Abstract
In many organisms, a synaptonemal complex (SC) intimately connects each pair of homologous chromosomes during much of the first meiotic prophase and is thought to play a role in regulating recombination. In the yeast Saccharomyces cerevisiae, the central element of each SC contains Zip1, a protein orthologous to mammalian SYCP1. To study the dynamics of SCs in living meiotic cells, a functional ZIP1::GFP fusion was introduced into yeast and analyzed by fluorescence video microscopy. During pachytene, SCs exhibited dramatic and continuous movement throughout the nucleus, traversing relatively large distances while twisting, folding, and unfolding. Chromosomal movements were accompanied by changes in the shape of the nucleus, and all movements were reversibly inhibited by the actin antagonist Latrunculin B. Normal movement required the NDJ1 gene, which encodes a meiosis-specific telomere protein needed for the attachment of telomeres to the nuclear periphery and for normal kinetics of recombination and meiosis. These results show that SC movements involve telomere attachment to the nuclear periphery and are actin-dependent and suggest these movements could facilitate completion of meiotic recombination.
Keywords: actin, meiosis, recombination, synaptonemal complex, yeast
Sporulation of diploid cells in the ascomycete Saccharomyces cerevisiae is accompanied by a typical meiotic cell cycle that culminates with the production of four haploid ascospores. Chromosomes pair, undergo recombination, and then segregate from each other in two successive divisions. Reciprocal recombination between homologous chromosomes takes place during prophase of the first meiotic division (prophase I) and is essential for proper segregation. In S. cerevisiae, prophase I can be divided into leptotene-, zygotene-, pachytene-, and diplotene-like substages defined by the state of chromosome pairing and condensation. During leptotene, chromosomes organize on proteinaceous axial elements, and double-strand breaks (DSBs) begin to appear in DNA, initiating recombination. At the same time, perinuclear telomeres begin to cluster near the spindle pole body (the yeast centrosome), pushing chromosomes into a bouquet-like configuration at the leptotene/zygotene transition. At zygotene, the axial elements pair or synapse at the sites of DSBs, and tripartite proteinaceous structures called synaptonemal complexes (SCs) begin to form between homologous chromosomes (1–3). At pachytene, the SCs have matured by a zipper-like mechanism into ribbon-like structures that are embedded at each end in the nuclear envelope and intimately connect each pair of homologues from end to end (3–5). Mature SCs are present for at least 1 h in strain SK1 and longer in other strains, so that pachytene accounts for a relatively large part of prophase I (6–8). In S. cerevisiae, DNA strand invasion and Holliday junction recombination intermediates are observed throughout zygotene and pachytene (9–12), suggesting that recombination is completed during pachytene, presumably when chromosomes are still bound within SCs. At the diplotene-like stage, the SCs are dismantled, but homologues are still joined by mature chiasmata at or near sites where reciprocal recombination has taken place. The profound changes that occur to chromosomes during prophase I have only begun to be characterized by live cell imaging. In an early study in rat spermatocytes, heterochromatin could be seen moving during leptotene and zygotene. However, most movement ceased when cells reached pachytene (13). In hamsters, SCs appeared to twist or rotate during the progression of pachytene (14). In yeast, telomeres have been observed to move about the nuclear periphery (15), suggesting the possibility of more extensive chromosomal movements, but the process of chromosome pairing and how chromosomes behave once they are paired could not be discerned.
Zip1 is an integral and essential protein component of the central element of the SC (16) and is believed to be the ortholog of the mammalian protein SYCP1 (17). A ZIP1::GFP fusion that enabled the visualization of SCs in live sporulating yeast cells has been used to show that SCs often locate near the nuclear periphery during pachytene (8). This report describes an improved ZIP1::GFP construct that was used to study the dynamics of SCs during prophase I of S. cerevisiae meiosis. Pachytene was found to be characterized by dramatic, extensive, and continuous chromosome gyrations, shape changes, and migrations, as well as nuclear movements that all appear actin-dependent and involve the attachment of telomeres to the nuclear periphery.
Results
Construction of Additional ZIP1-Fluorophor Fusions.
