Abstract
A growing body of literature has examined and implicated DNA methylation as a critical epigenetic modification in T helper (Th) cell differentiation. The absence of DNA methyltransferases or methyl-binding proteins derepresses many cytokine loci, allowing their ectopic expression, while methylation of specific CpG residues is sufficient to prevent expression. Here, we characterize demethylation events of the Th2 cytokine locus control region (LCR). rad50 hypersensitive site 7 (RHS7), a hypersensitive site within this LCR, becomes demethylated in a STAT6-dependent manner and only in cells stimulated under type 2 conditions. Robust demethylation appears to require signaling contributions from both IL-4 receptor, via STAT6, and CD28, but it cannot be effected by GATA3. Finally, RHS7 is demethylated independently of cell division, consistent with an “active,” rather than passive, mechanism. Taken together, these findings firmly connect RHS7 demethylation and Th2 LCR activation in the type 2 differentiation program.
Keywords: methylation, chromatin, costimulation, cytokine, epigenetics
Methylation of CpG DNA is generally associated with transcriptional inactivity within a locus (1–3). Conversely, removal of the methyl group from cytosine signals a shift from inert chromatin to active or “open” loci. It is therefore thought that DNA methylation is an important mechanism through which transcriptional activity is controlled.
This concept has become apparent in the study of T cell differentiation, where mounting data single out DNA methylation, of all chromatin modifications, as a significant regulator of lineage commitment and cytokine production (4). Mice deficient in Dnmt1, a maintenance methyltransferase, or MBD2, a methyl-CpG binding protein, experience ectopic cytokine production in T cells, where aberrant expression of IL-4 is attributed to inappropriate demethylation and derepressed silencing of the il4 gene (5, 6). In a higher-resolution study of promoter-targeted demethylation, Bruniquel and Schwartz (7) identified key residues in the il2 promoter whose unmethylated status was both necessary and sufficient to drive IL-2 expression on T cell activation.
Whereas most methylation analyses have focused on promoter regions of genes, methylation may also play a role in nonpromoter loci such as enhancers and locus control regions (LCRs). Recently, we identified a T helper 2 (Th2) cytokine LCR and showed that changes in DNA methylation and histone acetylation within this region mirror those seen in promoters of the cytokine genes (8). This pattern of simultaneous epigenetic changes is consistent with a model of locus control whereby the cytokine gene promoters and the LCR form an active chromatin hub via intrachromosomal interactions (9). We postulate that LCR demethylation may enable trans-factor recruitment necessary for its regulatory activity in the locus. There are many other examples of lineage-specific nonpromoter locus demethylation, including the T cell receptor α (TCR-α) LCR and CNS1 of the il4 locus (10, 11).
The mechanistic details of DNA demethylation associated with gene activity have yet to be clarified. In the passive model of demethylation, a fully methylated allele, that is, an allele methylated on both strands of DNA, undergoes DNA replication during S phase to yield two hemimethylated alleles. Normally, Dnmt1 is preferentially targeted to such hemimethylated sites and in this way preserves the overall genetic pattern of methylation (12). Instead, during passive demethylation, Dnmt1 recruitment is inhibited, presumably by steric hindrance from a locus by other DNA-binding factors. Hemimethylated alleles further divide once more to give rise to fully demethylated DNA, and the methylation pattern is unable to be imprinted from parent to daughter cell.
In contrast, the active model of demethylation proposes catalytic removal of the methyl group by enzymatic activity, such as that observed in the il2 promoter upon T cell activation (7). Despite the rare evidence implicating an active mechanism, no enzyme capable of such catalytic activity in mammals has yet been identified (12). Recent reports have demonstrated that catalytic demethylation occurs through base excision repair by the DNA glycosylase/lyases DEMETER and ROS1 in Arabidopsis (13–15). A similar mechanism has been shown to be directed by the stress-responsive gene Gadd45a in Xenopus, although the enzyme directly responsible for demethylation was not identified (16).
Previously, we determined that the third CpG residue in rad50 hypersensitive site 7 (RHS7) of the Th2 LCR undergoes the most dramatic increase in demethylation in the entire IL-4 locus upon Th2 cell differentiation, from 4% of alleles demethylated in naïve T cells to 47% and 100% at days 2 and 5, respectively (8). In this article, we further characterize demethylation of RHS7. RHS7 is demethylated in a STAT6-dependent manner, but GATA3 is unable to effect this demethylation. In addition to determining the upstream factors and pathways involved in this demethylation, we find that RHS7 is demethylated via an active mechanism. Lastly, we implicate IL-2 signaling as a major determinant of Th2 LCR demethylation, providing one mechanism by which IL-2-driven Th2 differentiation may occur.
Results
Correlation of RHS7 Demethylation and IL-4 Expression.
