Abstract
Terminase enzyme complexes, which facilitate ATP-driven DNA packaging in phages and in many eukaryotic viruses, constitute a wide and potentially diverse family of molecular motors about which little dynamic or mechanistic information is available. Here we report optical tweezers measurements of single DNA molecule packaging dynamics in phage T4, a large, tailed Escherichia coli virus that is an important model system in molecular biology. We show that a complex is formed between the empty prohead and the large terminase protein (gp17) that can capture and begin packaging a target DNA molecule within a few seconds, thus demonstrating a distinct viral assembly pathway. The motor generates forces >60 pN, similar to those measured with phage φ29, suggesting that high force generation is a common property of viral DNA packaging motors. However, the DNA translocation rate for T4 was strikingly higher than that for φ29, averaging ≈700 bp/s and ranging up to ≈2,000 bp/s, consistent with packaging by phage T4 of an enormous, 171-kb genome in <10 min during viral infection and implying high ATP turnover rates of >300 s−1. The motor velocity decreased with applied load but averaged 320 bp/s at 45 pN, indicating very high power generation. Interestingly, the motor also exhibited large dynamic changes in velocity, suggesting that it can assume multiple active conformational states gearing different translocation rates. This capability, in addition to the reversible pausing and slipping capabilities that were observed, may allow phage T4 to coordinate DNA packaging with other ongoing processes, including viral DNA transcription, recombination, and repair.
Keywords: bacteriophage T4, molecular motor, optical tweezers, single-molecule, viral DNA packaging
A critical step in the assembly of many viruses is the packaging of the viral genome into a preassembled prohead shell by the action of an ATP-powered molecular motor (1, 2). Systems in which this mode of assembly occurs include numerous tailed dsDNA and dsRNA phages and certain animal viruses, including adenoviruses and herpesviruses. Viral DNA packaging complexes thus constitute a wide and potentially diverse family of molecular motors that are considerably understudied compared with cellular molecular motors such as myosins, kinesins, and helicases.
In a typical phage assembly pathway, a prohead shell of precise dimensions co-assembles with a scaffolding core. One of the vertices of the prohead is unique, containing a dodecameric portal ring structure (3). When the scaffolding leaves, a defined space is created inside the capsid. A packaging ATPase complex then docks onto the outer end of the portal, inserting one end of the viral genome into the 3.5- to 4-nm channel, and translocates the DNA by using ATP hydrolysis energy (2, 4). After genome packaging, the ATPase dissociates, leaving the portal with the head, the outer surface of which provides a platform for the assembly of tail components. When the virus infects a cell, the densely packed DNA exits rapidly through the portal channel and tail tube into the host (5).
Recently we found that the phage φ29 packaging motor translocates DNA at up to ≈165 bp/s and exerts surprisingly large forces, estimated to be as high as 80 pN (6, 7). A strong motor was shown to be necessary to overcome high internal forces that oppose dense DNA confinement in the prohead and which were proposed to play an important role in driving DNA ejection during infection (6). Recent studies of phage λ DNA ejection also deduced high forces of ≈20 pN (8). The dynamic and mechanistic characteristics of the packaging motors, however, are poorly understood. Considering that phages are the most abundant life forms on earth (9), these characteristics may vary widely with capsid size, shape, and structure; genome length; packing density; biochemical properties; and host cell environment.
T4 is an important model for large tailed phages that exhibits distinct differences from phage φ29 (10). Most notably, T4 has a much larger capsid (120 × 86 nm) and must package a 9× longer genome than φ29 during a shorter overall time window during infection (11, 12). The measured φ29 DNA translocation rate is too slow to explain the full packaging of the 171-kb T4 genome in this time window. T4 is also the prototype for a majority of viruses that package DNA by a headful mechanism (4) and lacks the unusual RNA component found in the φ29 motor (1).
Several recent advances make T4 an attractive system for studying the packaging motor function. A defined in vitro T4 packaging system consisting of only three components [empty proheads, the large terminase protein (gp17, the packaging ATPase), and DNA] has recently been developed. In the presence of ATP and under carefully optimized reaction conditions in bulk assays, as much as 50–100% of the input DNA has been packaged (13–15). T4 is also the only virus for which an atomic structure of the packaging ATPase has been determined. Specifically, in recent work the N-terminal ATPase domain of gp17 was determined at up to 1.8 Å resolution (16). The structure was found to most closely resemble monomeric helicases, which are proposed to translocate along DNA by an inchworm mechanism (17).
