Abstract
Budding yeast undergo robust oscillations in oxygen consumption during continuous growth in a nutrient-limited environment. Using liquid chromatography-mass spectrometry and comprehensive 2D gas chromatography-mass spectrometry-based metabolite profiling methods, we have determined that the intracellular concentrations of many metabolites change periodically as a function of these metabolic cycles. These results reveal the logic of cellular metabolism during different phases of the life of a yeast cell. They may further indicate that oscillation in the abundance of key metabolites might help control the temporal regulation of cellular processes and the establishment of a cycle. Such oscillations in metabolic state might occur during the course of other biological cycles.
Keywords: gas chromatography-mass spectrometry, liquid chromatography-mass spectrometry, metabolic cycle, metabolite profiling
Circadian rhythms are driven by biological clocks found in virtually all kingdoms of life (1, 2). Genome-wide expression studies in plants, flies, and mice have shown that many genes are expressed periodically as a function of the circadian cycle (3–6). It is now predictable from studies of oscillating gene expression that the circadian regulatory apparatus should differentially control metabolic state as a function of the circadian cycle (7, 8). By contrast, direct and comprehensive measurements of changes in sentinel metabolites have not been performed as a function of the circadian cycle or of biological cycles of other temporal dimensions.
The budding yeast Saccharomyces cerevisiae exhibits various modes of oscillatory behavior during continuous growth (9–12). We recently established a continuous culture system that reveals an ≈4- to 5-h yeast metabolic cycle (YMC) in which over half of the genome is periodically expressed as a function of robust recurring oscillations in oxygen consumption (12). Expression profiling studies have led to the prediction that a variety of cellular and metabolic processes are temporally orchestrated around three major phases of the YMC [oxidative (Ox), reductive/building (RB), and reductive/charging (RC)] (12). Cells undergoing the YMC are furthermore synchronized with respect to the cell cycle, because the YMC strictly gates cell division to the RB phase when respiration decreases significantly (12).
A prediction of temporal compartmentalization is that the concentrations of critical metabolites might exhibit periodic fluctuation and, in turn, play a reciprocal role in regulating the YMC (8). To determine whether cyclic changes in metabolic state might occur during the YMC, we used both liquid chromatography (LC)/tandem mass spectrometry (LC-MS/MS) and comprehensive 2D gas chromatography (GC)/time-of-flight mass spectrometry (GC×GC-TOFMS) to monitor the intracellular concentrations of ≈150 common metabolites at regularly spaced intervals throughout the YMC. The combined use of LC-MS- and GC-MS-based methods provides a means to evaluate the consistency of results for metabolites detected by both methods as well as complementary insights for many metabolites not found in common by the two methods. The results of these surveys show that many metabolites oscillate in abundance with a periodicity precisely matching that of the YMC. From analysis of these profiling data, the logic of metabolite oscillation largely matches that predicted by the previously reported transcript array dataset (12). Furthermore, metabolite profiling extends this logic in ways not inherently obvious by inspection of gene expression profiles. Such cyclic changes in metabolic state might also occur during other biological cycles.
Results
To observe the YMC, the prototrophic yeast strain CEN.PK (13) was grown in a chemostat to a high cell density (≈1 × 108 cells per ml). After a short starvation period, upon feeding the culture continuously with low concentrations of glucose, the cell population becomes highly synchronized, as judged by robust oscillations of oxygen consumption. To determine whether the intracellular concentrations of metabolites might change during the YMC, we prepared metabolite extracts from the cell population at 24 evenly spaced time intervals collected over two consecutive metabolic cycles (≈23–25 min per interval) (Fig. 1A) [see supporting information (SI) Appendix].
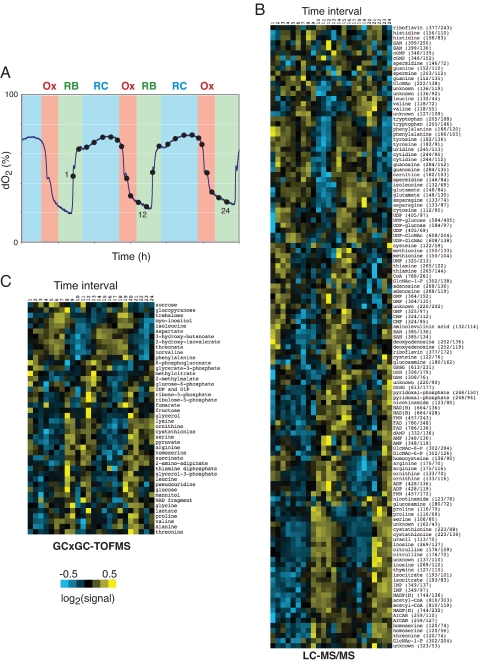
Fig. 1.
