset demonstrates Western blot analysis of ER-α and GAPDH for the corresponding samples. (D) Effect of tamoxifen on ERE-luc responsiveness. T1-GFP and T1-HOXB13 cells were cotransfected with pERE-luc and pRL and then treated with vehicle or tamoxifen at the doses shown for 48 h. Luciferase assays were performed, and results are representative of three independent experiments. (E) Quantitation of apoptosis in T1-GFP and T1-HOXB13 ovarian cancer cell lines in the indicated doses of tamoxifen. Apoptotic cell death was determined by FACS analysis of phycoerythrin-conjugated annexin V staining. The error bars represent standard deviation in triplicate experiments.
Taken together, our results indicate that HOXB13 has a pro-proliferative and pro-survival function in ovarian cancer cells and that HOXB13 may play an important role in the development of tamoxifen resistance in patients with ER-positive ovarian cancer. Although the majority of primary ovarian tumors express ERs (26), the efficacy of endocrine therapies has not been adequately evaluated in ovarian cancer. Clinical studies of tamoxifen in a small number of chemoresistant ovarian cancer patients have demonstrated that a small subset of unselected patients responds to tamoxifen treatment (26–28). However, a recent study has demonstrated that preselection of ovarian cancer patients according to ER status results in a significantly higher percentage of patients responding to antihormonal therapy (29). This observation is analogous to that of human breast cancer in which there is an endocrine-sensitive subgroup among ER-positive ovarian cancers. In view of the observation that HOXB13 is expressed in a subset of human ovarian cancers and that HOXB13 is associated with tamoxifen resistance, it is possible that HOXB13 may serve not only as a biomarker but also as a functional regulator of antihormonal resistance in human ovarian cancer. Our observations and the recent resurgence of interest in endocrine therapies for ovarian cancer (29) merit further investigation of these hypotheses at the clinical and basic science levels.
Materials and Methods
Cell Lines.
SKOV-3, CAOV-3, OVCAR-3, OV-30, OV-90, ES-2, and TOV112D cell lines were obtained from American Type Culture Collection (Manassas, VA). IOSE80 cells were provided by Nelly Auersperg (University of British Columbia, Vancouver, BC, Canada), and OVCAR-5 and -8 cells were provided by Michael Seiden (Fox Chase Cancer Center, Philadelphia, PA). The genetically defined mouse ovarian epithelial cell lines were generated and maintained as described in refs. 15–17.
Analyses of Cell Proliferation, Cell Cycle, Apoptosis, and Growth in Soft Agar.
To monitor cell proliferation, cells were plated in triplicate at 4.4 × 104 cells/cm2, media replaced daily, harvested on days 2, 3, 4, 6, and 8, and directly counted by using trypan blue. For cell cycle profiling analysis, an aliquot (1 × 106 cells) of cells from a proliferation assay was washed with PBS, resuspended in 70% ethanol (20°C), stored at 4°C for 24 h, and incubated in propidium iodide staining solution (1 mg/ml RNase in 20 mg/ml propidium iodide), and DNA content was analyzed by flow cytometry. The presence of apoptosis in confluent cells was determined by TUNEL assay (Chemicon, Temecula, CA). For the soft agar assay, 3 × 104 cells were added to growth media containing 0.4% agar and layered on top of solidified 0.5% agar beds. Cells were counted and photographed after a 14-day incubation; assays were conducted in duplicate triplicates in two independent experiments. Quantitation of apoptosis by annexin V/propidium iodide staining was performed as described previously in ref. 30.
RT-PCR.
Total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA), DNaseI treated for 30 min at 37°C, phenol extracted, and ethanol precipitated, and then the cDNA was synthesized from total RNA by using SuperScriptII Reverse Transcriptase (Invitrogen). The following primer sequences were used: 5′-mouse HOXB13, ATGCCTGCGGTCCCTGGTGA; 3′-mouse HOXB13, GGCTCCAACGCTGATGCCAA; 5′-human HOXB13, CCGCCCCACTGAGTTTGCCTTCTA; 3′-human HOXB13, TTGCGCCTCTTGTCCTTGGTGATG; 5′-β-actin, AGCGCAAGTACTCCGTGTG; and 3′-β-actin, GACTGGGCCATTCTCCTTAG.