Here we report direct measurements of single DNA molecule packaging in T4. First, we demonstrate a distinct assembly pathway for the prohead–motor complex, leading to rapid initiation of packaging. Second, we find that the motor generates very high forces, suggesting that high force generation is a universal property of viral DNA packaging motors. Third, we find that the prohead–gp17 complex can, by itself and without any other components, translocate DNA processively at strikingly high rates, consistent with the packaging of the enormous T4 genome in vivo in a limited time window during infection. Fourth, we find that the motor has the capability to translocate DNA at variable rates and to reversibly pause and slip, capabilities that may be needed to regulate and coordinate DNA packaging with ongoing DNA transcription, recombination, and repair processes.
Results and Discussion
Initiation of Packaging.
We found that single T4 prohead–motor complexes could be triggered to initiate DNA packaging in vitro by manipulation with optical tweezers. This approach represents an improvement over earlier studies of phage φ29 that assessed partly packaged, prestalled complexes (6, 18) because it enables us to study the initiation and earliest stages of DNA packaging. This approach was made possible by our finding that prohead–gp17 complexes could be preassembled with a nonhydrolyzable ATP analog (γ-S-ATP) and immobilized on microspheres by using antibodies. DNA molecules were tethered to a separate preparation of microspheres, and packaging was initiated by bringing the DNA and prohead–motor complexes into proximity, as shown schematically in Fig. 1. A microsphere carrying DNA was captured in one trap, and a microsphere carrying prohead–gp17 complexes was captured in a second trap. The two microspheres were quickly brought into near contact for 1 s in the presence of packaging buffer containing ATP and then quickly separated. This procedure was repeated until we detected single DNA binding, inferred by the measured rise in force when the DNA was stretched. We found that binding could occur rapidly, within seconds of bringing DNA into the proximity of the complexes, and that DNA translocation (packaging) initiated within 1 s after binding.
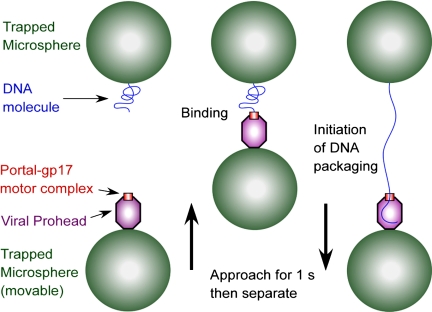
Fig. 1.
Schematic of the experiment. T4 prohead–gp17 complexes were attached to antibody-coated microspheres and captured in one optical trap (lower left). Biotinylated DNA molecules were tethered to streptavidin-coated microspheres and captured in a second optical trap (upper left). The bottom trap was moved with respect to the top trap while measuring the force. To initiate packaging, the microspheres were brought into near contact for 1 s (middle) and then quickly separated to detect DNA binding and translocation (right).
Our study thus defines a distinct assembly pathway for the packaging machinery. In previous studies, the pathway was unclear because three components (proheads, gp17, and DNA) were simply mixed together. It was proposed that the terminase complex first binds the DNA and then binds to the prohead to initiate packaging (10). Our present experiment demonstrates an alternate pathway in which a prohead–gp17 motor assembles first and then binds DNA. We note that this competent prohead–gp17 complex does not retain activity very long after a large dilution in the buffer containing ATP. After injection of the microspheres carrying the prohead–gp17 complexes into the measurement chamber (an essentially infinite dilution), packaging was only initiated when the DNA was presented within ≈20 s. Similar behavior was also observed with the phage φ29 motor (J. Rickgauer, D.N.F., and D.E.S., unpublished data). Inclusion of BSA (0.1 mg/ml) and DTT (1 mM), commonly used stabilizers, had no effect (n = 14 recorded events). The success of our assay thus depended on our ability to quickly load the microspheres into the traps and to quickly probe for DNA binding. After initiating packaging, the complex becomes very strongly bound to the prohead, as evidenced by the fact that large forces can be applied (as described below). One interpretation is that threading of the DNA through the portal is necessary to stabilize the active translocating conformation.
Data were collected in two modes (6). In the “fixed-traps” mode, after the DNA was stretched and packaging was detected, we held both traps stationary and measured a progressive increase in DNA tension as the motor reeled in the DNA. In the “force–clamp” mode, the DNA tension was held constant by using a feedback loop to control the separation of the traps.
Force Generation.
