Abstract
New genes with novel functions arise by duplication and divergence, but the process poses a problem. After duplication, an extra gene copy must rise to sufficiently high frequency in the population and remain free of common inactivating lesions long enough to acquire the rare mutations that provide a new selectable function. Maintaining a duplicated gene by selection for the original function would restrict the freedom to diverge. (We refer to this problem as Ohno's dilemma). A model is described by which selection continuously favors both maintenance of the duplicate copy and divergence of that copy from the parent gene. Before duplication, the original gene has a trace side activity (the innovation) in addition to its original function. When an altered ecological niche makes the minor innovation valuable, selection favors increases in its level (the amplification), which is most frequently conferred by increased dosage of the parent gene. Selection for the amplified minor function maintains the extra copies and raises the frequency of the amplification in the population. The same selection favors mutational improvement of any of the extra copies, which are not constrained to maintain their original function (the divergence). The rate of mutations (per genome) that improve the new function is increased by the multiplicity of target copies within a genome. Improvement of some copies relaxes selection on others and allows their loss by mutation (becoming pseudogenes). Ultimately one of the extra copies is able to provide all of the new activity.
Keywords: gene amplification, gene divergence, gene duplication, natural selection
Gene duplications are the principal source of new genes (1–4). Early ideas on origins of new genes were developed and popularized by Ohno (5). As described by him, duplication creates a redundant gene copy that is free from the “relentless pressure of natural selection” and can, while off selection for its initial function, accumulate previously “forbidden mutations,” eventually leading to a new function. Later Kimura and Ohta incorporated the statement, “gene duplication must always precede the emergence of a gene having a new function,” as one of the five principles governing molecular evolution (6). This classical model for the origin of genes with new functions has been called the mutation during nonfunctionality (MDN) model (7) or the neo-functionalization model (8).
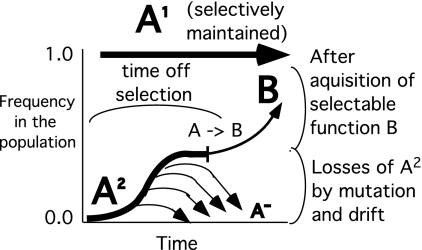
A problem with the MDN model is that the newly duplicated gene is supposed to be neutral and therefore subject to loss by drift and by common inactivating mutations (deletions, frameshifts, nonsense mutations). Thus, the extra copy must drift to high frequency in the population and remain functionally intact long enough to acquire a new selectable function by rare beneficial mutations. The MDN process is diagrammed in Fig. 1.
Fig. 1.
Changes in the frequency of the extra (duplicate) allele (A2). According to the MDN model, the gene (A) duplicates at t = 0, and one copy (A1) is maintained by selection for its original function. The extra allele (A2) is subject to loss from the population by drift and inactivation by common mutations. Frequent loss of A2 will continue until a rare mutation provides a new selectable function.
The Dilemma.
The process described above poses a formidable problem. A new gene copy must acquire the rare mutations that provide a new selectable function. These rare mutations can be acquired only if the gene copy remains in the population for a sufficient time and at a sufficient allele frequency. The standard solution would be to maintain the extra copy by selection. However, such selection would restrict the ability of the copy to lose its old activity and gain a new function.
The Magnitude of the Problem.
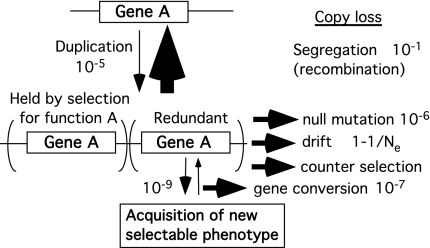
Fig. 2 shows the fate of tandem duplications in bacteria. To assure retention of the extra copy, some form of selection must overcome opposing drift, mutation, recombinational segregation, and gene conversion. Despite the general assumption of the MDN model that duplications are neutral, it seems likely that they are often counterselected due to metabolic cost or deleterious alteration of gene dosage ratios (9–11). In bacteria, the dominant problems are likely to be segregational loss (up to 10% per generation) and counterselection, which varies from undetectable to 15% depending on the size and location of the duplication (M. Pettersson, S. Sun, D.I.A., and O. G. Berg, unpublished results; A. B. Reams, E. Kugelberg, and J.R.R., unpublished results; R. Dawson and J.R.R., unpublished results). Drift will be more important in organisms with smaller populations. However, regardless of population size, loss is the expected fate of the overwhelming majority of duplicated genes (5, 7, 12–14).
Fig. 2.
Comparing rates at which an extra copy either is lost or acquires a new function. The magnitude of the several routes of loss will be different for various organisms. Given numbers are estimated as they might affect bacteria.
Results and Discussion
Previous Models for Maintenance of Multiple Identical Genes.
Several ways of resolving the dilemma have been suggested.
Redundancy could be beneficial.