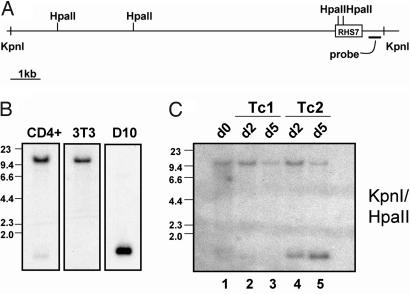
In our initial study of the Th2 cytokine LCR, we described a highly Th2-specific pattern of demethylation in one of its hypersensitive sites, RHS7 (8). Given the importance of RHS7 in il4 enhancer activity, we postulated that demethylation of this hypersensitive site would occur most strongly in cell types that expressed IL-4. We previously showed that RHS7 is fully methylated in naïve CD4+ T cells (Fig. 1B Left) and undergoes little demethylation in effector Th1 cells, in which no IL-4 is expressed. Methylation analysis conducted with the methyl-CpG sensitive restriction enzyme HpaII revealed that RHS7 also remains completely methylated in 3T3 fibroblasts, an IL-4 nonexpressor (Fig. 1B Center).
Fig. 1.
RHS7 demethylation in IL-4-expressing cell types. (A) Schematic diagram of the genomic fragment containing RHS7 of the Th2 LCR. HpaII and KpnI sites are indicated as vertical hash marks. Probe is represented by thick horizontal bar. (B) Methylation analysis of CD4+, fibroblasts, and D10 cells by Southern blotting. (C) Methylation analysis of Tc1 and Tc2 development. CD8+ T cells were stimulated under type 1 and 2 conditions for 2 and 5 days. In B and C, genomic DNA was double-digested with KpnI and HpaII, transferred, and probed as indicated. The parental band in all Southern blots is 11.5 kb, and the cleaved fragment denoting RHS7 demethylation is ≈1.2 kb.
In contrast, the Th2 clone D10 possesses the same pattern of demethylation as primary Th2 cells; that is, RHS7 is fully demethylated, indicative of long-term, stable Th2 identity (Fig. 1B Right). Finally, we assessed RHS7 methylation patterns in the T cytotoxic (Tc) CD8+ subsets Tc1 and Tc2. RHS7 undergoes very little demethylation in unactivated CD8+ and Tc1 cells (Fig. 1C, lanes 1–3). However, in the same manner as Th2 cells, Tc2 cells also demethylated at roughly the same kinetics (Fig. 1C, lanes 4 and 5), although not to the same extent, as evidenced by maintenance of the 11.5-kb parental fragment (see Fig. 1A for map) contrasting with demethylation in Th2 cells, where the parental band is extinguished at 5 days (8). Thus, RHS7 is demethylated strongly in cells that express or are activated by IL-4, consistent with its function as an important regulatory element in the Th2 LCR.
GATA3-Independent IL-4 Signaling Requirement for RHS7 Demethylation.
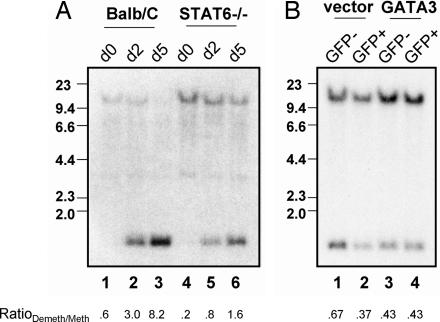
The transcription factor STAT6 is largely indispensable for optimal Th2 differentiation and IL-4 production (17). We therefore asked what role STAT6 plays in RHS7 demethylation. STAT6-deficient Th2 cells were cultured for 5 days, and genomic DNA was harvested and subjected to methylation analysis by Southern blotting. BALB/c Th2 cells underwent strong demethylation of RHS7, with complete extinction of methylated alleles occurring by day 5 (Fig. 2A, lanes 1–3). (Crude ratios of demethylated signals to methylated signals, herein called RatioDemeth/Meth, were calculated by densitometry to provide rough quantitative measurements of the extent of demethylation.)
Fig. 2.
Involvement of STAT6 but not GATA3 in RHS7 demethylation. (A) Time course of RHS7 demethylation in BALB/c and STAT6−/− Th2 cells. Methylation analysis was performed as described in Fig. 1. (B) Retroviral transduction of GATA3 in Th1 cells. Developing Th1 cells were introduced with empty vector or GATA3 after 24 h and allowed to culture for an additional 4–5 days. GFP-positive and -negative cells were sorted and analyzed for RHS7 demethylation. The RatioDemeth/Meth of densitometric values is listed below each lane.
In contrast, demethylation of RHS7 in STAT6-deficient Th2 cells was impaired compared with wild-type cells, as indicated by maintenance of the uncut 11.5-kb parental fragment and lower RatioDemeth/Meth values (Fig. 2A, lanes 4–6). This result implicates a partial, albeit important, role of STAT6 and, therefore, IL-4 signaling in effecting complete RHS7 demethylation. Furthermore, as a note of interest, our initial study of RHS7 demethylation was performed in C57BL/6 mice (8); that RHS7 also became demethylated in BALB/c Th2 cells suggests a pan-strain generality of this finding.