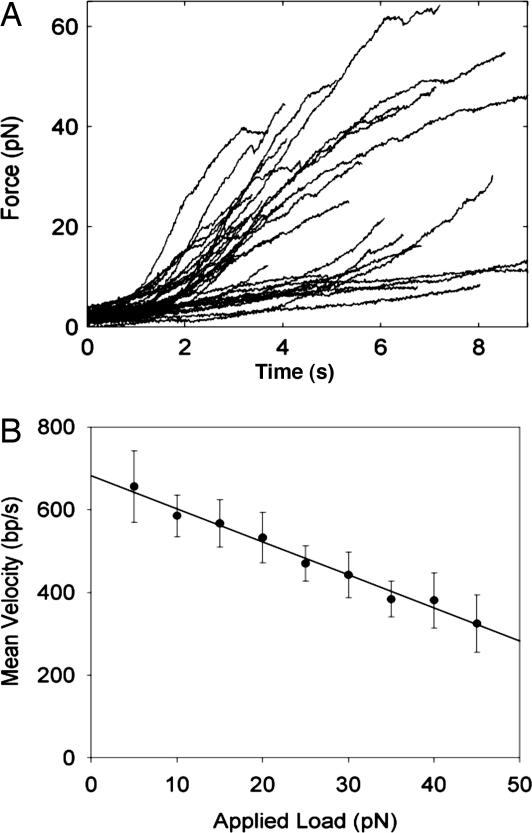
Measurements in the fixed-traps mode on n = 33 individual packaging complexes are shown in Fig. 2A. These measurements were made with <10% of the full T4 genome length of DNA packaged, where the internal forces resisting DNA confinement are expected to be small (7, 19). These data show that the T4 motor can generate large forces. The maximum force recorded was 62 pN, which is close to the force at which dsDNA becomes strongly distorted under tension (20). This value is a lower bound on the force generation because most measurements ended with the tether breaking, rather than the motor stalling. These breakage events are consistent with forced disruption of the antibody–prohead link (21), although they could also correspond to release of the DNA from the motor–prohead complex. The wide variability in the plots in Fig. 2A reflects differences in the starting forces imposed during the experiment, as well as inherent differences in the rates of packaging by different complexes, as discussed further below.
Fig. 2.
Force and velocity measurements. (A) Force measurements on n = 33 complexes in the fixed-traps mode. As DNA packaging proceeds, the DNA tension and load opposing the motor increases. Each measurement ends with breakage of the tether (see text). (B) Analysis of the data in A yields the mean velocity vs. applied load. The error bars are standard error of the mean, calculated as SD divided by the square root of the number of measurements (n = 33). The solid line is a linear fit.
The free energy release of ATP hydrolysis may be calculated as ΔG = ΔG0 + RT ln([ADP][Pi]/[ATP]) ≅ −73 kJ/mol, where ΔG0 ≅ −30 kJ/mol is the standard free energy change; R is the gas constant; T is the temperature; and [ATP], [ADP], and [Pi] are the concentrations of ATP, ADP, and Pi in the reaction mixture (22). Expressed in energy units of force × displacement per ATP hydrolyzed, 73 kJ/mol ≅ 120 pN·nm per ATP. The observation that the motor can translocate the DNA against a force, F, >60 pN implies that the length of DNA translocated per ATP hydrolyzed (ΔL) must be <6 bp because the work done (FΔL) must be less than the free energy release of ≈120 pN·nm per ATP hydrolysis (120/60 = 2 nm ≅ 6 bp). The lower bound on force implies an upper bound on step size because energy = force × step size. Any higher value of force or energy conversion efficiency <100% would imply an even smaller step size.
The force generated by the T4 motor is at least 20× higher than that generated by skeletal muscle myosin, 8× higher than that generated by conventional kinesin motors, and 8× higher than that generated by RecBCD helicase (23, 24). In our most recent studies of φ29 (7), we showed that combination of an externally applied load and an internal load at high capsid filling implies that the motor can generate >80 pN. In the present measurements, we put a bound of >62 pN through direct measurement by applying external load alone. We conclude that both motors generate similarly high forces in the sense that >80 pN and >62 pN are both much higher than the forces generated by classical motors such as myosin II, which generates only ≈3 pN (23). That both the T4 and φ29 motors produce very high forces suggests that high force generation may be a general feature of viral DNA packaging motors. This finding is sensible given that most dsDNA viruses package DNA to similarly high density, and this packing is expected to require overcoming large opposing forces resulting from DNA bending rigidity, entropic change, and electrostatic self-repulsion.
Dependence of Motor Velocity on Load.
Analysis of the data in Fig. 2A reveals the dependence of the mean velocity (averaged over all complexes) vs. applied force (Fig. 2B). In the limit of low load, the average velocity is very high, ≈700 bp/s, which is ≈5× higher than that of the φ29 motor. As with φ29, the velocity decreases with increasing load, indicating that a step in the mechanochemical cycle of the motor involving DNA translocation is an important rate-limiting step in the kinetics (25). In addition to being faster than φ29, we find that the T4 motor has a lower sensitivity to force. When the load was increased from 5 to 30 pN, the T4 motor velocity decreased by ≈1/3, whereas the φ29 motor velocity decreased by ≈1/2 (7), suggesting that the T4 motor may be capable of generating even higher forces than the φ29 motor. This was somewhat surprising because the forces resisting packaging of the T4 genome are actually predicted to be ≈40% lower than those in φ29 (because of the larger capsid size, different shape, and slightly lower packing density) (19).