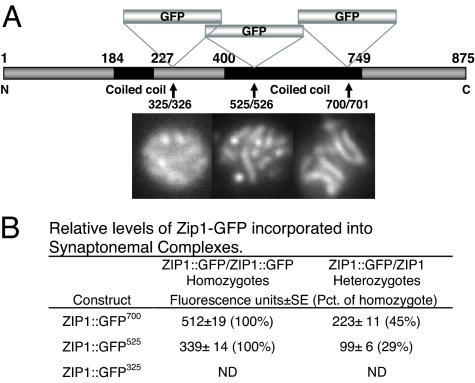
To produce a better fluorescent tag for observing SCs, GFP was inserted at several additional locations within the ZIP1 coding region (Fig. 1A). Insertion at the end of the second coiled-coil domain at position 700, ZIP1::GFP700, produced a protein that behaved more like WT than the original ZIP1::GFP525 construct, which caused a short delay (Fig. 2). Insertion in the globular domain at residue 325 (ZIP1::GFP325) gave weakly fluorescent SCs and no significant complementation of the sporulation or spore lethality phenotypes and was not used. ZIP1::GFP700 produced spore viability virtually equal to WT (97 ± 2% vs. 95 ± 3%) compared with ZIP1::GFP525, which produced marginally fewer viable spores (92 ± 3%). Neither functional construct affected total percent sporulation [supporting information (SI) Fig. 10]. Based on fluorescence intensity measurements comparing ZIP1::GFP homozygotes and ZIP1::GFP/ZIP1 heterozygotes, Zip1-GFP700 protein was incorporated into SCs almost as efficiently as WT ZIP1 protein and more efficiently than the Zip1-GFP525 protein (Fig. 1B). ZIP1::GFP700 heterozygotes displayed about half the fluorescence intensity of homozygotes, whereas ZIP1::GFP525 heterozygotes were only ≈30% as bright as homozygotes. ZIP1::GFP700 homozygotes also produced 1.5- to 2.2-fold brighter SCs. Surface-spread chromosomes showed the same qualitative differences but could not be quantitated because of rapid bleaching.
Fig. 1.
Structure of ZIP1::GFP constructs and relative efficiency of their incorporation into SCs. (A) ZIP1 gene showing location of GFP inserts. Constructs were introduced into S. cerevisiae as described in Materials and Methods. Representative fluorescent images of pachytene nuclei for each construct are shown below. (B) Relative efficiency of incorporation into SCs in heterozygotes and homozyotes. Strains HW122, HW123, EW102, and EW103 were incubated in sporulation medium and analyzed when the percentage of cells in pachytene was highest. Zip1 fluorescence was quantitated as described in Materials and Methods. ND, not determined.
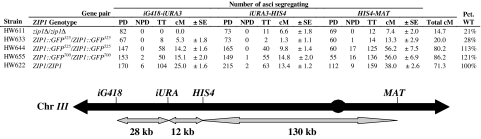
Fig. 2.
Meiotic reciprocal recombination in strains containing different ZIP1::GFP constructs. The percent recombination (cM ± SE) was calculated from the number of parental ditype (PD), nonparental ditype (NPD), and tetratype (TT) asci, as described in Materials and Methods. Pct. WT, total recombination relative to WT. iG418 is YCL056C::kanmx, and iURA is an insertion of URA3 in YCL040W.
Examination of reciprocal recombination indicated that ZIP1::GFP525 and ZIP1::GFP700 strains both exhibited approximately the same total rate that was marginally greater than WT and four to five times greater than either ZIP1::GFP325 or zip1 deletion strains (Fig. 2). Crossovers in the fluorescently labeled strains exhibited minor differences in crossover distributions. However, this type of variation is common (18), often compensated for in adjacent intervals as was found here, and thus was not further investigated.
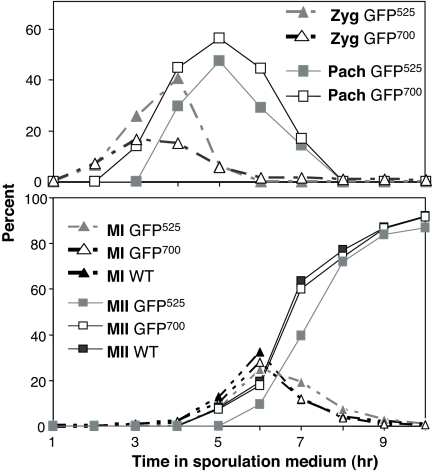
Most important, ZIP1::GFP700 did not alter the kinetics of sporulation and the incorporation of Zip1-GFP700 into mature SC appeared more rapid than Zip1-GFP525, as demonstrated by the smaller fraction of cells in zygotene (Fig. 3).