GATA3, the master Th2 transcription factor, has recently been implicated in regulating chromatin remodeling activity of the Th2 LCR (9). To assess the requirement of GATA3 for RHS7 demethylation, we ectopically introduced a GATA3-IRES-GFP construct into primary Th1 cells by retroviral transduction. At 4–5 days after transduction, GFP-positive cells were sorted, and DNA from these cells was analyzed by Southern blotting.
As expected, RHS7 in vector-transduced cells was demethylated to the same extent as in normal Th1 cells (Fig. 2B, lanes 1 and 2). However, GATA3 failed to restore full demethylation, because DNA from the GATA3-positive, GFP-positive population (Fig. 2B, lane 4) shared the same HpaII cleavage profile and RatioDemeth/Meth values as the uninfected and GFP-negative controls. Therefore, simply overexpressing GATA3 was not sufficient to induce RHS7 demethylation.
Critical Role for IL-2 in RHS7 Th2 LCR and il4 Gene Accessibility.
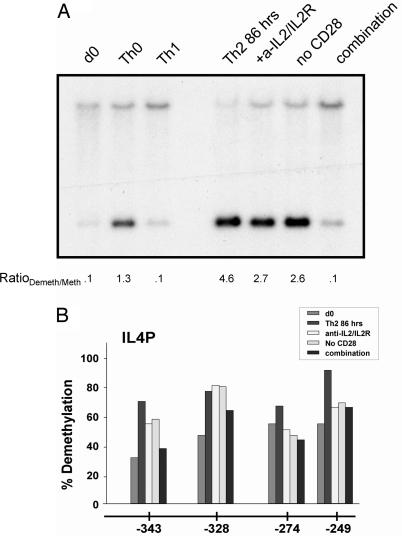
In a recent report, Paul and colleagues (18) revealed a novel requirement of IL-2 in priming T cells for IL-4 production and enabling chromatin accessibility. Therefore, we investigated whether IL-2 signaling shared any role in demethylating RHS7. Using the same experimental system as that study, we cultured Th2 cells in the presence of anti-IL-2 and anti-IL-2R (CD25) antibodies for 86 h. As shown in Fig. 3A, neutralization of IL-2 and blockade of IL-2R appeared to have a minor effect on RHS7 demethylation; these samples became demethylated to a slightly lesser extent than the Th2 control.
Fig. 3.
Effect of IL-2 neutralization and absence of costimulation on RHS7 and il4 demethylation. (A) Methylation analysis of RHS7 from 86-h cultures. T cells were stimulated and analyzed for RHS7 demethylation. Anti-IL-2 and anti-IL-2R (10 mg/ml each) were used. For Th0 conditions, only anti-CD3 and anti-CD28 stimulation were used, without neutralizing antibodies to IL-4 or IFN-γ. The RatioDemeth/Meth of densitometric values is listed below each lane. (B) Methylation analysis of the proximal il4 promoter. DNA from cells used in A were modified with bisulfite and analyzed for methylation status. Each bar represents 25–31 clones sequenced.
In a previous study, Rulifson et al. (19) found that CD28 costimulation augmented IL-4 production in T cells, although the mechanism for this action was not established. Therefore, in the same culture prepared above, we stimulated Th2 cells for 86 h without anti-CD28 antibodies to prevent costimulation. Interestingly, although blockade of CD28 costimulation did not completely abolish RHS7 demethylation, the partial inhibition mirrored that of conditions under which IL-2 was neutralized (Fig. 3A). Finally, we cultured Th2 cells under a combination of these conditions. As shown in Fig. 3A (rightmost lane), stimulation of Th2 cells in the presence of anti-IL-2 and anti-IL-2R and in the absence of CD28 costimulation reduced RHS7 demethylation to levels similar to that of unactivated and Th1 cells; that is, the Th2 cytokine LCR underwent minimal demethylation, correlating with conditions nonpermissive for IL-4 expression.
Furthermore, we observed a steadily decreasing amount of demethylation at the il4 promoter under increasingly nonpermissive culture conditions (Fig. 3B). Under normal Th2 conditions, the four proximal-most CpG residues in the minimal il4 promoter exhibited 66–90% demethylation at 86 h. Either with the addition of IL-2- and IL-2R-neutralizing antibodies or in the absence of CD28 costimulation, the demethylation profile decreased slightly to 46–80%. In combination, however, these conditions caused an even greater decrease to 37–65%, roughly the same levels of demethylation seen in naïve and Th1 cells, i.e., the basal level of demethylation in IL-4 nonexpressors. Taken together, blockade of both IL-2 signaling and CD28 costimulation produced a concurrent reduction in RHS7 and il4 promoter demethylation, conditions previously shown to be nonpermissive for IL-4 production (18, 19).