The T4 motor also generates 7× higher power (a product of average force and velocity) than the φ29 motor. The maximum power occurred with an applied load of 40 pN, where the velocity was 380 bp/s (Fig. 2B), yielding 40 pN × 380 bp/s = 15,200 pN·bp/s = 5.2 × 10−18 W. If the motor step per ATP is similar to that of φ29 (2 bp) (18), T4 must be capable of hydrolyzing ATP at least 7× faster than φ29. The power generation may seem small, but the motor is a nanoscale device, occupying a volume of only approximately (10 nm)3. The power density is thus ≈5,000 kW/m3, approximately twice that generated by a car engine. Given a free energy release of 120 pN·nm per ATP, the power value implies an ATP hydrolysis rate of at least 5,200 pN·nm/s ÷ 120 pN·nm per ATP ≅ 40 ATP molecules per second (ATP/s) per complex (or higher if the energy conversion efficiency is <100%). If each motor step is tightly coupled to one ATP hydrolysis, and the step size is independent of load, the upper bound of 6 bp on the step size, and the measured maximum velocity of 1,840 bp/s, imply an even higher bound on the hydrolysis rate of at least 1,840 bp/s ÷ 6 bp per ATP = 300 ATP/s; higher than the highest bound previously reported of ≈400 ATP per gp17 per minute (26) (≅ 33 ATP/s per complex, assuming a pentameric gp17 ring).
Force–Clamp Measurements.
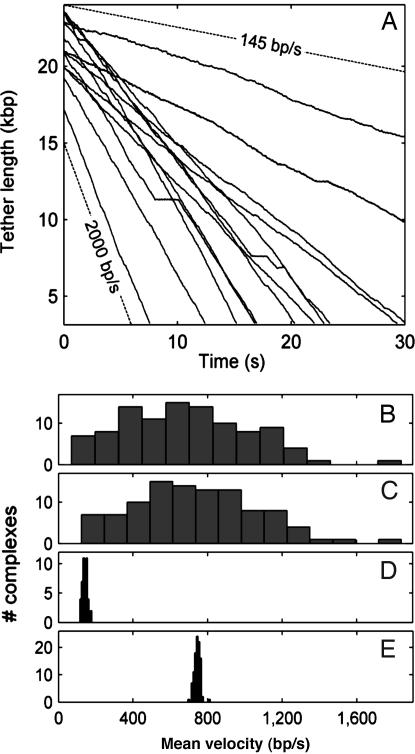
These measurements allowed us to follow the packaging dynamics over longer DNA lengths (Fig. 3A). A low, 5-pN DNA stretching force was applied to facilitate accurate measurement while not decreasing the motor velocity substantially. A 25.3-kb DNA construct, much shorter than the full T4 genome, was used, such that negligible internal force was expected. Indeed, the change in velocity with filling was negligible (≈1 bp/s on average per percentage of the genome packaged), in accord with theoretical predictions (19). Technical limitations in the optics currently prevent us from manipulating the entire T4 genome. The present measurements focus on characterizing inherent motor function aside from internal load effects. Two immediately evident features are the high packaging rates and the high variability in the rates. The average rate was 690 bp/s; however, some complexes were slower and some substantially faster, ranging up to ≈2,000 bp/s.
Fig. 3.
DNA translocation dynamics and static disorder. (A) DNA tether length vs. time, measured after initiation of packaging and application of a 5-pN force clamp. Each line is a recording on a different packaging complex. Thirteen of the n = 102 recorded data sets are shown. The lower dashed line indicates a packaging rate of 2,000 bp/s, and the uppermost indicates 145 bp/s, the average rate of phage φ29. (B) Distribution of average motor velocities measured for each of the recorded packaging events with n = 102 complexes in the force–clamp mode. (C) Same distribution after editing pauses and slips from the records (see text). (D) Corresponding distribution recorded with φ29 (n = 50). (E) Distribution of rates expected for simulated data sets having the same mean velocity, using the simple kinetic model (see text).
Reversible Pausing and Slipping.
Occasional pauses in translocation were evident in the force–clamp measurements, appearing as plateaus in the length vs. time plots (Fig. 3A). Plateaus of duration >0.2 s, preceded and followed by active packaging, during which the standard deviation (SD) in length was statistically indistinguishable (within 2 SD) from that in negative control experiments, were scored as pauses. Negative control experiments were done by tethering the DNA template between microspheres, as described in ref. 21, in the absence of packaging proteins. A total of 633 pauses were identified in the 102 recorded events; an average of 2.3 kb of DNA was packaged per pause. The mean pause duration was 0.66 s (SD = 0.66 s), and the maximum was 5.4 s. Within experimental resolution, pauses occurred at random positions and did not depend strongly on the length of DNA packaged (correlation coefficient = −0.04). On average, the pauses were ≈6× shorter in duration than those observed with φ29 (6), a difference that mirrors the ≈5× higher average packaging rate with T4 and suggests a similar mechanism of pause recovery.