Cyclic changes in metabolic state during the YMC. (A) The YMC. Shown are several metabolic cycles exhibited by a WT diploid strain and intervals of the dissolved oxygen trace that correspond to the Ox, RB, and RC phases. Intracellular metabolites were extracted for LC-MS/MS analysis at the 24 time points indicated (dots). For GC×GC-TOFMS sampling time points (which are similar to those for LC-MS/MS but not the same), see SI Appendix. x-axis tick marks denote 2-h intervals. (B) Hierarchical clustering analysis of metabolite profiles obtained from LC-MS/MS analysis. Rows correspond to metabolites (parent ion mass/daughter ion mass), and columns correspond to 24 time intervals over two consecutive metabolic cycles as depicted in A. Metabolite data were log2-transformed, centered about the mean, normalized, and clustered by using Spearman–Rank correlation (36). (C) Hierarchical clustering analysis of metabolite profiles obtained from GC×GC-TOFMS analysis. Shown are the ≈40 metabolites identified to have robust oscillations (>1.5-fold amplitude) in concentration. Metabolite data were log2-transformed, centered about the mean, normalized, and clustered by using Spearman–Rank correlation (36). For a heat map of the data, which shows more depth of modulation, see SI Appendix.
We first used a directed LC-MS/MS-based technique called multiple reaction monitoring for metabolite profiling of the extracts. A library of ≈130 common metabolites was constructed, and parameters for the detection of the two most abundant daughter ions for each metabolite upon collision-induced fragmentation were optimized (see SI Appendix). Many multiple reaction monitorings were run simultaneously throughout an entire chromatographic run, resulting in a specific and sensitive method to quantify a large number of metabolites of interest.
As a complementary approach, we used an unbiased, GC-based method (GC×GC-TOFMS) to analyze similarly prepared extracts from cells undergoing the YMC (see SI Appendix). After derivatization, metabolite extracts were separated and detected by GC×GC-TOFMS (14). The resulting 3D data required chemometric “multivariate” data reduction tools to locate metabolites exhibiting periodic behavior. Initial identification of the metabolites of interest was achieved by using a software package developed in-house, followed by an identification confirmation and relative signal quantification based on the full mass spectral parallel factor analysis deconvolution (see SI Appendix).
We observed that numerous metabolites (≈60% of those targeted by LC-MS/MS), including amino acids, carbohydrates, and nucleotides, exhibited robust oscillations in abundance (P < 0.05) as a function of the YMC (Fig. 1 B and C) (see SI Appendix). The median amplitude of oscillation was ≈2.4-fold. For any single metabolite, a similar pattern of oscillation was frequently observed between the two consecutive metabolic cycles, and also between different metabolic cycle experiments, demonstrating that our extraction and profiling methods were accurate and reproducible. Moreover, in 8 of 11 cases, the LC-MS/MS and GC×GC-TOFMS profiles for a given metabolite were highly similar, further validating our approach. We also discovered 29 additional metabolites by GC×GC-TOFMS and ≈12 additional metabolites by LC-MS/MS that displayed robust oscillations as a function of the YMC, but we could not assign their identities. Thus, we conclude that dynamic changes in the metabolic state of a yeast cell take place during continuous growth under nutrient-limited conditions.
Upon inspection of the metabolite profiles, we observed that different metabolites peaked at entirely different times of the YMC. Using hierarchical clustering analysis, we could group metabolites by their temporal profiles and identify those that exhibited similar changes in concentration during the YMC (Fig. 1 B and C). For example, clustering analysis revealed that the amino acids containing aromatic side chains (tyrosine, tryptophan, and phenylalanine) displayed highly similar profiles, presumably because they are derived from a common biosynthetic pathway. Many of the nucleotide monophosphates (UMP, CMP, and GMP) and nucleosides (uridine, cytidine, and guanosine) clustered in a similar temporal compartment. Moreover, other metabolites already known to be in common biochemical pathways tended to have similar temporal abundance profiles (Fig. 1 B and C) (e.g., ornithine, citrulline, arginine).
The periodic metabolites each peaked during particular phases of the YMC (Ox, RB, or RC) (Table 1). Numerous amino acid precursors and amino acids, nucleotide precursors (e.g., IMP), and tricarboxylic acid cycle intermediates (e.g., isocitrate, succinate, pyruvate) increased significantly during the Ox phase. These findings are consistent with the YMC gene expression data, because many genes encoding amino acid and nucleotide biosynthetic enzymes are copiously expressed during the Ox phase (12). The increase in concentration of tricarboxylic acid cycle intermediates indicates that flux through the electron transport chain is significantly enhanced during the Ox phase. Together, the metabolite and gene expression data suggest that the Ox phase represents a temporal window dedicated to respiration for the production of energy.