An Active Mechanism for RHS7 Demethylation.
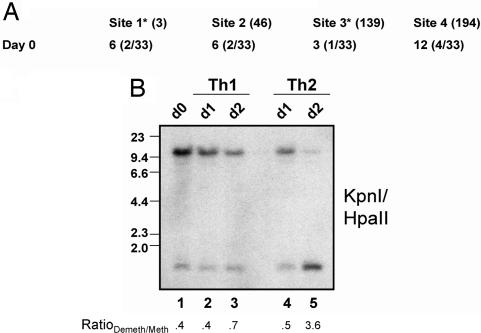
One of the most elusive features of demethylation is whether it occurs via an active or passive process. To address this issue, we first examined the state of methylation in unactivated cells. Previously, we showed that RHS7 was completely methylated in naïve T cells by Southern blotting analysis (8). We confirmed this result by sequencing the sense strand of RHS7 from bisulfite-treated naïve T cell DNA; in Fig. 4A, we show the results of such an analysis of the antisense strand of RHS7. Just as in the sense strand, only 3–6% of the four CpG motifs analyzed were unmethylated on the antisense strand in all clones tested. Notably, the two HpaII sites assayed by Southern blotting were almost completely methylated, at 6% and 3%, respectively, consistent with HpaII analysis.
Fig. 4.
Basal methylation and early kinetics of RHS demethylation. (A) Methylation analysis of the antisense strand of RHS7. Naïve CD4+ T cell DNA was treated with bisulfite and analyzed for methylation status. HpaII sites are indicated with an asterisk. The number of clones sequenced is indicated in parentheses. (B) Early time points of RHS7 demethylation. Th1 and Th2 cells were harvested at 24 and 48 h, and their DNA was subjected to methylation analysis by Southern blotting as performed in Fig. 1. The RatioDemeth/Meth of densitometric values is listed below each lane.
Previously, we demonstrated that no significant demethylation of RHS7 occurred before 12 h (8). To determine exactly when RHS7 demethylates, we cultured primary Th1 and Th2 cells for 24 and 48 h. Genomic DNA was harvested and subjected to Southern analysis. As shown in Fig. 4B, lanes 1–3, no appreciable differences in demethylation were seen between unactivated and early Th1 time points. At 24 h, the demethylation profile and RatioDemeth/Meth values of Th2 cells also appeared to be equivalent to that of unactivated T cells (Fig. 4B, lane 4). After 48 h, however, a significant amount of demethylation was observed, as evidenced by induction of the cleaved 1.1-kb HpaII fragment (Fig. 4B, lane 5). Thus, unlike the rapid kinetics of IL-2 promoter demethylation (20 min), (7) RHS7 demethylation occurs between 24 and 48 h postactivation.
We then tested the dependence of RHS7 demethylation on cell division. Cleavage of DNA by HpaII requires the CpG in its recognition site, CCGG, to be unmethylated on both the sense and antisense strands (20). Thus, any RHS7 allele cleaved by Southern blotting analysis must transit from methylation on both strands in naïve cells to demethylation on both strands in effector cells. As shown above, we do indeed find both strands to be fully methylated.
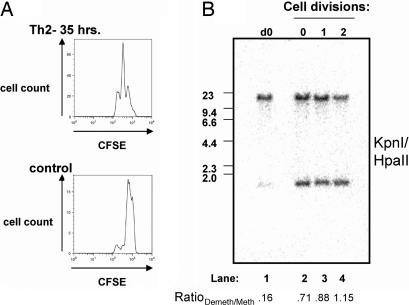
Experimentally, CD4+ T cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) and cultured under Th2 conditions. After 35 h, CFSE peaks were sorted and assayed for methylation status by Southern analysis; at this time point, T cells have divided zero, one, or two times (Fig. 5A Upper) (21). Control CFSE-labeled CD4+ cells were cultured with antigen-presenting cells and without stimulating antibodies or cytokines for reference (Fig. 5A Lower). As shown in Fig. 5B, lane 2, there is a striking increase of demethylated RHS7 alleles in the CFSE peak containing undivided cells compared with alleles in unactivated T cells (Fig. 5B, lane 1); by densitometry analysis, the RatioDemeth/Meth value increased from 0.16 in unactivated T cells to 0.71 in activated, undivided Th2 cells (Fig. 5B). These results indicate that one or both alleles of RHS7 become demethylated on both strands independent of cell division, strongly suggesting an active mechanism for this process.
Fig. 5.
Active demethylation of RHS7. (A) CFSE profiles of 35-h Th2 cultures. CFSE-labeled Th2 cells were cultured for 35 h and sorted. (Upper) The resulting histogram. (Lower) The histogram of CFSE-labeled control cells that have not undergone any proliferation. This undivided cell peak was used as a point of reference for the CFSE profile in Upper. (B) Methylation analysis of Th2 cells in A. Individual CFSE peaks were sorted and analyzed for RHS7 demethylation by Southern blotting. See Fig. 1 legend for the experimental procedure and fragment sizes. The image was generated by PhosphorImaging. The RatioDemeth/Meth of densitometric values is listed below each lane.