Slips in which the DNA came partially out of the capsid were occasionally observed (Fig. 2A). A total of 112 slips >50 bp were tabulated. The mean slip was 150 bp (SD = 186 bp), and the maximum was 1.6 kb. The slips occurred evenly throughout packaging. Approximately half were faster than the average rate (690 bp/s), and half were slower. This observation of slow slips differs from our finding with φ29, where all slips were faster than the packaging rate (6). The mean rate of the slow slips was 400 bp/s (SD = 185 bp/s), mean size was 135 bp (SD = 98 bp), and mean duration was 0.48 s (SD = 0.48 s). The longest duration was 2.0 s for a slow slip of 440 bp. Given that the T4 virus can eject its DNA within seconds (12), the observed rate of slipping must be limited by friction between the motor and DNA. On the basis of its structure in φ29 (3), the portal has been proposed to function during packaging as a “one-way valve” to restrict backward slipping (G. Oster, personal communication). Our observation of many slips slower than the ejection rate is consistent with this proposal.
We also conclude that the motor is highly processive: On average, 12.8 kb was packaged per slip, similar to our finding with φ29 (6). Although 12.8 kb is considerably less than the 171 kb genome length of T4, most slips were short and did not cause the DNA to completely exit the capsid. Thus, packaging as a whole is highly processive. Overall, pauses and slips had only a minor effect on the packaging rate, slowing it by ≈10% on average, although some complexes were slowed by up to 33%.
Variability in Motor Velocity.
A histogram of the mean packaging rates for all packaging events is shown in Fig. 3B. The mean is 690 bp/s, and the SD is very large (340 bp/s). The slowest complex averaged 70 bp/s and the fastest averaged 1,840 bp/s. Pauses and slips had a significant effect on certain records but were not the primary cause of the overall variability. Fig. 3C shows the velocities after editing of the pauses and slips as described above. The mean rate increased slightly to 770 bp/s, and the SD remained high (320 bp/s). The strikingly high velocity of the T4 motor is evident when the velocity distribution is compared with that of φ29 (Fig. 3D) (mean = 142 bp/s, SD = 14 bp/s). The T4 velocity is also much more variable. The finding of such different behavior is perhaps not surprising because there are several distinct differences between the two motors, as noted in the Introduction.
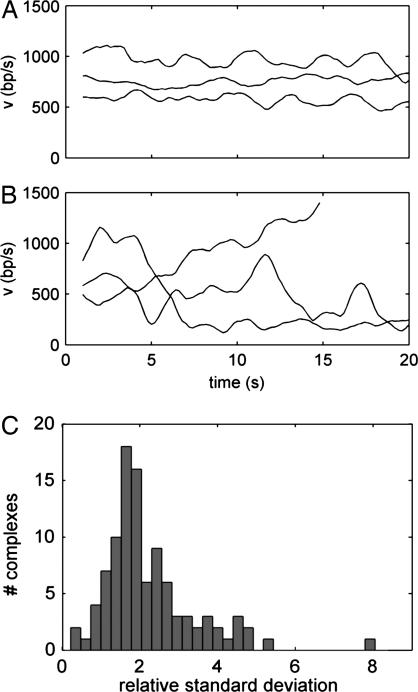
The wide velocity variations observed are rather surprising and cannot be explained by simple kinetics models (25, 27). A simple class of models applied to many motors assumes that discrete stepping is tightly coupled to the ATP hydrolysis cycle, occurring at a well defined average rate. The φ29 motor has been described by such a model, in which the motor makes 2-bp steps at a rate limited by phosphate release (18). In such a model, velocity variations occur because Brownian fluctuations play an important role in driving the motor transitions. In the most random possible case, referred to as a “Poisson stepper,” one expects an exponential distribution of waiting times between steps (27). Larger steps produce larger velocity fluctuations, but we have shown above that the step size of the T4 motor must be very small (<6 bp). A Poisson stepper moving at the measured rate, with a step size of 6 bp and with added noise equal to the measurement noise, cannot explain the large variations we observe (Fig. 3E). Some individual complexes also exhibited large velocity fluctuations in time (Fig. 4 A and B), which were different for each complex and were not correlated with position on the DNA. These fluctuations also could not be explained by a simple kinetic model (Fig. 4C) (42% had SD >2-fold greater than that of a Poisson stepper, and 19% had SD >3-fold greater).
Fig. 4.