Redundancy might be positively selected because it protects the genome from negative fitness consequences of degenerative mutations (15, 16). The suggested benefit would seem to be small and to cease as soon as mutants lacking one of the new paralogues become prevalent.
Duplications may be selectively stabilized by subfunctionalization.
Duplicate copies may be free of selection at the moment of duplication, but can soon be stabilized by mutations that inactivate one subfunction of each copy (8). These mutations leave two genes that complement to provide the function of the first. Whereas the two parent gene copies are not selectively maintained initially, this model minimizes their time off selection by using a frequent class of mutations (degenerative mutations that lead to partial loss of function) to create separate genes that can be selectively maintained together. Support for this model has focused on cases in which a single gene gives rise to two copies that perform the same function at different times or in different locations because of alteration of regulatory regions (14).
This model explains how the number of genes (i.e., coding sequences) might increase, but it does not explain how a gene with a totally novel function might evolve. The two stabilized copies are not free to acquire a new function, because both are under selection to provide the original function.
Conversion of alternative alleles to paralogues.
If two alternative alleles at a single locus show a heterotic interaction (overdominance), the heterozygous combination is selected. This advantageous genotype can be stabilized in the population if the locus is duplicated (taking it off selection) and recombination moves the alleles to different loci in the same haploid genome (5, 17). As seen for subfunctionalization, the two copies are maintained by their heterotic effect but neither is free to assume a new function.
Selection for increased gene dosage.
Amplification has long been recognized as a way to maintain multiple copies of a gene in population (5). Whereas there are many examples of gene duplications and amplifications with selective value, theoretical work on early steps in the evolution of new genes has neglected selected amplification as a way to maintain extra copies before divergence. This neglect may reflect the common assumption that selection pressure for multiple copies of a gene is always for “more of the same” in the context of the gene's primary function. An example of this viewpoint is Ohno's discussion of evolution of multiple copies of rRNA genes (5). Similarly, gene duplications and amplifications have generally been treated as temporary responses to selection for higher levels of the original function (18, 19), rather than as intermediates in a process leading to a new gene. Even after amplification was recognized as likely to be “common in the evolution of new enzyme specificities” (19), the nature of their contribution was not defined. Dismissal of amplification neglects the possibility that selection may act on a secondary minor activity of the original gene as proposed here.
Previous Models for Functional Divergence Before Duplication.
The problems inherent in the MDN model led to the idea that the new function was acquired before duplication of the parent gene. The functions of a preexisting multifunctional gene are then partitioned between duplicate copies during the period off selection (7, 20–22).
Jensen (20) advocated the idea that new genes arise by subdividing the function of a preexisting gene having a broad range of activities. After duplication, the copies are off selection until each of them either loses or specializes a different activity yielding two genes with more limited functionality. Similarly, Hughes (7) suggested that an existing gene might acquire a new secondary function that improves fitness. That is, rather than having broad functionality, the parent gene acquires a discrete, selectively valuable second function before duplication.
As in the subfunctionalization model, duplication takes a copy of the (already bifunctional) parent gene off selection, allowing common loss-of-function mutations to remove different activities of the separate genes. These mutations leave two genes that can be selectively maintained for their distinct functionalities (7). The time off selection is minimized by the fact that the specializing mutations are common ones that impair one of the preexisting functions rather than creating a new one.
Origin of New Genes by Selective Amplification (IAD).
The model proposed here differs from previous models in that selection operates at all stages of the process. The model involves innovation, amplification, and divergence (IAD). The IAD model is presented here in detail with supporting experimental observations. Preliminary versions have been outlined (23, 24) and discussed (25).
Innovation.
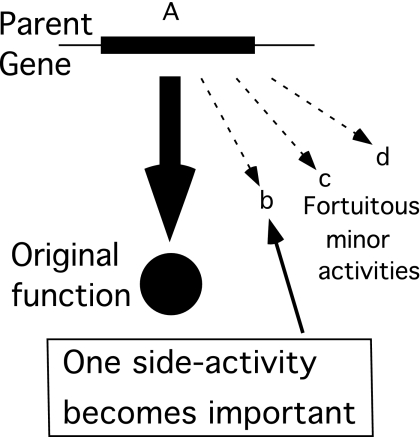
The parent gene encodes a protein, which (like many proteins) provides not only its primary selected function, but also a variety of minor activities that are neither beneficial nor deleterious before the process starts (Fig. 3, activities “b,” “c,” and “d”).
Fig. 3.
Innovation before duplication. It is proposed that the parent gene possesses a minor side activity that becomes selectively valuable before the parent allele is duplicated.
The process of forming a new gene is initiated when a change in the ecological niche (e.g., availability of a novel nutrient or presence of a toxic compound) makes one of these minor activities valuable and imposes a selection for an increase in its level. Alternatively selection could be imposed first and a new mutation could confer a trace of the beneficial side activity before duplication of the parent gene. In either case, the parent allele possesses a trace of the new valuable activity. Selection will then favor any increase in the level of this trace activity.