Fig. 3.
Kinetics of SC formation and meiosis in ZIP1::GFP strains. (Upper) Initiation and completion of SCs were monitored by determining the percentage of cells showing punctate (Zyg-zygotene) and mature worm-like (Pach-full-length pachytene) ZIP1::GFP fluorescent structures in strains EW102 (GFP525) and HW122 (GFP700). (Lower) Meiotic divisions were monitored in strains EW102 (GFP525), HW122 (GFP700), and NKY279 (WT nonfluorescent control) by DAPI staining samples taken at the times noted from cultures incubated in liquid sporulation medium and scoring the number of cells containing one (not shown), two (MI, meiosis I), and four nuclei (MII, meiosis II). At least 200 cells were scored per time point sample.
Live Cell Imaging.
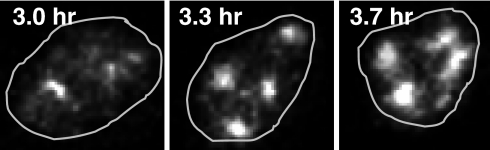
Analysis of strain HW122 cells by fluorescence video microscopy using low-intensity illumination revealed similar kinetics to that shown above (Fig. 3), starting with diffuse Zip1-GFP700 fluorescence at ≈2 h, followed by punctuate dots and a few rods (zygotene), and culminating with full-length ribbon-like structures that became brighter with time (Fig. 4 and SI Movie 1). These “mature” SCs persisted for 1–2 h and were most abundant from 4 to 6 h (Fig. 5). It was possible to see cells transitioning from zygotene to pachytene and from pachytene to the diplotene-like stage by visualizing the growth or diminution of fluorescent structures (Figs. 4 and 5 and SI Movie 2). Cells also completed sporulation during and after video microscopy (SI Movie 2).
Fig. 4.
Zip1-GFP700 containing nucleus undergoing transition from zygotene into pachytene. Time elapsed after introduction into sporulation medium and the outline of the nucleus is displayed.
Fig. 5.
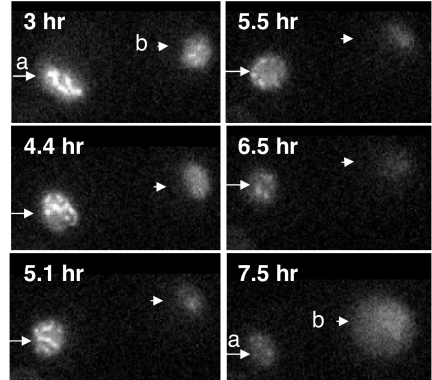
Zip1-GFP700 containing nuclei undergoing transitions during imaging. Images were captured at the noted times. a, Rearrangements of pachytene SCs (3–5.1 h) followed by diplotene SC disassembly (5.5–6.5 h) and loss of most fluorescence (7.5 h). b, Early diplotene nucleus with last remnants of SC (3–4.4 h) followed by loss of most fluorescence (5.1 h). This nucleus began to autofluoresce (7.5 h) and formed a four-spored ascus ≈1 h after recording was stopped (not shown).