Plasmid Constructs and Transfection.
HOXB13 was cloned into pcDNA3.1(+) by using standard PCR from pDNR-HOXB13 provided by Joshua LaBaer (Harvard Medical School, Boston, MA). Gateway technology (Invitrogen) was used for the cloning of HOXB13 into the retroviral RCAS vector (31). HOXB13 without homeobox domain (1–215 aa) and HOXB13 homeobox domain (216–284 aa) were cloned into pcDNA3.1(+) into BamHI and EcoRI sites by using standard PCR method. The RCAS-HOXB13, RCAS-GFP, and pBabe-puro-K-rasG12D and -H-rasV12 viral supernatants were prepared as described in ref. 15.
In Vitro Infection of Cells and Mouse Injections.
MEFs from β-actin-TVA and β-actin-TVA/p53−/− mice, and the ovarian cancer cell lines, were infected as described in ref. 15. The infection efficiencies were determined by GFP expression of infected cells. The T22+K-ras and T22+H-ras cell lines were generated by transfecting T22 cells with pBabe-puro-K-rasG12D and -H-rasV12, respectively, followed by selection with 2.5 μg/ml puromycin. Mice were injected as described in ref. 16.
Antibodies and Western Blotting.
A KLH fusion protein containing 16 aa (EPGNYATLDGAKDIEC) from the N terminus of human HOXB13 was generated in the MGH peptide core facility and used to produce antisera under contractual agreement with Cocalico Biologicals (Reamstown, PA). The antisera recognized the KLH-HOXB13 fusion peptide in ELISAs and ectopically expressed HOXB13 in T1 cells by Western blot analysis.
Immunofluorescense Microscopy.
Cells grown on glass chamber slides (Nalge, Naperville, IL) were fixed with 4% paraformaldehyde at room temperature for 15 min. The cells were then washed with PBS, permeabilized for 4 min with 0.1% Triton X-100 in PBS, and blocked with 3% goat serum for 30 min at room temperature. Subsequently, the cells were incubated with the HOXB13 antisera (1:300) in PBS with 1.5% goat serum at 4°C overnight. After washing with PBS, the cells were incubated with fluorescence-labeled secondary antibodies for 60 min at room temperature. The slides were embedded in mounting medium with DAPI (Vector Laboratories, Burlingame, CA) and analyzed with a Carl Zeiss (Oberkochen, Germany) fluorescence microscope.
Lentiviral Production, Infection, and Colony Formation Assay.
The shRNA lentiviral constructs were purchased from Sigma–Aldrich (St. Louis, MO), and high-titer amphotrophic lentiviral stocks were generated as described in ref. 32. The targeted shRNA sequences for HOXB13 were CGCCAGATTACCATCTGGTTT and CTGTGGACAGTTACCAGTCTT, and the nontarget shRNA control sequence was CAACAAGATGAAGAGCACCAA. Infected cells were selected in 0.5 μg/ml puromycin for 4 days, counted, plated at 0.5 × 104 or 1 × 104 cells per plate, maintained in DMEM–FCS with 0.5 μg/ml puromycin for 14–21 days, and stained with crystal violet as described in ref. 15.
Luciferase Assays.
T1-GFP and T1-HOX were seeded at six-well plate at 2 × 105/well and incubated at 5% CO2 and 37°C for 24 h before cells were transfected with pERE-luc (33) and pRL by using FuGene 6 following manufacturer's instruction. Cells were collected in passive cell lysis buffer (250 μl/well), and luciferase activities in 20 μl of lysate were determined by using dual-luciferase reporter assay kit (Promega, Madison, WI) following the manufacturer's instruction. All luciferase assays were performed and reporter activities were normalized to the internal Renilla luciferase control. Faslodex and tamoxifen were obtained from Acros (Geel, Belgium) and Sigma–Aldrich, respectively.