Amplification.
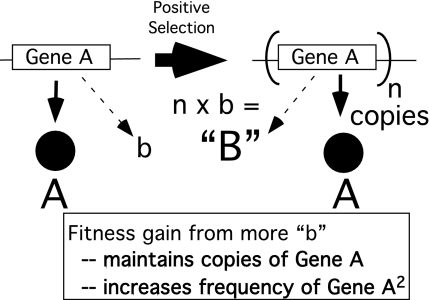
Because duplication and amplification events are four to eight orders of magnitude more common than improving point mutations (see below), an increase in the level of the side activity is likely to occur by amplification of the original gene rather than by point mutations. The number of added gene copies is not limited to duplication. Thus amplification is likely to precede divergence of the parent and nascent gene. The parent gene amplification is raised in frequency in the population and maintained by selection for an increase in the minor activity (Fig. 3, activity “b,” and Fig. 4). This amplification solves the basic problem (Ohno's dilemma).
Fig. 4.
Amplification increases level of side-function. When selection favors a higher level of side-function “b,” a frequent response is amplification of gene A. This amplification leaves a source of the original function “A” and increases the level of the novel function “b.” Selection for increase in the novel function can selectively maintain multiple additional copies of the gene in the genome and increase the frequency of this amplification in the population.
Divergence.
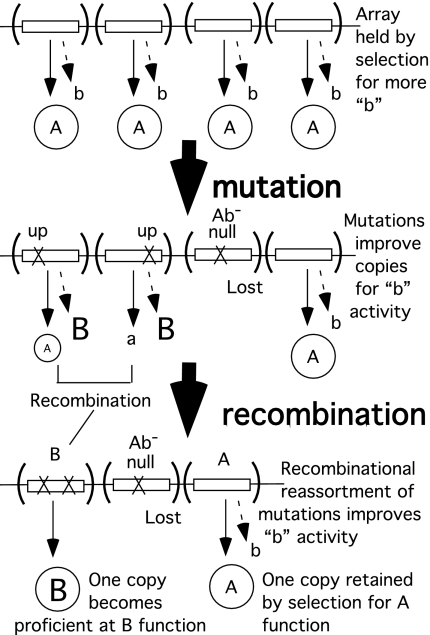
The same selection that favored amplification can favor improvement in the amount of the new activity provided by any single gene copy, all of which are targets for improvement. The number of extra copies enhances the probability of an improving mutation because more mutational targets are available (Figs. 4 and 5). Mutations that improve different gene copies can be assorted by recombination between copies to make new combinations that improve the functionality of some individual copy.
Fig. 5.
Improvement of new function by mutation and recombination. The multiple copies of gene A that are held under selection can be improved for “b” by point mutations in any copy and reassortment of the improving mutations between copies. When any one allele becomes fully proficient at providing “B” function, selection to maintain the other extra copies is relaxed.
A partially improved extra copy is subject to selective amplification if further increases improve fitness. Such secondary amplification could start a new cycle of selected amplification and divergence.
As one copy improves, selection is relaxed on remaining copies, allowing them to be removed from the population by inactivating mutations and drift. Finally one of the extra copies improves sufficiently to provide the new function alone.
Selection will maintain the original function in at least one of the copies as other copies diverge. This activity is likely to be lost by the extra copies in the process of improving their ability to perform the new function. If an excess of the original function imposes a fitness cost, a loss of that function may be positively selected in the course of improving the new activity in the extra copies (Fig. 5).
The IAD Model Is a Darwinian Explanation for the Cairns System.
The IAD model for evolution of new genes was suggested by the behavior of a bacterial genetic system developed to test the idea that selection might cause an increase in mutation rate (26, 27). In this system, a strain of Escherichia coli with a partial defect in its lacZ gene (β-galactosidase) is plated on lactose, conditions that select for improved function of the mutant lacZ allele. The original mutation is leaky and reverts at a rate of ≈10−8 per cell per division during unrestricted growth. However after exposure to selection conditions, 108 cells give rise to 100 Lac+ revertant clones over several days. During this period, the population as a whole shows no growth. Thus valuable mutations seem to be induced by stress in a nongrowing population.
The increase in mutant yield under selection seems to be due almost entirely to selection favoring growth of a subpopulation with an amplification of the weakly functional mutant lac gene. Whereas the plated lac population as a whole grows very little, preexisting cells with two copies of the partially functional mutant lacZ gene initiate clones within which growth is progressively improved by further amplification under selection. Selection increases the number of mutation targets (more cells and more lac copies per cell) rather than by increasing the mutation rate per target copy (28, 29). Ultimately one copy of the mutant allele in one cell of the colony is corrected by a rare mutation and the resulting lac+ strain overgrows the colony. Selection contributes to the creation of a rare highly functional mutant gene by favoring growth of cells with more copies of the partially functional parent allele.