Most startling were observations that nascent SCs during zygotene and full length SCs during pachytene exhibited vigorous and continuous movements that persisted until SCs were no longer visible (SI Movie 2). SC movements were so rapid that they were visible in real time by direct observation with a conventional fluorescence microscope as well as in consecutive images (Fig. 6). Tracking single bivalents relative to immobile ones in the same focal plane revealed an average speed of 0.43 ± 0.27 μm/sec (n = 53). SCs appeared to move independently of each other, and the movements frequently occurred along with gross changes in the shape of the nucleus (SI Movies 2 and 3). Changes in the shape of the nucleus, quantitated as described in Materials and Methods, occurred rapidly and were virtually equal whether video capture rates were 5.0 (Fig. 7B) or 0.7 sec per frame (data not shown). Nuclear deformations became most prominent when one or a few “maverick” chromosomes moved away from and then returned to the bulk of the chromosomes (Fig. 8A and SI Movie 3). Based on 30 cells that were videotaped, mavericks were observed within 5 min in 26% of the nuclei. Maverick movements that could be timed lasted an average of 6.2 ± 2.5 sec (n = 15). Thus, maverick chromosomes are likely to be observed three to six times per nucleus during a 60- to 120-min pachytene stage (60–120 min/5 min × 0.26). In some cells, SCs could be seen moving between the perinuclear and more uniformly distributed arrangements that appeared to fill the nuclear volume (Fig. 9 and SI Movie 3) (8). SC and nuclear movements were observed by using either ZIP1::GFP700 or ZIP1::GFP525, and their behavior was indistinguishable.
Fig. 6.
Consecutive images showing rapid mobility of Zip-GFP-labeled SCs in WT strains and impaired mobility in ndj1 mutants. (Upper) WT nucleus (strain HW122), Arrowhead points to a single SC that remains immobile providing a reference point near fixed line. Upper Left arrow points to a single more elongated SC that comes into the plane of focus, extends, and then slides along the nuclear periphery. Arrows on Lower Right point to three U-shaped SCs that change shape. (Lower) An ndj1 mutant (strain EW105) nucleus where all SCs appear almost immobile and relatively extended except one which slides into the image plane (arrowhead). Elapsed time is shown at top. Nuclei are outlined.
Fig. 7.
Quantitation of chromosomal movement and nuclear deformations. (A) Paths of the ends of randomly chosen individual fluorescent SCs were followed in WT and ndj1 mutant nuclei as described in Materials and Methods by using images captured every 0.5 sec. The effects of LatB and formaldehyde (Formald) addition are shown. (B) Changes in nuclear shape (Δ[l/w]) were quantitated as described in Materials and Methods by using images captured every 5 sec. Error bars denote SEM.
Fig. 8.
Zip1-GFP containing nuclei showing extensive chromosome movement and nuclear deformations in WT and impaired movement in LatB-treated and ndj1 mutant cells. Video frames captured at the noted intervals with nuclei outlined. (A) WT. Arrows point to deforming nucleus and movement of “maverick” chromosomes out of and then back to the bulk of the chromosomal mass. (B) WT + LatB. WT nucleus treated with LatB where all SC appear virtually immobile. (C) Mutant ndj1 where SCs and nucleus show limited mobility and arrow points to a stationary reference chromosome.
Fig. 9.
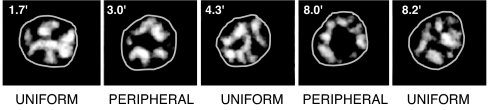
SCs shuttling between the peripheral and uniformly distributed arrays. Frames from videos taken at the noted intervals after 3 h of incubation in sporulation medium. Nuclei are outlined.
Bivalent and Nuclear Mobility Requires Polymerized Actin but Not Microtubules.
To examine whether any components of the cytoskeleton were required, the effects of the actin-depolymerizing drug Latrunculin B (LatB), and the microtubule-binding drugs nocodazole and benomyl were studied. Drugs were added directly to pachytene cells and video microscopy continued. LatB immediately eliminated the vigorous SC movements, leaving some locally restricted movements that were likely to be caused by Brownian motion (Figs. 7A and 8 and SI Movie 4). Full SC movement was restored when LatB was washed from the cells (SI Movie 5). Neither, benomyl nor nocodazole added at concentrations known to inhibit microtubule function (15, 19) had an observable effect on SC movement nor did addition of DMSO used to dissolve some of the drugs. Nuclear deformations were also reduced significantly after the addition of LatB (Fig. 7B). These results indicate that both chromosomal and nuclear movements are actin-dependent and suggest they might be related.
Next, cells were observed after addition of formaldehyde. Neither gross nor Brownian-type SC movements were seen, and the GFP fluorescence of the immobile SCs faded upon prolonged recording (SI Movie 6). Single bivalent tracking revealed slightly less movement than LatB-treated cells (Fig. 7A), and nuclear shapes did not appear to change [Δ[l/w]≈0 (l, length; w, width); SI Fig. 11]. Paralysis by formaldehyde indicates the movements are biological and depend on the presence of native proteins.