[3H]Thymidine Incorporation Assays.
Cells were seeded at 5 × 105 cells per well in six-well plates in triplicate. On the next day, the cells were washed and placed in phenol-free medium with 5% charcoal-stripped FBS for 48 h. Estradiol (E2) or the vehicle (ETOH) was added for 16 h, followed by addition of [3H]thymidine (5 μCi/well; DuPont/NEN, Boston, MA) for 8 h. DNA was harvested as described in ref. 34.
Acknowledgments
We thank Daniel Haber and Leif Ellisen for helpful discussions and reading of the manuscript; Stacie Loftus (National Institutes of Health, Bethesda, MD), Yi Li (Baylor College of Medicine, Houston, TX), Brian Lewis (University of Massachusetts Medical School, Worcester, MA), David Tuveson (University of Pennsylavnia, Philadelphia, PA), and Gregory Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for viral plasmid constructs; and Kristy Daniels for assistance in the preparation of the manuscript. This work was supported by National Institutes of Health Grants R01-CA103924 (to S.O.) and R01-1CA112021-01 (to D.C.S.); Department of Defense Grants W81XWH-04-1-0606 (to D.C.S.) and W81XWH-04-1-0485 (to S.O.), and Ovarian Cancer Research Training Program Award DAMD17-03-1-0161 (to J.M.); Susan G. Komen Breast Cancer Foundation Grant BCTR0402932 (to D.C.S.); a Pilot Project Award under Ovarian Specialized Program of Research Excellence P50CA105009 (to S.O.); and the Avon Foundation (D.C.S.). Support from the Avon Foundation was restricted to the human in vitro-based experiments.
Abbreviations
- ER
estrogen receptor
- ERE
estrogen response element
- MEF
mouse embryonic fibroblast
- RCAS
replication-competent avian leukosis virus long terminal repeat with splice acceptor
- shRNA
small hairpin RNA.
Footnotes
The authors declare no conflict of interest.
References
- 1.Abate-Shen C. Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 2.Aberdam D, Negreanu V, Sachs L, Blatt C. Mol Cell Biol. 1991;11:554–557. doi: 10.1128/mcb.11.1.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E, et al. Proc Natl Acad Sci USA. 2000;97:12776–12781. doi: 10.1073/pnas.97.23.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, Schirripa O, Lucia MS, Nordeen SK. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 5.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 6.Tiberio C, Barba P, Magli MC, Arvelo F, Le Chevalier T, Poupon MF, Cillo C. Int J Cancer. 1994;58:608–615. doi: 10.1002/ijc.2910580426. [DOI] [PubMed] [Google Scholar]
- 7.Vider BZ, Zimber A, Hirsch D, Estlein D, Chastre E, Prevot S, Gespach C, Yaniv A, Gazit A. Biochem Biophys Res Commun. 1997;232:742–748. doi: 10.1006/bbrc.1997.6364. [DOI] [PubMed] [Google Scholar]
- 8.Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Nat Med. 2005;11:531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- 9.Naora H, Montz FJ, Chai CY, Roden RB. Proc Natl Acad Sci USA. 2001;98:15209–15214. doi: 10.1073/pnas.011503998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, et al. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Lopez R, Garrido E, Pina P, Hidalgo A, Lazos M, Ochoa R, Salcedo M. Int J Gynecol Cancer. 2006;16:329–335. doi: 10.1111/j.1525-1438.2006.00350.x. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita T, Tazawa S, Yawei Z, Katayama H, Kato Y, Nishiwaki K, Yokohama Y, Ishikawa M. Int J Oncol. 2006;28:931–938. [PubMed] [Google Scholar]