In drawing parallels between behavior of the Cairns system and the process of evolving a new gene, the residual functionality of the mutant lac gene is analogous to the minor side activity of a parental gene, and the rare revertant allele (lac+) is analogous to the new gene. Selection acts on even very small increases in the original (residual) activity and favors growth of cells with such an increase. Small progressive increases are most frequently provided by common events that add copies of the mutant allele. The constant low probability of mutations that improve a single copy provides more mutants when multiple copies of the target are provided by amplification and growth. The cells within a developing bacterial colony are analogous to the individuals in natural population that carry the developing new gene.
Previous work in Drosophila demonstrates the general applicability of the events proposed to explain the Cairns system. The leaky eye color allele (wi) was amplified 4-fold (eight copies in the diploid) by visually selecting individuals with more pigment. Flies with this amplification and a block in forming brown eye pigment had yellow eyes, in which red spots arose as a result of somatic mutations that restored w+ function to any wi copy. Mutation frequency increased linearly as target dosage was increased from one (hemizygous) to eight copies (diploid for the quadruplication) (30). These results demonstrate how selection can increase a function by increasing the dosage of a low-activity gene and how the reversion rate of a mutation in that gene can be enhanced by increasing the number of target sequences in the genome.
Evidence That Many Genes Have Minor Secondary Activities.
The minor activities possessed by many normal enzymes has been referred to as enzyme promiscuity, enzyme moonlighting and underground enzymes (31–35). These secondary activities are likely to be initially neutral. The IAD model suggests that whenever new conditions make a side activity valuable, selection will favor increases in the level of that activity. These quantitative increases are most often achieved by adding copies of the parent gene (see next section).
A widely used laboratory method (high-copy suppression) provides evidence that amplification of a side activity of one protein often serves to supply a novel function. In this method, a null mutation eliminates a particular function and a clone library is screened for genes that (when present in high copy number) can supply the missing function and suppress the phenotype of the original mutation. This method reveals genes with side activity that can supply the missing function (36–38); these activities go unnoticed when the gene is in single copy. For example, a recent study of 104 single gene knockouts in E. coli found that 20% could be rescued by overexpressing noncognate genes (89). The importance of these auxiliary functions in the evolution of new genes has been noted before (35, 39, 89).
Even small dosage increases can reveal secondary activities. In yeast, 8% of a set of 300 null mutants constructed in haploids, became aneuploid for a different chromosome in the process of their isolation, suggesting that the missing function is being selectively replaced at low level by duplication of a distinct gene (40). The same phenomenon has been observed in bacteria, where tandem duplications are selectively stabilized by mutations whose growth defect is relieved by more copies of a distant chromosomal region (R. Dawson and J.R.R., unpublished results). This ability of frequent duplications to provide a missing activity demonstrates the efficacy of a small dosage increase and the frequency with which it can arise.
Evidence That Amplification Is More Likely than Point Mutations to Increase Levels of a Growth-Limiting Activity.
Qualitative improvements of a protein or increases in promoter strength are provided by particular changes at particular sites. Protein alteration is especially restrictive when the parent gene (before duplication) is still under selection to maintain its original function. If one takes 10−10 per cell per division as a frequency of base substitutions per base pair and assumes that 10 such sites per gene can improve function, the expected frequency of improving mutations would be 10−9 per cell per division. A similar substitution rate is seen in metazoan cells and in bacteria (41).
In contrast, in an unselected bacterial population, the frequency of cells with a duplication of various chromosomal regions ranges between 10−2 and 10−5 depending on the region tested (ref. 42 and A. B. Reams and J.R.R., unpublished results). The frequency of duplications approaches a steady-state level that reflects the relative rates at which each duplication forms and is lost by recombination between copies (A. B. Reams and J.R.R., unpublished results). Typical rates of formation and loss in Salmonella enterica are 10−5 and 10−2 per cell per generation respectively, giving duplication-bearing cells a steady state frequency of 10−3. Rates of forming and losing duplications are less well known for eukaryotes, but tests in Drosophila melanogaster and Caenorhabditis elegans suggest a duplication rate in the range of 10−4 to 10−6 (ref. 43 and U.B., unpublished results). The variation in rates is likely to reflect the position of flanking direct-order sequence repeats that allow unequal recombination to form the duplication (42). Recent discoveries of frequent copy number polymorphisms in humans suggest similarly high rates of amplification (44). After duplication, further amplification is expected to be very frequent because the duplicated sequence is a large substrate for unequal sister-strand recombination. Whereas the rate of duplication is lower than that of individual steps of amplification, both are much more frequent than point mutations that improve gene functionality or increase promoter strength. Thus amplification rather than mutational improvement is expected to be the initial response to selection.