NDJ1 Is Required for Bivalent Mobility.
NDJ1 encodes a meiosis-specific telomere protein required for telomere clustering and attachment to the nuclear periphery, WT levels of recombination and normal sporulation kinetics (20–23). A ZIP1::GFP containing ndj1 mutant, strain EW105, was investigated to determine how SCs were affected. Ribbon-like SCs formed, but a single bright structure indicative of a polycomplex (24) was found in ≈10% of the pachytene cells, whereas similar structures were rarely seen in WT pachytene cells. SC mobility was dramatically reduced (Figs. 6–8 and SI Movies 7 and 8). The average SC velocity was approximately half that found in WT nuclei (0.24 ± 0.09 μm/sec; n = 53 vs. 0.43 ± 0.27 μm/sec; n = 53), a significant difference (P < 0.001). Distances traveled by single-tracked SCs during a 5-sec period were reduced compared with WT but were clearly greater than LatB- or formaldehyde-treated samples and were further reduced by LatB to that found in similarly treated WT cells (Fig. 7A). Most prominent was that SCs appeared as more rigid rod-like structures in ndj1 cells (Fig. 6). The typical bending, folding, and twisting observed in WT nuclei was virtually absent, as were maverick chromosomes. These observations were confirmed by using maximum projection videos of 3D-recorded nuclei that clearly showed diminished movement compared with WT, eliminating any possibility that mobility effects in 2D recordings could be related to focal plane slippage (SI Movies 9 and 10). Nuclear deformations in ndj1 mutants were similarly reduced to about half that found in WT cells and further reduced by LatB to that observed in similarly treated WT cells (Fig. 7B and SI Fig. 11). Reduced chromosomal movement in ndj1 mutants indicates that chromosomes must be tethered to the nuclear periphery to achieve maximum movement, especially with respect to the observed contortions. Nevertheless, normal-appearing although quantitatively reduced nuclear deformations are still possible in ndj1cells. These results led to the suggestion that virtually all mobility is actin-based, and the motive force moves either a section of the nucleus or something attached to the nuclear periphery, so that tethered chromosomes are dramatically propelled as well as contorted.
Untethered ndj1 chromosomes move, but the movement appears more passive and is therefore likely to be the indirect result of the continuation of the actin-propelled nuclear movements. Moreover, tethered WT chromosomes can span large parts of each nucleus, and their distal ends could pull on other sections of the nuclear envelope. The absence of these connections in ndj1 cells might be likely to lead to reduced nuclear deformations. Alternatively, some chromosomal movements might not depend on tethering, but these movements must also depend on actin polymerization. Still another possibility for reduced movement is that the nuclear periphery itself is affected in ndj1 cells. Finally, no differences in the distribution of actin patches and cables in WT and ndj1 mutant cells containing Zip1-GFP were observed (see SI Fig. 12). Therefore, any absence of mobility was not because of ndj1 affecting the actin cytoskeleton.
Discussion
An improved functional ZIP1::GFP fusion and sensitive video microscopy were used to investigate SC behavior in live meiotic cells. Fluorescent SCs were observed to form, mature, and disassemble. Throughout prophase I, the SCs displayed dramatic and continuous movement traversing relatively large distances throughout the nucleus while bending, twisting, folding, and unfolding. These movements were accompanied by changes in the shape of the nucleus. Normal chromosome movement and some level of nuclear deformation depended on the NDJ1 gene. In addition, all movements were reversibly inhibited by LatB but were insensitive to microtubule drugs. These results suggest that the SC and nuclear movements depend on actin polymerization and the NDJ1-dependent association of telomeres with the nuclear periphery. They are consistent with the ideas that the chromosomal movements are propelled by the nuclear deformations, and that these movements aid and abet the latter stages of meiotic recombination that take place during pachytene.
The movements described here appear to be distinct from those seen in mammals, which were restricted to leptotene and zygotene (13). Similarly, they appear to be different from the “horsetail” motions observed during Schizosaccharomyces pombe meiosis (25). Although both yeasts use an analogous protein complex to connect telomeres to the nuclear periphery (26, 27), S. pombe “horsetail” movements are microtubule-dependent, whereas the S. cerevisiae SC and nuclear movements are actin-dependent. In addition, S. cerevisiae chromosomes appear to move independently of each other, a phenomenon not yet observed in S. pombe.