Table 1.
Metabolites that peak during a particular phase of the YMC
| Ox | RB | RC |
|---|---|---|
| Acetyl-CoA | ADP | 2-Hydroxy-isovalerate |
| Arginine (RB) | Aminolevulinic acid | 6-Phosphogluconate |
| Citrulline | AMP | Asparagine |
| Cystathionine | CMP (RC) | Aspartate |
| Glycerol-3-P | FAD | Carnitine |
| Homocysteine (RB) | GMP (RC) | CoA |
| Homoserine (RB) | Methyl-citrate | Cytidine (Ox) |
| IMP | NAD(H) (Ox) | Cytosine |
| Inosine | Nicotinamide (Ox) | GlcNAc (Ox) |
| Isocitrate | Pyridoxal-phosphate | GlcNAc-6-P (RB) |
| Lysine | Pyruvate (Ox) | Glutamate (Ox) |
| NADP(H) | S-adenosylhomocysteine | Glutathione |
| Ornithine | Succinate (Ox) | Guanine |
| Proline | Guanosine (Ox) | |
| Riboflavin | Histidine (Ox) | |
| Serine | Isoleucine (Ox) | |
| Threonine | Leucine (Ox) | |
| Thymine | Methionine | |
| Uracil | Myo-inositol | |
| Valine (RC) | Phenylalanine (Ox) | |
| Thiamine | ||
| Trehalose | ||
| Tryptophan (Ox) | ||
| Tyrosine (Ox) | ||
| UDP | ||
| UDP-GlcNAc (RB) | ||
| UDP-glucose (Ox) | ||
| UMP | ||
| Uridine (Ox) |
Metabolites were assigned to phases of the YMC based on the time intervals when their concentrations were at a maximum. Some metabolites show secondary peaks during an additional phase (in parentheses).
Metabolites associated with high glycolytic flux (e.g., pyruvate and glucose-6-phosphate) increased in abundance during the RB phase (Table 1). We and other researchers have reported that ethanol and acetate, additional hallmarks of glycolytic metabolism, rise significantly in the temporal window corresponding to early RB phase (12, 15). These findings reinforce the notion that yeast cells become highly glycolytic and less respiratory upon entry into this reductive phase of the YMC. Because cell division initiates in a fraction of the cell population (≈40–50%) in each RB phase, a shift to a highly glycolytic metabolism might occur as a means to minimize oxidative damage to DNA during replication (11, 12, 16).
The abundance of carnitine, storage carbohydrates [e.g., trehalose, and glycogen (see SI Appendix)], and certain amino acids peaked during the RC phase of the YMC (Table 1). Genes involved in fatty acid oxidation and the synthesis and breakdown of storage carbohydrates are all up-regulated in the RC phase (12), which is largely consistent with the metabolite data. The strong oscillation in trehalose and glycogen suggests cycles of carbohydrate storage and breakdown during each metabolic cycle, which has been observed during yeast oscillations (15, 17, 18). Such catabolism of polymeric sugars in the late RC and early Ox phases is anticipated to fuel energy production for the preparation of cell division, perhaps in the form of a metabolic or energetic burst (17, 19, 20).
These metabolite profiling data gathered over the YMC and the reported transcript array dataset confirm dynamic metabolic changes that occur as yeast self-synchronize their growth under continuous, nutrient-limited conditions. We next asked whether some of these changes in metabolic state might be important for establishment of a metabolic cycle. As an example, from the YMC gene expression data, cellular metabolism during the RC phase of the YMC can be predicted to enhance the production of acetyl-CoA and NADPH (12). Numerous genes involved in fatty acid oxidation, glycolysis, ethanol utilization, and the breakdown of storage carbohydrates are highly expressed during the RC phase (8, 12). The collective result of these reactions is the enhanced production of acetyl-CoA, which is vital for optimal fueling of the tricarboxylic acid cycle. Several enzymes of the pentose phosphate pathway, the primary pathway for the production of NADPH, are also highly up-regulated during the RC phase (12). Because these included the transketolase and transaldolase enzymes, which convert five-carbon sugars back to glycolytic intermediates, it was predicted that, during this temporal window, the main purpose of the pentose phosphate pathway is for the synthesis of NADPH and not pentose sugars (8, 12). Furthermore, the genes encoding NAD+ and NADH kinases (UTR1, YEF1, POS5), which phosphorylate NAD(H) to NADP(H) (21), are also highly up-regulated during RC phase (Fig. 2B).