Experimental evidence supports the above considerations. Populations can be placed under selection for increasing the level of one of their gene products and such selections yield variants in which the gene for the growth-limiting function is amplified. In bacteria, gene amplifications have been shown to facilitate growth on limited carbon sources (45–50), during infection (51), in the presence of antibiotics (52), and under novel thermal regimes (53, 54). In yeast, gene amplification mediates copper resistance (55) and has been seen in many experimental selection experiments (56–60). In insects amplification provides insecticide resistance (61–64) and metal tolerance (65). In plant (66, 67) and mammalian cell cultures and in tumors, amplifications have been shown to provide resistance to cytotoxic drugs (68). It was recently discovered that gene duplication in humans contributes to resistance to HIV (69).
Applicability of the IAD Model.
The amplification model for origin of new genes was suggested by behavior of bacteria and much of the supporting evidence is from microbes. Whereas we believe that the model described here applies to bacteria, the magnitude of the opposing forces in these organisms (Fig. 2) is large and several lines of evidence suggest that most new eubacterial genes are acquired by horizontal transfer (70). However, on a longer time scale, it seems clear that bacteria have evolved new genes internally (71, 72), whereas horizonal acquisitions are ultimately lost over extended periods. When new genes have formed internally, it seems likely to have involved positive selection and a subset of duplications with low selective cost and segregation rates.
In populations of multicellular organisms, weaker selection may be sufficient because duplications can form there by mechanisms that do not involve the inherently unstable tandem repeats. Furthermore, in these organisms even tandem repeats seem less unstable than in bacteria. Finally, in smaller populations initial duplications will be less subject to selective loss. This weaker counterselection could be the basis for the observed correlation between increases in number of genes in a genome and decreases in the effective population size (73). However, there is no evidence that smaller populations are causally related to increased gene number and, in any event, the measurable cost of duplications suggests that they are not neutral as classically assumed. Thus the process of new gene evolution is likely to require positive selection for the new activity as outlined in the model presented here.
A Role for Amplification in Acquiring New Genes by Horizontal Transfer.
Even acquisition of genes by horizontal transfer may benefit from amplification of the sort described here. In the rare situation that a horizontally acquired gene is beneficial, it is likely to be initially weak at performing the particular function because of problems optimizing gene expression and integration of the new function into a foreign system. When an increase in the new function is required, selective amplification is likely to be the first step in the process. Support for this idea is the observation that foreign genes are more likely to be present in multiple homologous copies in E. coli (74–76).
Predictions of the IAD Model (and Some Examples).
Below are predictions of the amplification model for evolution of new genes and some published evidence that seems to fulfill them. Only a few particularly illustrative examples are presented.
The model predicts that selectable levels of a novel function can be provided by amplification of a parent gene.
An experimental demonstration of a selectable phenotype is the bacterial resistance to novel third-generation cephalosporins by amplification of the chromosomal gene (TEM-1) for β-lactamase. In single copy, this parent allele confers no detectable resistance to these antibiotics (M. Petterson, S. Sun, D.I.A., and O. G. Berg, unpublished results) but is inferred to possess an amplifiable low-level activity. Thus, degradation of a novel substrate is provided by amplification of a gene not known to possess any activity toward this substrate.
The evolution of a new gene may be accompanied by appearance of paralogues in the genome.
After appearance of a new gene, one may find paralogues, some of which are identical to the parent and others that represent transition forms intermediate between the parent and the new gene. After formation of the new gene, these intermediate paralogues can be lost by mutation (become pseudogenes) and ultimately be lost entirely.
Plant defensive genes confer resistance to various pathogens and are found in multiple copies in the genome, frequently clustered on a single chromosome. These genes are thought to have been generated by a “birth and loss” model in response to a succession of slightly variant pathogens (77, 78). Most variation arises by point mutations and exchanges between alleles at a single locus rather than gene conversion between distantly positioned loci. It seems likely that this process is enhanced by local tandem duplications and exchanges between linked paralogues. In support of this idea, clusters with the most closely related homologues show the highest Ka/Ks ratios, suggesting that such clusters are under strong selection and amplification may be an early event in the process of genetic adaptation.
Resistance of the malarial parasite Plasmodium falciparum to certain anti-malarial drugs is sometimes caused by a 2- to 5-fold increase in the copy number of genes for an energy-dependent efflux pump (90). The amplified gene encodes a transmembrane protein homologous to the mammalian mdr gene, which is involved in resistance to several anti-cancer drugs. The model predicts that after sufficient exposure to this selection the pathogen might improve one copy of this array such that the other extra copies could be lost, leaving a new gene.
Some pseudogenes may be found among the paralogues appearing during or after evolution of a new gene.