Temporal fluctuations in the velocity determined in a 2-s sampling window. (A and B) Examples of complexes exhibiting relatively small changes (A) and relatively large changes (B). (C) Histogram of SD in velocity for all complexes relative to the corresponding SDs for simulated data sets generated by a simple kinetic model (see text).
We emphasize that two types of variability were observed: (i) variation in the average velocities of different complexes, referred to in the literature as “static disorder,” and (ii) variations in the velocities of single complexes in time, referred to as “dynamic disorder” (28–30). Although it is difficult to rule out protein degradation or instability as a contributing factor to static disorder, such effects cannot fully explain dynamic disorder because the velocity was observed to vary both up and down in time. Degradation would be expected to only cause decreases. In the case of φ29, loss of even a single motor subunit is expected to completely stop packaging because coordinated, successive firing of the ATPase subunits has been shown to be essential to motor function (18).
To further confirm our findings, we made n = 33 measurements using an independently prepared batch of gp17. Both static (SD = 370 bp/s) and dynamic (mean relative SD = 2.3 bp/s) disorder were consistent with the other measurements. Because one notable difference between T4 and φ29 is the endonuclease activity in T4, we conducted n = 22 additional measurements with a gp17 mutant that lacks nuclease activity but has full DNA packaging efficiency (D401N) (31). These measurements again revealed significant static (SD = 490 bp/s) and dynamic (mean relative SD = 1.6 bp/s) disorder, not statistically different than with the wild type, suggesting that nuclease function does not explain the variability.
Similar levels of static and dynamic disorder in enzyme kinetics have been reported in several previous single-molecule experiments, including studies of lactate dehydrogenase, cholesterol oxidase, λ exonuclease, and RecBCD helicase (24, 32–34). Our data show that such behavior can also occur in a more complex, multicomponent (gp17, gp20) motor. Xie and coworkers (28–30) have provided evidence that such variability can be attributed to the existence of multiple active conformational states of the enzyme complexes and slow interconversion between them. In the case of the T4 motor, such an interpretation is consistent with recent biochemical studies that suggest multiple gp17 conformers and assembly states exhibiting different levels of ATPase activity. The findings of these studies were as follows: (i) gp17 has very weak intrinsic ATPase activity (Kcat = 1–2 ATP hydrolyzed per gp17 per minute), stimulated 50- to 100-fold by the small terminase protein, gp16 (35); (ii) three stimulated ATPase states were observed, and the one with the highest activity, ≈400 ATPs hydrolyzed per gp17 per minute, was attributed to an oligomerized gp17 (26); (iii) gp17 is modestly stimulated by the portal (20%) and by the phage T4 ssDNA binding protein, gp32 (100%) (26); and (iv) ≈8- to 10-fold stimulation was observed when gp17 is part of the complete motor during active DNA translocation (14).
Evidence suggests that T4 packaging may begin while transcription, recombination, and repair are still under way on the same DNA substrate (36). The products of T4 DNA replication and recombination are complex concatenated and branched structures that must be resolved while the DNA is “fed” to the packaging machine. Gene products involved in transcription, including gp32 and gp55, have been shown to interact with gp17, suggesting that packaging is coordinated with other enzymatic processes (37). Given the high packaging rate, we suggest that the capability of the motor to reversibly pause, slip, and change velocity may be necessary to coordinate packaging with transcription, recombination, and repair processes on the same DNA substrate.
We conducted additional experiments in which the purified small terminase (gp16) was added to the prohead–gp17 complexes and found that it severely inhibited packaging (no packaging was recorded in >200 trials). These experiments show that gp16 does interact strongly with the prohead–gp17 complex; however, simply mixing these proteins together in vitro does not result in the formation of an active complex, consistent with the recent findings of bulk assays (13, 14).
Implications of High Motor Velocity.
The high packaging rate is a distinct feature of T4. The motor, which has structural similarities to helicases (16), translocates DNA faster than the fastest known helicase (RecBCD, which moves at speeds up to ≈1,000 bp/s) (38). Only FtsK, a helicase-like complex that transports dsDNA across a membrane in dividing Escherichia coli, has been reported to move faster (up to 5,000 bp/s) (39). The packaging rates we measured are consistent with known time constraints for viral DNA packaging in vivo. T4 has an ≈9× longer genome than φ29, yet it completes assembly slightly faster (≈20 min vs. ≈30 min at 37°C). Because essential packaging proteins are not expressed until the latter half of the infection cycle, only an ≈5–10 min window is available to complete packaging in vivo (36). Pulse–chase and temperature shift experiments with mutants indicate that the DNA is packaged in vivo in as little as ≈3 min at 37°C (11, 12). Our measurements were made at room temperature (≈23°C). Preliminary studies of φ29 (M. White and D.E.S., unpublished data) indicate that the packaging rate increases several fold when the temperature is increased from 23° to 35°C; a trend similar to that found with many enzymes. Thus, if packaging takes ≈3 min in vivo at 37°C, it may be expected to take on the order of 10 min at ≈23°C.