Not unexpectedly, RHS7 alleles in once- and twice-divided cells (Fig. 5B, lanes 3 and 4, respectively) also exhibited substantial demethylation (RatioDemeth/Meth values were 0.88 and 1.15, respectively), but here it may be caused by an active process, a passive one, or both. While cells in the undivided peak may have undergone one round of replication, passage through S phase would result in two hemimethylated strands, neither of which would have been able to be cleaved by HpaII. We therefore submit that the HpaII cleavage we observe is not merely caused by replication, but is a direct result of biochemical demethylation.
Discussion
We report the characterization of the demethylation of a specific hypersensitive site in the Th2 LCR. Within 48 h after stimulation under type 2 conditions, RHS7 demethylated completely on both sense and antisense strands in CD4+ T cells. IL-2 and IL-4 signaling appear to be required for efficient demethylation, whereas GATA3 appears to be insufficient for this remodeling.
That the Th2 LCR also demethylated in cytotoxic CD8+ Tc2 cells indicates il4 locus regulation may also be governed by LCR activity in them. However, the Th2 LCR did not demethylate to completion as seen in Th2 cells, and this observation is consistent with the predisposition of CD8+ T cells toward the type 1 phenotype, producing 100-fold less IL-4 than Th2 cells (22, 23). In fact, Tc2 cells, although cultured in the presence of IL-4, are able to up-regulate IFN-γ (24). One mechanism for this bias may be the lower levels of GATA3 and higher levels of repressor of GATA (ROG) expressed in Tc2 cells, leading to il4 promoter hypoacetylation levels relative to Th2 cells (25). We speculate that optimal function of the Th2 LCR is impaired in developing Tc2 cells, as evidenced by less than complete demethylation of RHS7, a setting that may explain the lack of robust IL-4 expression in Tc2 cells.
Demethylation is tightly associated with accessibility, and incomplete demethylation of the Th2 LCR may inhibit its ability to coordinate chromatin changes and confer enhancer activity on cytokine promoter regions in the Th2 locus. Interestingly, dnmt1-deficient CD8+ Tc2 cells are able to produce equivalent amounts of type 2 cytokines as their CD4+ counterparts, further implicating a refractory Th2 cytokine locus in CD8+ cells (26).
Here, we provide evidence that there is a third signaling component, IL-2, required for efficient demethylation and activation of the Th2 LCR and il4 promoter. Moreover, we argue that the signals emanating from IL-4R stimulation are secondary to IL-2 and IL-2-induced IL-4 production (18) and are not a consequence of IL-4 directly, despite the involvement of STAT6. IL-2 production is largely a result of CD28 costimulation, and when both IL-2 and CD28 signaling are extinguished, demethylation of il4 and RHS7 decreased dramatically, even in the presence of exogenous IL-4. This progression of events is consistent after CD28-inducible histone acetylation and DNA demethylation at the il2 promoter (27). It remains to be seen, however, whether IL-2-mediated demethylation is direct, such as through STAT5 binding to RHS7, or indirect. It is interesting to note that a single signaling component is not sufficient to effect optimal demethylation. Moreover, the mere absence of just one component is enough to reduce demethylation to levels seen in Th1 cells.
These conclusions represent insights gleaned from a minimalist experimental system used to activate and differentiate T cells, ex vivo (i.e., an antibody-triggered method). It is unclear whether the demethylation-abrogating effects of the lack of CD28 costimulation and antibodies to IL-2 would occur in a peptide/MHC system, or in vivo, where additional cell–cell (e.g., LFA–ICAM, Notch–Jagged) or cytokine–cell signals could compensate, thereby negating the effects on demethylation. An examination of this phenomenon in an in vivo setting, presumably reflecting the true epigenetic and cellular development of T cells during an immune response, should enable the identification of the necessary or redundant signals leading to locus activity via this mechanism.
The existence of a demethylating enzyme continues to intrigue us, because the implication of such a protein has been based almost solely on circumstantial evidence (12). Here, we show cell division-independent demethylation of the Th2 LCR despite full methylation of both strands in the unactivated T cell. In contrast to the il4 locus and RHS5 of the Th2 LCR, where passive demethylation appears to occur, RHS7 became demethylated in a manner suggestive of an active mechanism. Although this demethylation is rapid relative to these other important cis-elements, it is nonetheless preceded by il2 promoter demethylation.