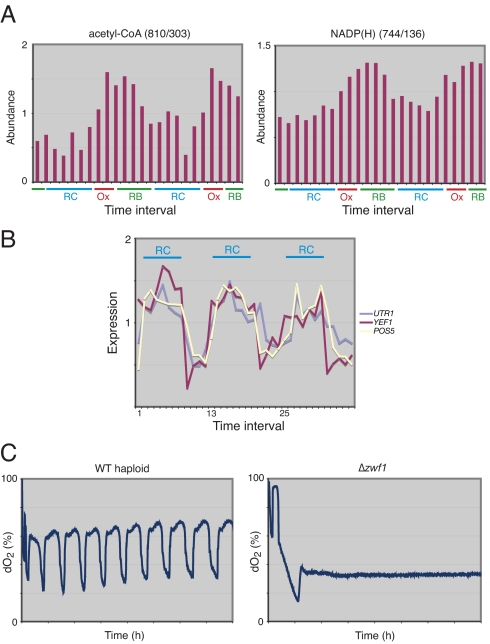
Fig. 2.
Acetyl-CoA and NADP(H) levels are highly periodic during the YMC. (A) Acetyl-CoA and NADP(H) concentrations over two consecutive metabolic cycles. Note that both acetyl-CoA and total NADP(H) periodically increase in concentration at the end of the RC phase and into the Ox phase. (B) Temporal expression profiles of the three NAD(H) kinases in yeast. Expression data are from ref. 12. (C) Cells lacking glucose-6-phosphate dehydrogenase (Δzwf1, which cannot use the pentose phosphate pathway to synthesize NADPH) do not exhibit metabolic cycles during continuous growth. Shown is the dissolved oxygen trace of a WT and Δzwf1 strain for ≈40 h after the start of continuous mode. x-axis tick marks denote 5-h time intervals.
Our metabolic profiling confirms that both acetyl-CoA and total NADP(H) levels are highly periodic, peaking at the end of RC and the beginning of Ox phase exactly as predicted (Fig. 2A). Because it is known that the intracellular NADPH:NADP+ ratio is consistently maintained quite high, often ≈20:1 or greater (22), we can infer that the majority of the NADP pool will be in the reduced form. NADPH is the major source of cellular reducing equivalents and a substrate for many reductive biosynthetic reactions. Thus, NADPH may be up-regulated during the Ox phase to help protect cells against oxidative stress associated with respiration and to function in reductive biosynthetic reactions necessary for the preparation of cell division (see also below).
To test the importance of NADPH for metabolic cycling, we created a strain disrupted in glucose-6-phosphate dehydrogenase (Δzwf1), the first enzyme of the pentose phosphate pathway. Strikingly, cells lacking ZWF1 did not undergo a metabolic cycle, even though they displayed a growth rate similar to a WT strain when studied in nutrient-rich log phase growth (Fig. 2C). Thus, entry into the pentose phosphate pathway is absolutely required for cells to properly undergo the YMC. We specifically suggest that cyclic changes in the output of NADPH-dependent reactions or the redox molecule itself might be fundamentally important to temporal orchestration of respiratory oscillations.
We further noted that some amino acids and amino acid-like metabolites tended to oscillate with a peak during the Ox phase, whereas others peaked during the RC phase (Table 1). Examination of the biosynthetic pathways for these molecules reveals that the majority of those that increase in abundance during the Ox phase (e.g., ornithine, proline, homoserine) tend to depend on NADPH as a substrate in their biosynthesis, unlike those that increase during RC phase (e.g., aspartate, asparagine, glutamate). These observations suggest that cells have tuned these reactions to occur when the level of a key substrate (i.e., NADPH) is maximally available. Such orchestration of biosynthetic pathways argues that cells have extensively organized metabolic output according to existing levels of NADPH. Taken together, it appears that oscillation of NADP(H) abundance is of central importance for metabolic oscillation.
Metabolic profiling revealed that the levels of aminolevulinic acid (ALA) oscillated with a high amplitude during the YMC (Fig. 3A). The synthesis of ALA is the first and rate-limiting step of the heme biosynthesis pathway, suggesting that the production of heme is also periodic during the YMC. ALA levels began to increase just as the cells were completing the respiratory phase and peaked when oxygen consumption was at a minimum (Fig. 3A). The synthesis of heme might be directed to this reductive, nonrespiratory temporal window because it is known that heme is susceptible to oxidative damage as well as capable of causing oxidative damage (23). We hypothesize that the heme prosthetic groups in cytochromes might need to be regenerated after a round of intense respiration in the Ox phase of the YMC.