Maverick chromosomes were commonly observed moving away from and then back to the bulk of the chromosomes. Because SCs are associated with the nuclear periphery at their ends (5) and perhaps at points along their lengths as well (8), we suggest mavericks are the result of actin-driven nuclear movements and protrusions that drag chromosomes. SC folding and unfolding found in WT cells but absent in the ndj1 mutant might be similarly propelled.
Based on static images, we previously described two SC distributions, one where the SCs were arranged near the nuclear periphery and another where they uniformly filled the entire volume of the nucleus. Kinetic investigation indicated the perinuclear distribution predominated earlier suggesting two distinct temporal pachytene substages (8). Nevertheless, static images could not distinguish between two temporal stages and the possibility that SCs oscillated between perinuclear and uniform distributions where the perinuclear distribution predominated early on. The live cell imaging suggests that SCs are able at least to shuttle back and forth between the perinuclear and uniform distributions.
The actin-dependent movement of telomeres into a cluster has been suggested to aid meiotic pairing and recombination by helping to align chromosomes, colocalizing homologous sequences to a smaller volume, thus hastening the search for homology (reviewed in refs. 15 and 28). The accompanying nuclear movements might further aid pairing by stirring up chromatin and diffusible proteins so they find their targets faster. The continuation of these movements well beyond the telomere clustering stage into pachytene when chromosomes are fully paired suggests this mobility may also be involved in the ensuing processes of recombination such as Holliday junction formation and resolution, which are believed to involve the SC. Based on studies of S. cerevisiae ndt80 mutants, which arrest in meiotic pachytene with unprocessed Holliday junctions, it has been suggested that SCs are involved in processing these recombination intermediates into mature chiasmata (8, 9). The observed SC movements might therefore facilitate the efficient completion of recombination.
Full chromosome mobility depended on a WT NDJ1 gene, which is required to tether meiotic telomeres to the nuclear periphery (20, 21). Mutants in NDJ1 show a 2-h meiosis I delay, much of it spent during pachytene and 25–50% less meiotic recombination than WT (21–23). The ndj1 delay might therefore be the result of both delayed pairing and a defect in a subsequent stage of recombination that requires the movements described here (20–22).
How tethered telomeres are linked to chromosome mobility is poorly understood, and the locations of any actin-based motors involved are unknown with respect to both the telomeres and the nuclear periphery. Because most S. cerevisiae actin seems to be cytoplasmic, and actin filaments appear to be highly dynamic during sporulation (29), it is possible the motive force stems from cytoplasmic actin filaments that interact with the outside of the nucleus. Such a mechanism might use fibrils similar to those observed in higher eukaryotes that are attached to the telomeres and span the nuclear membrane (30). Alternatively, reports of nuclear actin-like proteins make it possible that similar yeast proteins could be involved in mediating chromosome mobility (31).
Altogether, the dramatic mobility of SC-bound chromosomes demonstrated here suggests the process of recombination and prophase I progression in S. cerevisiae is more dynamic than first thought, involving dramatic chromosome movements both during and well after pairing has taken place. These movements are accompanied and likely propelled by complex nuclear movements that also last throughout prophase I. Their nature and function certainly deserve further investigation.
Materials and Methods
S. cerevisiae Strains, Cell Growth, Genetic Analysis, and Sporulation.
Strains listed in Table 1 were all constructed from heterothallic genetically marked derivatives of strain SK1 (32), generously provided by Nancy Kleckner (Harvard University, Cambridge, MA). Cells were grown and sporulated on standard media and asci dissected and analyzed as described (8, 33). Sporulation in liquid medium was carried out with vigorous rotary shaking at 30°C. The extent of sporulation was assayed by monitoring cells stained with DAPI in the fluorescence microscope. Percent recombination was calculated using the Tetrads program (34).
Table 1.