In Arabidopsis thaliana, the first four enzymes of the synthetic pathway for gibberellic acid (GA) are each encoded by a single gene, but the genome includes multiple paralogues of each one (91). The family (KS) that includes the gene for the second enzyme has nine paralogues, three located in tandem and the rest scattered on four different chromosomes. The one gene active in GA synthesis is located within the cluster. The scattered paralogues do not contribute to GA synthesis but seem to have acquired a distinct function, synthesis of polycyclic diterpenes, made in response to pathogen infection and UV irradiation. One paralogue is a pseudogene. If the genes of the GA pathway have been used as precursors for catalysts in a new pathway, then the multiplicity of new paralogues, the location of some paralogues in tandem and the inclusion of pseudogenes among the paralogues are all predictions of the model.
New genes (and possibly pseudogenes) may be clustered with the parent gene.
This prediction is expected when duplications arise as tandem repeats. There are numerous examples that support this expectation, including the hox genes (79), globins (80), and human red-green opsin genes (81, 82). Perhaps the most striking example is the genome of Trypanosoma cruzi, which contains ≈50% repetitive sequence, consisting mostly of surface proteins, retrotransposons, and subtelomeric repeats (83). The genome contains 1,052 paralogous clusters of ≈2 genes and as many as 46 clusters or ≈20 genes. Approximately 15% of the total number of genes are pseudogenes. Whereas duplicates may often be in tandem, the IAD model does not require this direct-order clustering because alternative mechanisms of gene duplication in eukaryotes can generate copies in inverse order or on different chromosomes (58, 84, 85).
The possibility of creating new genes under selection suggests that new genes could arise rapidly.
Positive selection opens the possibility of greater increases in copy number and increased rate (per genome) of mutationally improved copies. New genes might arise during speciation under selection. A recent example is in evolution of group-I phospholipase in elapids, which seems associated with speciation events (86). An alternative role for duplications in speciation has been suggested (87).
Sequences of new genes should show evidence of continuous selection.
Classical models (MDN) and subfunctionalization predict that the sequence of a new gene will show evidence of a period off selection. In contrast, the IAD model described here predicts that the new genes (with new functions) arise under continuous positive selection. Direct tests support continuous selection during evolution of new genes (13, 22). A particular example of selection during divergence is the Drosophila gene jingwei, which acquired eight replacements and no synonymous substitutions over the estimated 30 million years during which it arose (88).
Many homologue families, pseudogenes, and copy-number polymorphisms may reflect operation of the IAD model.
The model posits that the frequency of copy number variants will increase in response to selection and that (after appearance of a highly functional new gene) the excess copies will be lost by mutation or segregation. Many of the large gene families and pseudogenes observed in genomes may reflect the operation of this process. Alternatively, copy-number polymorphisms may be maintained because they compensate for deleterious mutations in genes within the amplified region.
Summary.
It is suggested that new genetic functions arise when selection is imposed on a minor side function of a preexisting gene. This activity is increased by duplication and higher amplification of the original gene with extra copies held by selection for the new activity. As extra-copies improve their specific activity for the new function, they can assort variability by recombination and diverge from the parent gene.
Acknowledgments
The germ of the idea presented here was suggested by Frank Stahl upon first hearing the amplification model for adaptive mutation and while watching Galapagos tortoises in the San Diego Zoo. The idea followed a weekend of discussions of the new gene problem with Stahl, Russell Lande, and J.R.R. We thank Mel Green for discussions of duplications in Drosophila. This work was supported by National Center for Research Resources Grant P20 RR18754 (to U.B.), the Swedish Research Council (D.I.A.), and National Institutes of Health Grant GM27068 (to J.R.R.).
Abbreviations
- MDN
mutation during nonfunctionality
- IAD
innovation, amplification and divergence
- GA
giberellic acid.
Footnotes
The authors declare no conflict of interest.