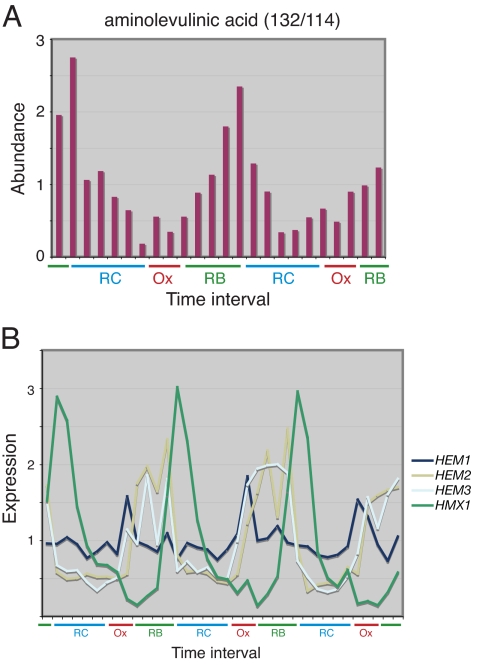
Fig. 3.
Cycles of heme synthesis during the YMC. (A) ALA levels oscillate with a large amplitude during the YMC. Note that ALA levels begin to increase as cells exit Ox phase and enter RB phase. (B) Temporal expression profiles of genes encoding enzymes of the heme pathway during the YMC. Note that HEM1 transcript levels are highest in Ox phase just before the observed increase in ALA concentration. HEM2 and HEM3 transcript levels peak shortly after HEM1. The gene encoding heme oxygenase (HMX1) is up-regulated strongly in the RC phase. Expression data are from ref. 12.
The expression of ALAsynthase, encoded by the yeast HEM1 gene, peaks in the Ox phase, shortly before the rise in ALA levels (Fig. 3B). The expression of HEM2 and HEM3, which execute the next two steps in the heme biosynthesis pathway, peak shortly after HEM1, illustrating the principle of just-in-time synthesis. Moreover, the heme oxygenase HMX1, which controls the breakdown of heme, is also highly periodically expressed (Fig. 3B), but during the later RC phase, suggesting that heme synthesis and breakdown are under exquisite control as a function of the YMC.
Several metabolites in the pathways regulating sulfur metabolism exhibited robust oscillations as a function of the YMC (Figs. 1 B and C and 4B). These included cystathionine, homocysteine, homoserine, serine, S-adenosylhomocysteine (SAH), and glutathione (GSH), with many of these peaking at slightly different temporal windows. The sulfur-containing amino acids methionine and cysteine are energetically costly to synthesize (24). For their de novo biosynthesis, four equivalents of NADPH are required for the reduction of sulfate to sulfide. Sulfide is then used for the biosynthesis of homocysteine, which can then be converted to either cysteine or methionine in the two branches of the pathway that ensue (Fig. 4A). In the GSH branch, homocysteine is converted to cystathionine and then to cysteine. Cysteine can subsequently be used to synthesize GSH, which is the primary reducing agent used by cells to buffer against oxidative stress. In the S-adenosylmethionine (SAM) branch, homocysteine acquires a methyl group and becomes methionine, which can then be converted into SAM, the major biological methyl donor. Once SAM donates its methyl group in an enzymatic reaction, it becomes SAH, which can then be hydrolyzed and recycled back to homocysteine. Thus, depending on the branch of the sulfur pathway, the resulting sulfur metabolites have entirely different functions and redox properties.
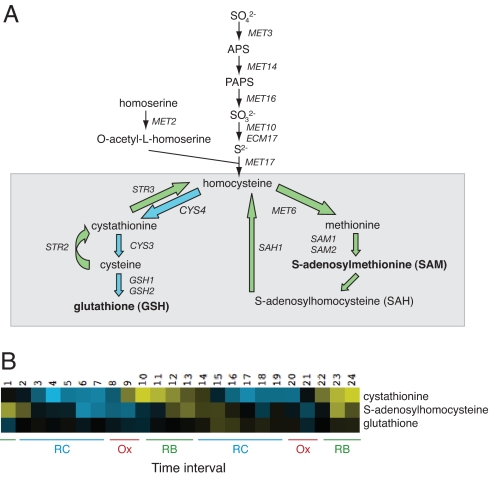
Fig. 4.
Sulfur metabolism in budding yeast. (A) Diagram of the sulfur metabolism pathway in budding yeast. Note that homocysteine lies at a critical juncture between two branches of the pathway. The GSH branch (blue arrows) is dedicated to the biosynthesis of cysteine and GSH, and the SAM branch (green arrows) is dedicated to the biosynthesis of methionine and SAM. (B) Several sulfur metabolites are highly periodic as a function of the YMC. Cystathionine, SAH, and GSH levels are all periodic and peak at slightly different time intervals of the YMC. Note that cystathionine peaks during the Ox and RB phases distinctly before SAH.