On the basis of our measured rates, the 171-kb T4 genome would be packaged in ≈4 min at the average of 700 bp/s or in ≈1.5 min at the maximum of 1,840 bp/s, if the rate did not decrease with capsid filling. These are lower limits because we expect the rate to decrease with filling due to of the buildup of internal forces resisting DNA confinement as packaging proceeds (19). If the dependence on filling in T4 follows the same trend as that for φ29, full packaging would take 7 min on average or 2.7 min at maximum velocity. These values are an upper limit because T4 motor velocity decreases more gradually with load (Fig. 2B) than does that of φ29, and the maximum internal force is predicted to be lower (19).
Approximately 80% of the complexes in our assay exhibited sufficient rates to package the full genome within 10 min. This implies that the gp17 complexes in our assay, operating in vitro with no other components, translocate DNA at a rate consistent with the measured rates of packaging in vivo. Only ≈20% of the complexes exhibited rates too slow to account for native packaging kinetics. Because T4 assembly is highly efficient, with few unfinished complexes, these “slow” motor conformations must not often form in vivo. Interaction of gp17 with various components not present in our experiments, including gp16, gp32, and gp55, may facilitate assembly of the most active conformation of the motor complex, consistent with recent biochemical findings (26).
Fundamentally, packaging rate = motor step size × stepping rate. Because the measured φ29 step size is ≈2 bp, and because our measurements show that the T4 step is <6 bp, we can conclude from our maximum measured rate of 1,840 bp/s that the T4 motor is capable of stepping at least 4× higher than the φ29 motor. The measured power generation at high load put an even higher bound on the ATP hydrolysis rate, as discussed above. Although the structure of the T4 ATPase has recently been determined, and an inchworm translocation mechanism proposed (16), the structure of the φ29 ATPase is not yet available for comparison. There may be differences in the catalytic sites of the two motors that would be of interest to structural biologists for future investigations.
Conclusions
We have developed an optical tweezers method for measuring single DNA packaging dynamics in phage T4. A defined complex consisting of only proheads and gp17 ATPase can package DNA very rapidly, reconciling the ability of the virus to package its large genome in a limited time window during the natural infection process. Packaging can initiate via a pathway in which prohead–gp17 complexes are formed and then rapidly bind and translocate DNA. The T4 motor can generate very high forces (>60 pN), suggesting that high force generation is a common property of viral motors. It can also translocate DNA at variable rates and reversibly pause and slip, capabilities that T4 and other viruses may need in order to regulate and coordinate packaging with transcription, recombination, and repair processes. The development of this single-molecule assay, combined with recent determination of the crystal structure of the gp17–ATPase domain (16), identification of critical functional residues of the motor and mutants, and recent development of a complementary fluorescence-correlation spectroscopy assay (15), make T4 an attractive model system for detailed structure/function investigations.
Experimental Procedures
Reagents.
T4 components were prepared as described in ref. 14. Bulk assays with ≈10-fold excess of linearized 4-kbp plasmid DNA showed that ≈20–30% of this DNA was packaged, consistent with at least one DNA packaged into every prohead (14). The biotinylated 25.3-kb DNA template was prepared and tethered to streptavidin-coated microspheres (2.1 μm in diameter), as described in ref. 21. Proheads (1.25 × 1010) were mixed with 0.15 nmol gp17 in 2 μl of 0.5 M Tris·HCl (pH 7.5)/50 mM MgCl2/10 mM spermidine/10 mM putrescine/1 M NaCl plus 10 μl of 10% (wt/vol) PEG 20,000 (#95172; Fluka, St. Louis, MO) and incubated for 2 min. Then, 2.5 mM γS-ATP was added to a final concentration of 0.4 mM, and the sample was incubated for 45 min at room temperature. Anti-T4 antibodies were attached to the protein G microspheres (2.1 μm in diameter) as described in ref. 21, and 2 μl were added to the prohead–gp17 complexes and incubated for 45 min. Measurements were carried out in 50 mM Tris·HCl (pH 7.5)/5 mM MgCl2/1 mM spermidine/1 mM putrescine/5% (wt/vol) PEG 20,000/100 mM NaCl/1 mM ATP/5 μM ADP/5 μM NaH2PO4.
Force–Clamp Measurements.