References
- 1.Muller HJ. Science. 1936;83:528–530. doi: 10.1126/science.83.2161.528-a. [DOI] [PubMed] [Google Scholar]
- 2.Lewis EB. Cold Spring Harbor Symp Quant Biol. 1951;16:159–174. doi: 10.1101/sqb.1951.016.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Sturtevant AH. Genetics. 1925;10:117–147. doi: 10.1093/genetics/10.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haldane JBS. The Causes of Evolution. Ithaca, NY: Cornell Univ Press; 1932. [Google Scholar]
- 5.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 6.Kimura M, Ohta T. Proc Natl Acad Sci USA. 1974;71:2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes AL. Proc R Soc London B Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 8.Force A, Lynch M, Bryan Pickett F, Amores A, Yan Y, Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veitia RA. BioEssays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- 10.Veitia RA. Genetics. 2004;168:569–574. doi: 10.1534/genetics.104.029785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papp B, Pal C, Hurst LD. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 12.Haldane JBS. Am Nat. 1933;67:5–19. [Google Scholar]
- 13.Lynch M, Conery JS. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 14.Lynch M, Force A. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark AG. Proc Natl Acad Sci USA. 1994;91:2950–2954. doi: 10.1073/pnas.91.8.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowak MA, Boerlijst MC, Cooke J, Smith JM. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 17.Spofford JB. Am Nat. 1969;103:407–432. [Google Scholar]
- 18.Romero D, Palacios R. Annu Rev Genet. 1997;31:91–111. doi: 10.1146/annurev.genet.31.1.91. [DOI] [PubMed] [Google Scholar]
- 19.Hartley BS. In: Microorganisms as Model Systems for Studying Evolution. Mortlock RP, editor. New York: Plenum; 1984. pp. 23–54. [Google Scholar]
- 20.Jensen RA. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 21.Piatigorsky J, Wistow G. Science. 1991;252:1078–1079. doi: 10.1126/science.252.5009.1078. [DOI] [PubMed] [Google Scholar]
- 22.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. research0008.1–0008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth JR, Benson N, Galitski T, Haack K, Lawrence JG, Miesel L. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F, Ingraham J, Low K, Magasanik B, Schaechter M, Umbarger H, editors. Vol 2. Washington, DC: Am Soc Microbiol; 1996. pp. 2256–2276. [Google Scholar]
- 24.Hendrickson H, Slechta ES, Bergthorsson U, Andersson DI, Roth JR. Proc Natl Acad Sci USA. 2002;99:2164–2169. doi: 10.1073/pnas.032680899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francino MP. Nat Genet. 2005;37:573–577. doi: 10.1038/ng1579. [DOI] [PubMed] [Google Scholar]
- 26.Cairns J, Overbaugh J, Miller S. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- 27.Cairns J, Foster PL. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kugelberg E, Kofoid E, Reams AB, Andersson DI, Roth JR. Proc Natl Acad Sci USA. 2006;103:17319–17324. doi: 10.1073/pnas.0608309103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Annu Rev Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- 30.Green MM, Todo T, Ryo H, Fujikawa K. Proc Natl Acad Sci USA. 1986;83:6667–6671. doi: 10.1073/pnas.83.18.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aharoni A, Gaidukov L, Khersonsky O, Mc QGS, Roodveldt C, Tawfik DS. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 32.Bornscheuer UT, Kazlauskas RJ. Angew Chem Int Ed Engl. 2004;43:6032–6040. doi: 10.1002/anie.200460416. [DOI] [PubMed] [Google Scholar]
- 33.Copley SD. Curr Opin Chem Biol. 2003;7:265–272. doi: 10.1016/s1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 34.D'Ari R, Casadesus J. BioEssays. 1998;20:181–186. doi: 10.1002/(SICI)1521-1878(199802)20:2<181::AID-BIES10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 35.O'Brian P, Herschlag D. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 36.Ueguchi C, Ito K. J Bacteriol. 1992;174:1454–1461. doi: 10.1128/jb.174.5.1454-1461.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg CM, Wang MD, Vartak NB, Liu L. Gene. 1988;65:195–202. doi: 10.1016/0378-1119(88)90456-8. [DOI] [PubMed] [Google Scholar]
- 38.Bender A, Pringle JR. Proc Natl Acad Sci USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller BG, Raines RT. Biochemistry. 2004;43:6387–6392. doi: 10.1021/bi049424m. [DOI] [PubMed] [Google Scholar]
- 40.Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, et al. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 41.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson P, Roth J. Proc Natl Acad Sci USA. 1981;78:3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shapira SK, Finnerty VG. J Mol Evol. 1986;23:159–167. doi: 10.1007/BF02099910. [DOI] [PubMed] [Google Scholar]
- 44.Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, Lam WL. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tlsty TD, Albertini AM, Miller JH. Cell. 1984;37:217–224. doi: 10.1016/0092-8674(84)90317-9. [DOI] [PubMed] [Google Scholar]