We observed that cystathionine levels accumulated significantly in Ox phase and into RB phase during the temporal window permissible for the initiation of cell division. The increase in cystathionine could be explained by either synthesis from homocysteine via cystathionine β-synthase (Cys4p) or synthesis from cysteine by cystathionine γ-synthase (Str2p) (Fig. 4A). On examination of the temporal expression profiles of sulfur pathway genes, STR2 and STR3 are clearly induced in late RC phase before all of the other enzymes (Fig. 5A). Because these genes are known to encode enzymes that convert cysteine back to homocysteine via cystathionine (24), sulfur metabolite flux appears to be directed away from the GSH branch and into the SAM branch during the RB phase (Fig. 4A, green arrows). We propose that, during this temporal window of the YMC, sulfur equivalents are shunted to SAM via homocysteine and the transsulfuration pathway.
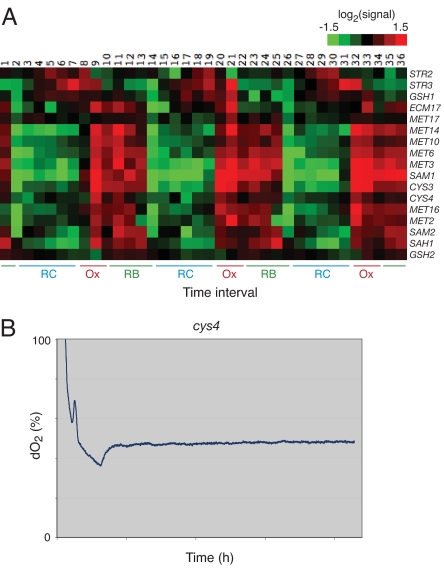
Fig. 5.
The logic of sulfur metabolism during the YMC. (A) Temporal expression profiles of sulfur metabolism pathway genes. Note that STR2 and STR3 are clearly up-regulated at the end of the RC phase before the other genes in the pathway, revealing a dynamic switch to the SAM branch of the pathway during RB phase. In the RC phase, sulfur flux is directed back to the GSH branch. Expression data are from ref. 12. (B) A cys4 mutant cannot undergo the YMC. Cells containing a disruption in the 3′ UTR of cystathionine β-synthase (CYS4) do not exhibit metabolic cycles during continuous growth. Shown is the dissolved oxygen trace of a cys4 mutant for ≈22 h after the start of continuous mode. x-axis tick marks correspond to 5-h intervals. Mutations in human cystathionine β-synthase have been linked to various disorders of the brain.
It is sensible that cells would increase the production of SAM during RB phase, because SAM is required for the methylation of newly synthesized histones, DNA, mRNA, and lipids, macromolecules that must be produced during cell division. Although we could not detect robust oscillations in SAM itself, the periodic increase in SAH levels during RB phase demonstrates that cells are in fact consuming more SAM during the RB phase (Fig. 4B). After this shift to the SAM branch during RB phase, sulfur metabolite flux might divert back to the GSH branch (Fig. 4, blue arrows), as evidenced by the decrease in SAH levels and subsequent increase in GSH levels during the RC phase. The dynamic switch between the two branches of the sulfur pathway can also be seen by examining the temporal profiles of the genes encoding the relevant rate-limiting enzymes. SAM1 (SAM synthetase) peaks sharply in Ox phase, whereas SAH1 (SAH hydrolase) and GSH2 (GSH synthetase) peak later in RB phase (Fig. 5A).
To determine whether oscillations in sulfur metabolites might be important for the YMC, we deleted various enzymes of the sulfur pathway or created partial loss-of-function alleles by disrupting the 3′ UTRs (25). Strikingly, cells containing a disruption in the 3′ UTR of CYS4, which encodes cystathionine β-synthase, the enzyme that synthesizes cystathionine from homocysteine, did not undergo a metabolic cycle (Fig. 5B). In contrast, other mutants of the sulfur pathway that we tested (cys3, sah1, met6, Δstr2, Δstr3, Δsam1, Δsam2) exhibited normal cycles.
This cys4 allele is a partial loss-of-function mutation, because it caused cells to become more sensitive to oxidative stress, perhaps due to its deficiencies in synthesizing cysteine and GSH (26) (see SI Appendix). These observations suggest that insufficient flux into the GSH branch of the transsulfuration pathway and/or excessive or inappropriate flux into the SAM branch could be the cause of the defects in metabolic cycling. Alternatively, a loss-of-function mutation in cystathionine β-synthase could compromise signaling by hydrogen sulfide (H2S), a biological gas known to be produced by this enzyme and reported to mediate population synchrony (27, 28). In any event, these results highlight the importance of proper temporal regulation of sulfur metabolite flux in the establishment of metabolic oscillation.
Discussion
By use of both LC-MS/MS and GC×GC-TOFMS metabolite profiling methods, we have demonstrated that budding yeast exhibit robust cyclic changes in metabolic state during continuous growth under nutrient-limited conditions. By monitoring and inspecting cyclic changes in sentinel metabolites, we have been able to formulate predictions and demonstrate that several metabolic pathways [i.e., NADP(H), sulfur] are important for proper establishment of metabolic oscillation. Because cells undergoing the YMC are also synchronized with respect to the cell cycle, these studies have revealed a set of metabolites that increase in abundance coincident with the initiation of cell division.