Mechanistically, it is sensible that Th2 LCR activation (as measured by RHS7 demethylation) follows IL-2 up-regulation chronologically. Studies performed by Paul and colleagues (18) implicate IL-2 as a major potentiator of IL-4 production and Th2 differentiation. We propose that TCR stimulation-induced IL-2 activates the Th2 LCR, as evidenced by its demethylation, to drive polarization of the Th2 phenotype. Consistent with this hypothesis is the observation that T-bet actively inhibits IL-2 production at the transcriptional level, further evidence of T-bet cross-regulation of the Th2 phenotype (28).
Although our CFSE experiments demonstrate a possible active mechanism for demethylation, they do not show conclusive evidence for such a process. Despite our best efforts to separate peaks of fluorescence by gating as narrowly as possible, we cannot rule out the possibility that the peak of undivided cells was slightly contaminated by the adjacent peak of cells that have divided once. Demethylated DNA is more often early replicating, and the 3–6% of demethylated alleles in the starting population may be predisposed for earlier passive demethylation, which could enhance the demethylated band in undivided cells, or accentuate the contributing signal in once-divided contaminants (by undergoing two rounds of passive demethylation).
Although both the il2 promoter and RHS7 demethylate by what appears to be an active mechanism, it is unlikely that they share the same putative demethylase machinery. The il2 promoter demethylates within 20 min of T cell activation, reminiscent of global genomewide demethylation observed in the preimplantation mouse embryo and paternal genome in the fertilized egg (7, 29–31). This time frame is far too rapid for the T cell to up-regulate cell-specific demethylases at the transcriptional level, let alone the translational level. Therefore, it has been proposed that general demethylases are constitutively expressed and act on the il2 promoter from signals stemming from TCR and CD28 ligation (7). We extend this hypothesis, arguing that initial RHS7 demethylation (such as the low levels seen in Th0 and Th1 cells) is also nonspecific and that its Th2 specificity is governed by cell-specific demethylases or demethylase recruitment to the LCR by Th2-specific factors.
This feature is yet another example of an emerging theme in T cell epigenetics, where ubiquitous chromatin remodeling machinery is engaged by TCR stimulation, followed by reinforcement of the resulting chromatin changes by lineage-specific factors up-regulated in the proper cytokine milieu. We and others have proposed a biphasic model of Th2 LCR and il4 demethylation through TCR/IL-2 ligation and the IL-4 receptor (8, 10), where TCR or IL-2 initiates basal epigenetic changes, and IL-4 signaling reinforces such changes in the proper Th2 lineage. As observed with acetylation of the Th2 cytokine promoters (32), there is nonspecific demethylation of RHS7 occurring after TCR stimulation, as evidenced by the partial phenotype of nonskewed T cells.
STAT6, which we have previously shown to be required for robust acetylation at the il4 promoter, is one such candidate that can up-regulate and reinforce lineage-specific epigenetic change. STAT6 appears to play an analogous role in RHS7 demethylation, where its presence effects Th2-specific demethylation and its absence allows only basal, lineage-nonspecific demethylation. That RHS7 demethylation persists after restimulation indicates active or passive demethylation is maintained at this locus (data not shown), although the extent to which this persistence in guided by STAT6, or other Th2-specific factors, is not known.
Although GATA3 is able to induce hypersensitivity and hyperacetylation at the il4 locus proper (33–35), RHS7 maintains its methylation pattern when GATA3 is overexpressed in Th1 cells. This finding is not surprising, because GATA3 appears to be less important for Th2 LCR activation and more critical for il4 locus-proximal events (36, 37). As we and others have shown, GATA3 can affect the structure around the il4 gene itself; although it has not been shown to bind the il4 promoter, localization of GATA3 has been observed at the conserved intronic regulatory element in the first intron of il4 (38). At this element, GATA3 is thought to preclude access of the il4 locus to maintenance methyltransferases, thereby propagating passive demethylation throughout the locus in much the same way GATA3 displaces MBD2 from intron 2 and CNS1 of the il4 locus (6). GATA3 likely induces a Th1 cell to secrete type 2 cytokines through these mechanisms, bypassing the Th2 LCR altogether.
Curiously, GATA3 also binds to RHS7 (9), but not only does it fail to induce hypersensitivity (36) and demethylation, it is entirely dispensable in terms of LCR function (37). It is possible that GATA3 acts in a similar manner as it does at the conserved intronic regulatory element in preventing methyltransferases from binding, thereby demethylating the LCR. But as we have shown here, demethylation appears to occur via an active mechanism. Thus, although GATA3 may be more critical in maintaining Th2 demethylation than in initiating it, the exact role of GATA3 binding in the Th2 LCR remains to be determined.
Materials and Methods
Mice, T Cells, and Antibodies.