- 46.Sonti RV, Roth JR. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horiuchi T, Horiuchi S, Novick A. Genetics. 1963;48:157–169. doi: 10.1093/genetics/48.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong S, Khodursky A, Dykhuizen DE, Dean AM. Proc Natl Acad Sci USA. 2004;101:11719–11724. doi: 10.1073/pnas.0404397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Straus DS, Hoffmann GR. Genetics. 1975;80:227–237. doi: 10.1093/genetics/80.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reams AB, Neidle EL. Mol Microbiol. 2003;47:1291–1304. doi: 10.1046/j.1365-2958.2003.03342.x. [DOI] [PubMed] [Google Scholar]
- 51.Mekalanos JJ. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 52.Edlund T, Normark S. Nature. 1981;292:269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- 53.Riehle MM, Bennett AF, Long AD. Proc Natl Acad Sci USA. 2001;98:525–530. doi: 10.1073/pnas.021448998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergthorsson U, Ochman H. J Bacteriol. 1999;181:1360–1363. doi: 10.1128/jb.181.4.1360-1363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fogel S, Welch JW. Proc Natl Acad Sci USA. 1982;79:5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansche PE. Genetics. 1975;79:661–674. doi: 10.1093/genetics/79.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams J, Puskas-Rozsa S, Simlar J, Wilke CM. Curr Genet. 1992;22:13–19. doi: 10.1007/BF00351736. [DOI] [PubMed] [Google Scholar]
- 58.Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Proc Natl Acad Sci USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Infante JJ, Dombek KM, Rebordinos L, Cantoral JM, Young ET. Genetics. 2003;165:1745–1759. doi: 10.1093/genetics/165.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bond U, Neal C, Donnelly D, James TC. Curr Genet. 2004;45:360–370. doi: 10.1007/s00294-004-0504-x. [DOI] [PubMed] [Google Scholar]
- 61.Wang JY, McCommas S, Syvanen M. Mol Gen Genet. 1991;227:260–266. doi: 10.1007/BF00259679. [DOI] [PubMed] [Google Scholar]
- 62.Newcomb RD, Gleeson DM, Yong CG, Russell RJ, Oakeshott JG. J Mol Evol. 2005;60:207–220. doi: 10.1007/s00239-004-0104-x. [DOI] [PubMed] [Google Scholar]
- 63.Devonshire AL, Field LM. Annu Rev Entomol. 1991;36:1–23. doi: 10.1146/annurev.en.36.010191.000245. [DOI] [PubMed] [Google Scholar]
- 64.Lenormand T, Guillemaud T, Bourguet D, Raymond M. Evolution (Lawrence, Kans) 1998;52:1705–1712. doi: 10.1111/j.1558-5646.1998.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 65.Maroni G, Wise J, Young JE, Otto E. Genetics. 1987;117:739–744. doi: 10.1093/genetics/117.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donn G, Tischer E, Smith JA, Goodman HM. J Mol Appl Genet. 1984;2:621–635. [PubMed] [Google Scholar]
- 67.Watanabe N, Takayama S, Yoshida S, Isogai A, Che FS. Biosci Biotechnol Biochem. 2002;66:1799–1805. doi: 10.1271/bbb.66.1799. [DOI] [PubMed] [Google Scholar]
- 68.Schimke RT. Gene Amplification. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1982. [Google Scholar]
- 69.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 70.Ochman H, Lawrence JG, Groisman EA. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 71.Yanai I, Camacho CJ, DeLisi C. Phys Rev Lett. 2000;85:2641–2644. doi: 10.1103/PhysRevLett.85.2641. [DOI] [PubMed] [Google Scholar]
- 72.Ge F, Wang LS, Kim J. PLoS Biol. 2005;3:e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lynch M, Conery JS. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 74.Hooper SD, Berg OG. J Mol Evol. 2002;55:734–744. doi: 10.1007/s00239-002-2369-2. [DOI] [PubMed] [Google Scholar]
- 75.Hooper SD, Berg OG. Genome Biol. 2003;4:R48. doi: 10.1186/gb-2003-4-8-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hooper SD, Berg OG. Mol Biol Evol. 2003;20:945–954. doi: 10.1093/molbev/msg101. [DOI] [PubMed] [Google Scholar]
- 77.Bergelson J, Kreitman M, Stahl EA, Tien D. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- 78.Michelmore RW, Blake CM. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 79.Wagner GP, Amemiya C, Ruddle F. Proc Natl Acad Sci USA. 2003;100:14603–14606. doi: 10.1073/pnas.2536656100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bunn HF, Forget BG. Hemoglobin: Molecular, Genetic and Clinical Aspects. Philadelphia: Saunders; 1984. [Google Scholar]
- 81.Hoffmann M, Tripathi N, Henz SR, Lindholm AK, Weigel D, Breden F, Dreyer C. Proc Biol Sci. 2007;274:33–42. doi: 10.1098/rspb.2006.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trezise AE, Collin SP. Curr Biol. 2005;15:R794–R796. doi: 10.1016/j.cub.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 83.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, et al. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 84.Lobachev KS, Shor BM, Tran HT, Taylor W, Keen JD, Resnick MA, Gordenin DA. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 86.Lynch VJ. BMC Evol Biol. 2007;7:2. doi: 10.1186/1471-2148-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lynch M, Force A. Am Nat. 2000;156:590–605. doi: 10.1086/316992. [DOI] [PubMed] [Google Scholar]
- 88.Long M, Langley CH. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 89.Patrick WM, Quandt EM, Swartzlander DB, Matsumura I. Mol Biol Evol. 2007 Sep 19; doi: 10.1093/molbev/mxm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nair S, Nash D, Sudimack D, Jaidee A, Barends M, Uhlemann AC, Krishna S, Nosten F, Anderson TJ. Mol Biol Evol. 2007;24:562–573. doi: 10.1093/molbev/msl185. [DOI] [PubMed] [Google Scholar]
- 91.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M, Matsuoka M. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]