- 13.Zhao Y, Yamashita T, Ishikawa M. Oncol Rep. 2005;13:721–726. [PubMed] [Google Scholar]
- 14.Jung C, Kim RS, Zhang HJ, Lee SJ, Jeng MH. Cancer Res. 2004;64:9185–9192. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- 15.Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, Varmus HE. Cancer Cell. 2002;1:53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing D, Orsulic S. Proc Natl Acad Sci USA. 2005;102:6936–6941. doi: 10.1073/pnas.0502256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing D, Orsulic S. Cell Cycle. 2005;4:1004–1006. doi: 10.4161/cc.4.8.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung C, Kim RS, Lee SJ, Wang C, Jeng MH. Cancer Res. 2004;64:3046–3051. doi: 10.1158/0008-5472.can-03-2614. [DOI] [PubMed] [Google Scholar]
- 19.Okuda H, Toyota M, Ishida W, Furihata M, Tsuchiya M, Kamada M, Tokino T, Shuin T. Oncogene. 2006;25:1733–1742. doi: 10.1038/sj.onc.1209200. [DOI] [PubMed] [Google Scholar]
- 20.Lohmann I, McGinnis W. Curr Biol. 2002;12:R514–R516. doi: 10.1016/s0960-9822(02)01025-4. [DOI] [PubMed] [Google Scholar]
- 21.Huang L, Pu Y, Hepps D, Danielpour D, Prins GS. Endocrinology. 2007;148:1235–1245. doi: 10.1210/en.2006-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Economides KD, Capecchi MR. Development (Cambridge, UK) 2003;130:2061–2069. doi: 10.1242/dev.00432. [DOI] [PubMed] [Google Scholar]
- 23.Daftary GS, Taylor HS. Endocr Rev. 2006;27:331–355. doi: 10.1210/er.2005-0018. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, Shioda T, Ma X-J, Sgroi DC. Clin Cancer Res. 2007 doi: 10.1158/1078-0432.CCR-07-0310. in press. [DOI] [PubMed] [Google Scholar]
- 25.Robertson JF. J Steroid Biochem Mol Biol. 2001;79:209–212. doi: 10.1016/s0960-0760(01)00138-8. [DOI] [PubMed] [Google Scholar]
- 26.Hatch KD, Beecham JB, Blessing JA, Creasman WT. Cancer. 1991;68:269–271. doi: 10.1002/1097-0142(19910715)68:2<269::aid-cncr2820680209>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 27.Markman M, Iseminger KA, Hatch KD, Creasman WT, Barnes W, Dubeshter B. Gynecol Oncol. 1996;62:4–6. doi: 10.1006/gyno.1996.0181. [DOI] [PubMed] [Google Scholar]
- 28.Ahlgren JD, Ellison NM, Gottlieb RJ, Laluna F, Lokich JJ, Sinclair PR, Ueno W, Wampler GL, Yeung KY, Alt D, et al. J Clin Oncol. 1993;11:1957–1968. doi: 10.1200/JCO.1993.11.10.1957. [DOI] [PubMed] [Google Scholar]
- 29.Smyth JF, Gourley C, Walker G, MacKean MJ, Stevenson A, Williams AR, Nafussi AA, Rye T, Rye R, Stewart M, et al. Clin Cancer Res. 2007;13:3617–3622. doi: 10.1158/1078-0432.CCR-06-2878. [DOI] [PubMed] [Google Scholar]
- 30.Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. J Clin Invest. 2007;117:1370–1380. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loftus SK, Larson DM, Watkins-Chow D, Church DM, Pavan WJ. DNA Res. 2001;8:221–226. doi: 10.1093/dnares/8.5.221. [DOI] [PubMed] [Google Scholar]
- 32.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 33.Catherino WH, Jordan VC. Cancer Lett. 1995;92:39–47. doi: 10.1016/0304-3835(95)03755-l. [DOI] [PubMed] [Google Scholar]
- 34.Voyno-Yasenetskaya TA, Pace AM, Bourne HR. Oncogene. 1994;9:2559–2565. [PubMed] [Google Scholar]