T cells used in these studies were obtained from C57BL/6, BALB/c, and STAT6−/− mice as described (32). Briefly, naive, splenic CD4+ CD62Lhi, CD44lo, and NK1.1lo T cells were isolated by FACS sorting. CD8+ T cells were purified by magnetic cell sorting. Effector Th1, Th2, Tc1, and Tc2 cells were derived by stimulation in vitro with anti-CD3 and anti-CD28 plus syngeneic, irradiated splenic antigen-presenting cells in the presence of 3.5 ng/ml IL-2 and 10 μg/ml anti-IL-4 for Th1 and Tc1 cultures, and 1,000 units/ml IL-4 and 10 μg/ml anti-IFN-γ for Th2 and Tc2 cultures (32, 39). In some experiments, T cells were cultured in 12-well plates coated with 2 μg/ml anti-CD3 and 2 μg/ml anti-CD28. For neutralization experiments, anti-IL-2 and anti-IL-2R were used at a concentration of 10 μg/ml. Upon harvesting at the time points indicated, cells were purified by centrifugation over Ficoll-Hypaque gradients for use in experiments.
CFSE Labeling.
CD4+ T cells were incubated in 3.5 μM CFSE in PBS for 15 min at 37°C. The reaction was halted with 10% FCS in Bruff's medium (Click's medium supplemented with 40 mM l-glutamine, 60 μM 2-mercaptoethanol, 0.7 mM sodium bicarbonate, and 58 mg/liter gentamycin; Gibco/BRL, Carlsbad, CA) (40), and cells were washed extensively and plated. As a reference point for undivided cells, CFSE-labeled T cells were cocultured with irradiated antigen-presenting cells without cytokines or stimulating antibodies, resulting in a single peak of undivided cells as assessed by flow cytometry.
Retroviral Transduction.
Retroviral transduction of T cells was performed as described (24). Retroviral vectors (provided by K. Murphy, Washington University, St. Louis, MO), allowed expression of GATA3 plus EGFP (24, 41). At 24 h after stimulation, cells were infected with retroviral supernatant. At days 5–6, cells were sorted into EGFP-negative and EGFP-positive populations, expanded for 4 days, and subjected to methylation analysis. The Phoenix-ECO packaging cell line was a gift of G. Nolan (Stanford University, Palo Alto, CA).
Methylation Analysis.
Genomic DNA was harvested by overnight proteinase K digestion in lysis buffer (50 mM Tris, pH 8.0/5 mM EDTA/0.2% SDS/200 mM NaCl), phenol-chloroform extraction, and isopropanol precipitation. Ten micrograms of DNA was double-digested with KpnI (50 units) or HindIII (60 units) and HpaII (50 units) or MspI (50 units) for 16 h. All restriction enzymes were obtained from New England Biolabs, Ipswich, MA. Reactions were run out on a 0.8% agarose gel and transferred to Hybond-N+ nylon membrane (Amersham Biosciences, Piscataway, NJ) with the Posi-Blotter transfer apparatus (Stratagene, La Jolla, CA). Southern hybridization was performed; the blots were probed with 32P-labeled DNA probes generated by PCR as described (8).
The crude ratio of signals from demethylated alleles to methylated alleles (RatioDemeth/Meth) was calculated by using the densitometry function on a gel documentation workstation (Alpha Innotech, San Leandro, CA), dividing the demethylated signal by the methylated signal, and then subtracting a background value. These ratios are provided as rough quantification of demethylation and do not represent a percentage of demethylated alleles.
For bisulfite analysis, T cell DNA was treated with bisulfite by using the CpGenome DNA Modification Kit according to the manufacturer's protocol (Chemicon International, Temecula, CA). Amplicons were cloned into the TOPO TA pCR2.1 vector. The numbers of clones sequenced for each site are as stated in the figures.
Primers used to amplify bisulfite-treated genomic DNA are as follows: RHS7 forward, 5′-CAAAACATTCTAAACTATC T A TATTTC-3′; RHS7 reverse, 5′-GATATTTTTGTTTATTTTTAGTATGTTG-3′; RHS7antisense forward, 5′-GGTTAAATTGTAGTTATGTGATTTTATTTA-3′; RHS7antisense reverse, 5′-AAAACATTTTTACTCATCCTCAACATAC-3′; IL-4 promoterproximal forward, 5′-CTTTCTTAATATTACTCTATCTTTCC-3′; IL-4 promoterproximal reverse, 5′-GGGTTGAGATTTATTAATAGTTTTG-3′; RHS4 forward, 5′-TTTATGGGGTAATTTTTGGATTTAA-3′; RHS4 reverse, 5′-ACAATACTTCACAATTCTACAACAAACA-3′; RHS5upstream forward, 5′-TGTGATTTTTAAATTTGTTTTTTTT-3′; RHS5upstream reverse, 5′-CACATTCTAATTTTATTAAACTATAAC-3′; RHS5downstream forward, 5′-TTATGGTATTTGGTTTTTTGTTTTTG-3′; RHS5downstream reverse, 5′-CCACAAACTCCACTTAAAAAAAATT-3′; RHS6upstream forward, 5′-TTTTATGAAAGAAGTATAAAGTATTAGGTA-3′; RHS6upstream reverse, 5′-CAACTATCACCATACAAAAAATATAC-3′; RHS6downstream forward, 5′-TTGTGATATTTTGAGTTTTTTTGTA-3′; and RHS6downstream reverse, 5′-ACCTCTCTACTTCACCAAATCTCCATTTT-3′.