The dual optical tweezers system was set up and calibrated as described in ref. 40. The force was monitored at 1 kHz, and if it was greater/less than the force set-point of 5 pN, the traps were moved closer/farther by 1 nm (<0.05% change in fractional extension). The DNA length was calculated knowing the separation between the traps, the compliance of the traps, and the force vs. fractional extension relationship measured in the packaging buffer. The velocity vs. length of DNA packaged was analyzed by determining the velocity in 1-kb-length bins by fitting the length vs. time data in each bin to a line. The average decrease in velocity for each complex was then determined by fitting the binned data to a line. The mean velocity of each complex was determined as the total change in tether length divided by the total elapsed time. The velocity vs. time was determined by fitting the length vs. time data in a 2-s sliding window to lines.
Fixed-Traps Measurements.
The motor velocity vs. load was determined with fixed-trap separations, such that the DNA tension rose as packaging proceeded. The tether length was calculated knowing the separation between the traps, force, compliances of the traps, and measured force vs. fractional extension relationship. Large, clearly discernable pauses (velocity <20 bp/s for >0.2 s) were removed before analysis. Velocities were calculated by fitting the length vs. time data in 0.5-s sliding windows (corresponding to certain average loads) to lines. All of the individual velocities, from all data sets, were averaged together in 5-pN bins.
Acknowledgments
We thank P. Rickgauer for assistance and P. Geiduschek and L. Black for discussions. Funding was provided by the National Institutes of Health, the Kinship Foundation, and The Beckman Foundation (D.E.S.) and by the National Science Foundation (V.B.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Jardine PJ, Anderson D. In: The Bacteriophages. Calendar RL, editor. New York: Oxford Univ Press; 2006. pp. 49–65. [Google Scholar]
- 2.Feiss M, Catalano CE. In: Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Catalano CE, editor. New York: Springer; 2005. pp. 5–39. [Google Scholar]
- 3.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black LW. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 5.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 6.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 7.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Proc Natl Acad Sci USA. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evilevitch A, Castelnovo M, Knobler CM, Gelbart WM. J Phys Chem B. 2004;108:6838–6843. [Google Scholar]
- 9.Hendrix R. Theor Popul Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 10.Rao VB, Black LW. In: Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Catalano CE, editor. New York: Springer; 2005. pp. 40–58. [Google Scholar]
- 11.Laemmli UK, Favre M. J Mol Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 12.Black LW, Silverman DJ. J Virol. 1978;28:643–655. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Black LW, Peng G. J Biol Chem. 2006;281:25635–25643. doi: 10.1074/jbc.M602093200. [DOI] [PubMed] [Google Scholar]
- 14.Kondabagil K, Zhang Z, Rao VB. J Mol Biol. 2006;363:786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 15.Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Biophys J. 2007;93:L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siyang S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. Mol Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 18.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 19.Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SB, Cui Y, Bustamante C. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 21.Fuller DN, Gemmen GJ, Rickgauer JP, Dupont A, Millin R, Recouvreux P, Smith DE. Nucleic Acids Res. 2006;34:e15. doi: 10.1093/nar/gnj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehninger AL, Nelson DL, Cox MM. Principles of Biochemistry. New York: Worth; 1993. [Google Scholar]
- 23.Mehta AD, Rief M, Spudich JA, Smith DA, Simmons RM. Science. 1999;283:1689–1695. doi: 10.1126/science.283.5408.1689. [DOI] [PubMed] [Google Scholar]
- 24.Perkins TT, Li HW, Dalal RV, Gelles J, Block SM. Biophys J. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller D, Bustamante C. Biophys J. 2000;78:541–556. doi: 10.1016/S0006-3495(00)76615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumann RG, Black LW. J Biol Chem. 2003;278:4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- 27.Svoboda K, Mitra PP, Block SM. Proc Natl Acad Sci USA. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie XS. J Chem Phys. 2002;117:11024–11032. [Google Scholar]
- 29.Min W, English BP, Luo G, Cherayil BJ, Kou SC, Xie XS. Acc Chem Res. 2005;38:923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- 30.English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 31.Rentas F, Rao VB. J Mol Biol. 2003;334:37–52. doi: 10.1016/j.jmb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 32.Xue Q, Yeung ES. Nature. 1995;373:681–683. doi: 10.1038/373681a0. [DOI] [PubMed] [Google Scholar]
- 33.Lu HP, Xun L, Xie XS. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 34.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 35.Leffers G, Rao VB. J Biol Chem. 2000;275:37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- 36.Mosig G, Eiserling F. In: The Bacteriophages. Calendar RL, editor. New York: Oxford Univ Press; 2006. pp. 225–267. [Google Scholar]
- 37.Malys N, Chang DY, Baumann RG, Xie DM, Black LW. J Mol Biol. 2002;319:289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- 38.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 39.Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, Bustamante C, Cozzarelli NR. Science. 2006;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 40.Rickgauer JP, Fuller DN, Smith DE. Biophys J. 2006;91:4253–4257. doi: 10.1529/biophysj.106.089524. [DOI] [PMC free article] [PubMed] [Google Scholar]