Such oscillations in metabolite concentration as seen during the life cycle of a yeast cell will undoubtedly be important for permitting allosteric activation or inhibition of enzymes critical to particular pathways. Moreover, it has become increasingly evident that many protein sensors have evolved to finely sense and respond to the metabolic state of a cell (29). As such, a variety of cellular and metabolic processes will appear to be intimately coupled to these cyclic changes in metabolic state.
Some of the principles of metabolic oscillation behind the YMC might pertain to other biological cycles (8). For example, cycles of heme synthesis and degradation appear to occur as a function of the mammalian circadian cycle (30). ALAsynthase is highly periodically expressed as a function of the mammalian circadian cycle (30), and heme has been shown to modulate expression of the PERIOD genes when injected into mice (30). Moreover, the CLOCK paralog NPAS2, which is centrally involved in the control of circadian rhythm, binds heme in its PAS domains to create a sensor for carbon monoxide (31). Thus, periodic cycles of heme synthesis and heme-based signaling might be fundamentally important for both the circadian cycle and the YMC.
We also speculate that oscillations in NADPH levels might occur during other biological cycles, especially those where phases of intense metabolic activity alternate with phases of reduced metabolic activity, such as the sleep–wake cycle (8). Furthermore, we found that reducing levels of the cystathionine β-synthase enzyme Cys4p by disrupting the 3′ UTR compromises the ability of cells to undergo to the YMC. Defects in cystathionine β-synthase might not only hinder the production of antioxidants, such as GSH, but also induce untimely and excessive production of SAM, which could lead to aberrant methylation of macromolecules. Interestingly, human patients carrying mutations in cystathionine β-synthase exhibit homocystinuria, symptoms of which include mental retardation, lens dislocation, vascular disorders, seizures, and other psychiatric disturbances (32, 33). Moreover, homocystinuria is a risk factor for various neurologic diseases, including Alzheimer's disease and other types of dementia (34). Thus, proper temporal control of sulfur metabolite flux between the two branches of the pathway (GSH and SAM), perhaps by means of a metabolic cycle, might be especially warranted in highly metabolically active cell types such as neurons. We close with the prediction that similar cyclic changes in metabolic or redox state as seen during the YMC might occur during the course of other biological cycles.
Methods
Yeast Protocols and Chemostat Growth.
All yeast manipulations and continuous culture experiments were performed as described in ref. 12. The CEN.PK122 strain was kindly provided by P. Kotter (Johann Wolfgang Goethe-University, Frankfurt am Main, Germany). The cys4 partial loss-of-function allele was created by disrupting the region 40–260 bases downstream of the stop codon (in the 3′ UTR of CYS4) with a G418 resistance marker (KanR).
Metabolite Extraction.
Intracellular metabolites were extracted from yeast, using a method adapted from Castrillo et al. (35). Briefly, at each time point, 1 ml of the continuous chemostat culture (OD ≈8–9) was rapidly quenched by addition into 4 ml of 60% methanol/10 mM Tricine, pH 7.4, that was maintained at −40°C to stop metabolism. After 5 min at −40°C, cells were spun at 1,000 × g for 3 min at −10°C, washed with 1 ml of the same buffer, and then resuspended in 1 ml of 75% ethanol/0.5 mM Tricine, pH 7.4. Intracellular metabolites were extracted by incubating at 80°C for 3 min, followed by incubation at 4°C for 5 min. Samples were spun at 20,000 × g for 1 min to pellet cell debris, and 0.9 ml of the supernatant was transferred to a new tube. After a second spin at 20,000 × g for 10 min, 0.8 ml of the supernatant was transferred to a new tube and stored at −80°C until analysis. For acidic extractions (to help preserve metabolites that might be sensitive to oxidation), the same procedure was used, but tricine was replaced with 0.1% formic acid.
LC-MS/MS and GC×GC-TOFMS.
Methods for LC-MS/MS and GC×GC-TOFMS analysis of metabolites are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank N. Stoilova for technical assistance, J. McDonald and S. Sadjadi for valuable advice on LC-MS, and A. Kudlicki for calculating the periodicities of metabolite oscillation. This work was supported by a National Institutes of Health Director's Pioneer Award (to S.L.M.), unrestricted funds provided by an anonymous donor (to S.L.M.), grants from the National Institutes of Health and National Institute of Diabetes and Digestive and Kidney Diseases (to E.T.Y.), funding support from LECO (to R.E.S.), a Helen Hay Whitney Foundation Postdoctoral Fellowship (to B.P.T.), a Sara and Frank McKnight Foundation Fellowship (to B.P.T.), and a Burroughs Wellcome Fund Career Award in Biomedical Sciences (to B.P.T.). This work was conducted in a facility constructed with support from Research Facilities Improvement Program Grant C06 RR-15437.