Acknowledgments
We thank Frances Manzo for preparation of this manuscript, Babis Spilianakis for help with figure preparation, Wyeth Laboratories for their generous contribution of IL-12, and Thomas Taylor for outstanding cell sorting.
Abbreviations
- Th
T helper
- Tc
T cytotoxic
- RHS
rad50 hypersensitive site
- LCR
locus control region
- TCR
T cell receptor
- CFSE
carboxyfluorescein diacetate succinimidyl ester.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Takai D. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 4.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 6.Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, Bird AP, Reiner SL. Mol Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 7.Bruniquel D, Schwartz RH. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 8.Fields PE, Lee GR, Kim ST, Bartsevich V, Flavell RA. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Spilianakis C, Flavell RA. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 10.Lee DU, Agarwal S, Rao A. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 11.Santoso B, Ortiz BD, Winoto A. J Biol Chem. 2000;275:1952–1958. doi: 10.1074/jbc.275.3.1952. [DOI] [PubMed] [Google Scholar]
- 12.Wilson CB, Makar KW, Shnyreva M, Fitzpatrick DR. Semin Immunol. 2005;17:105–119. doi: 10.1016/j.smim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 16.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 17.Mowen KA, Glimcher LH. Immunol Rev. 2004;202:203–222. doi: 10.1111/j.0105-2896.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 18.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. Proc Natl Acad Sci USA. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rulifson IC, Sperling AI, Fields PE, Fitch FW, Bluestone JA. J Immunol. 1997;158:658–665. [PubMed] [Google Scholar]
- 20.Bird AP, Southern EM. J Mol Biol. 1978;118:27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- 21.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Immunity. 2001;15:303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 22.Croft M, Carter L, Swain SL, Dutton RW. J Exp Med. 1994;180:1715–1728. doi: 10.1084/jem.180.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sad S, Marcotte R, Mosmann TR. Immunity. 1995;2:271–279. doi: 10.1016/1074-7613(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 24.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman GC, Glimcher LH. Cell. 2000;100:655–669. [Google Scholar]
- 25.Omori M, Yamashita M, Inami M, Ukai-Tadenuma M, Kimura M, Nigo Y, Hosokawa H, Hasegawa A, Taniguchi M, Nakayama T. Immunity. 2003;19:281–294. doi: 10.1016/s1074-7613(03)00210-3. [DOI] [PubMed] [Google Scholar]
- 26.Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Nat Immunol. 2003;4:1183–1190. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 27.Thomas RM, Gao L, Wells AD. J Immunol. 2005;174:4639–4646. doi: 10.4049/jimmunol.174.8.4639. [DOI] [PubMed] [Google Scholar]
- 28.Hwang ES, Hong JH, Glimcher LH. J Exp Med. 2005;202:1289–1300. doi: 10.1084/jem.20051044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li E. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 30.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 31.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 32.Fields PE, Kim ST, Flavell RA. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 33.Takemoto N, Kamogawa Y, Lee HJ, Kurata H, Arai K, O'Garra A, Arai N, Miyatake S. J Immunol. 2000;165:6687–6691. doi: 10.4049/jimmunol.165.12.6687. [DOI] [PubMed] [Google Scholar]
- 34.Takemoto N, Arai K, Miyatake S. J Immunol. 2002;169:4103–4107. doi: 10.4049/jimmunol.169.8.4103. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita M, Ukai-Tadenuma M, Miyamoto T, Sugaya K, Hosokawa H, Hasegawa A, Kimura M, Taniguchi M, DeGregori J, Nakayama T. J Biol Chem. 2004;279:26983–26990. doi: 10.1074/jbc.M403688200. [DOI] [PubMed] [Google Scholar]
- 36.Ansel KM, Djuretic I, Tanasa B, Rao A. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 37.Lee GR, Fields PE, Griffin TJ, IV, Flavell RA. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 38.Tykocinski LO, Hajkova P, Chang HD, Stamm T, Sozeri O, Lohning M, Hu-Li J, Niesner U, Kreher S, Friedrich B, et al. J Biol Chem. 2005;280:28177–28185. doi: 10.1074/jbc.M502038200. [DOI] [PubMed] [Google Scholar]
- 39.Lee GR, Fields PE, Flavell RA. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- 40.Koni PA, Flavell RA. J Exp Med. 1999;189:855–864. doi: 10.1084/jem.189.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorelik L, Fields PE, Flavell RA. J Immunol. 2000;165:4773–4777. doi: 10.4049/jimmunol.165.9.4773. [DOI] [PubMed] [Google Scholar]