Abbreviations
- ALA
aminolevulinic acid
- GSH
glutathione
- LC
liquid chromatography
- Ox
oxidative
- RB
reductive/building
- RC
reductive/charging
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- TOFMS
time-of-flight mass spectrometry
- YMC
yeast metabolic cycle.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708365104/DC1.
References
- 1.Lowrey PL, Takahashi JS. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wijnen H, Young MW. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 3.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 4.Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 5.McDonald MJ, Rosbash M. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 6.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 7.Rutter J, Reick M, McKnight SL. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 8.Tu BP, McKnight SL. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 9.von Meyenburg HK. Arch Microbiol. 1969;66:289–303. [Google Scholar]
- 10.Satroutdinov AD, Kuriyama H, Kobayashi H. FEMS Microbiol Lett. 1992;77:261–267. doi: 10.1016/0378-1097(92)90167-m. [DOI] [PubMed] [Google Scholar]
- 11.Klevecz RR, Bolen J, Forrest G, Murray DB. Proc Natl Acad Sci USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 13.van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin ML, Heijnen JJ, Hoare M, Lange HC, et al. Enzyme Microb Technol. 2000;26:706–714. doi: 10.1016/s0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 14.Mohler RE, Dombek KM, Hoggard JC, Young ET, Synovec RE. Anal Chem. 2006;78:2700–2709. doi: 10.1021/ac052106o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller D, Exler S, Aguilera-Vazquez L, Guerrero-Martin E, Reuss M. Yeast. 2003;20:351–367. doi: 10.1002/yea.967. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 17.Sillje HH, ter Schure EG, Rommens AJ, Huls PG, Woldringh CL, Verkleij AJ, Boonstra J, Verrips CT. J Bacteriol. 1997;179:6560–6565. doi: 10.1128/jb.179.21.6560-6565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Z, Tsurugi K. FEBS J. 2006;273:1696–1709. doi: 10.1111/j.1742-4658.2006.05201.x. [DOI] [PubMed] [Google Scholar]
- 19.Sillje HH, Paalman JW, ter Schure EG, Olsthoorn SQ, Verkleij AJ, Boonstra J, Verrips CT. J Bacteriol. 1999;181:396–400. doi: 10.1128/jb.181.2.396-400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Futcher B. Genome Biol. 2006;7:107. doi: 10.1186/gb-2006-7-4-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi F, Kawai S, Mori S, Kono E, Murata K. FEBS J. 2005;272:3337–3349. doi: 10.1111/j.1742-4658.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 22.Pollak N, Dolle C, Ziegler M. Biochem J. 2007;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atamna H. Ageing Res Rev. 2004;3:303–318. doi: 10.1016/j.arr.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Thomas D, Surdin-Kerjan Y. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 26.Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. Proc Natl Acad Sci USA. 2006;103:6489–6494. doi: 10.1073/pnas.0509531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sohn HY, Murray DB, Kuriyama H. Yeast. 2000;16:1185–1190. doi: 10.1002/1097-0061(20000930)16:13<1185::AID-YEA619>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 28.Kimura H. Antioxid Redox Signal. 2005;7:778–780. doi: 10.1089/ars.2005.7.778. [DOI] [PubMed] [Google Scholar]
- 29.Ladurner AG. Mol Cell. 2006;24:1–11. doi: 10.1016/j.molcel.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Kaasik K, Lee CC. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 31.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 32.Mudd SH, Skovby F, Levy HL, Pettigrew KD, Wilcken B, Pyeritz RE, Andria G, Boers GH, Bromberg IL, Cerone R, et al. Am J Hum Genet. 1985;37:1–31. [PMC free article] [PubMed] [Google Scholar]
- 33.Kraus JP, Janosik M, Kozich V, Mandell R, Shih V, Sperandeo MP, Sebastio G, de Franchis R, Andria G, Kluijtmans LA, et al. Hum Mutat. 1999;13:362–375. doi: 10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 34.Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D'Agostino RB, Wilson PW, Wolf PA. N Engl J Med. 2002;346:476–483. doi: 10.1056/NEJMoa011613. [DOI] [PubMed] [Google Scholar]
- 35.Castrillo JI, Hayes A, Mohammed S, Gaskell SJ, Oliver SG. Phytochemistry. 2003;62:929–937. doi: 10.1016/s0031-9422(02)00713-6. [DOI] [PubMed] [Google Scholar]
- 36.de Hoon MJ, Imoto S, Nolan J, Miyano S. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.