S. cerevisiae strains
| Strain | Genotype | Source/ref. |
|---|---|---|
| NKY278 | MATα/ MATa ho::LYS2/ho::LYS2 ura3/ura3 lys2/ lys2 | NKY278a × NKY278b Courtesy of N. Kleckner |
| EW102 | MATα/MATa lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 ZIP1::GPF525/ ZIP1::GPF525 | 8 |
| HW120 | MATα/MATa lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 ZIP1::GPF325/ ZIP1::GPF325 | HW120a × HW120b This study |
| HW122 | MATα/MATa lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 ZIP1::GFP700/ ZIP1::GFP700 | HW122a × HW122b This study |
| HW123 | MATα/MATa lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 ZIP1/ZIP1::GFP700 | NKY278a × HW122b This study |
| EW103 | MATα/MATa lys2 ho::LYS2/ho::LYS2 ura3/ura3 lys2/ lys2 ZIP1::GFP525/ ZIP1 | NKY278a × EW201 This study |
| EW105 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 ZIP1::GFP525/ZIP1::GFP525 ndj1::URA3/ndj1::URA3 | 8 |
| HW611 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 arg4/arg4 zip1::LEU2/zip1::LEU2 ycl056C::kanMX/YCL056C his4X::LEU2/leu2 iURA3 HIS4/his4 | NKY2515 (courtesy of N. Kleckner), transformed with ycl056C::kanMX |
| HW622 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 arg4/arg4 ZIP1/ZIP1 ycl056C::kanMX/YCL056C his4X::LEU2/leu2 iURA3 HIS4/his4 | This study |
| HW633 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 arg4/arg4 ZIP1::GFP325/ZIP1::GFP325 ycl056C::kanMX/YCL056C his4X::LEU2/leu2 iURA3 HIS4/his4 | This study |
| HW644 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 arg4/arg4 ZIP1::GFP525/ZIP1::GFP525 ycl056C::kanMX/YCL056C his4X::LEU2/leu iURA3 HIS4/his4 | This study |
| HW655 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 arg4/arg4 ZIP1::GFP700/ZIP1::GFP700 ycl056C::kanMX/YCL056C his4X::LEU2 iURA3 HIS4/his4 | This study |
DNA Manipulations.
Standard techniques were used for recombinant DNA plasmid construction and amplification in Escherichia coli (35). Recombinant sequences were introduced into S. cerevisiae by using 0.3 M lithium acetate, as described (36). Correct integration of all constructs was confirmed by PCR using appropriate primer pairs.
Construction of Additional ZIP1::GFP Fusions.
GFP was inserted after amino acids 325 and 700 by amplifying ZIP1 with primers listed in Table 2 using pEJW1 (ZIP1::GFP) (8) or pEJW3 [WT ZIP1 inserted as a KpnI-NotI fragment into pRS316 (37)] as template. GFP was obtained on an AscI-FseI fragment from pEJW2 and the three fragments assembled and inserted between the KpnI and NotI sites in integrating plasmid pRS306 to produce pHW120 (ZIP1::GFP325) and pHW122 (ZIP1::GFP700), respectively. GFP fusions include the short linker peptide and were integrated in place of the WT ZIP1 gene by two-step gene replacement as described (8, 38).
Table 2.
Primers for producing ZIP1::GFP fusions
| Primer | Sequence |
|---|---|
| ZIP::GFP325 | |
| 5'UTR-325 | Forward: 5' GCT AGG TAC CTA TAC AAC CGA TCG ACA AAT TAT 3' KpnI |
| Reverse: 5'CAT ACC TCC AGG CGC GCC ACC ACT GTT TTT GAT TTT TTC TTC3' AscI | |
| 326–3'UTR | Forward: 5'GGA TCC GGC TGC GGC CGG CCA TCC TTA ATA CAG GAA ATG GG3' FseI |
| Reverse: 5' CAT ACC TCC AGG CGC GCC ACC AAG AGA GCT AAT AAT CTG 3' NotI | |
| ZIP1::GFP700 | |
| 5'UTR-700 | Forward: 5' GCT AGG TAC CTA TAC AAC CGA TCG ACA AAT TAT 3' KpnI |
| Reverse:5'CAT ACC TCC AGG CGC GCC ACC TGT TAT ATC TTG TTC CTC CG 3' AscI | |
| 701–3'UTR | Forward: 5'GGA TCC GGC TGC GGC CGG CCA GCT GAA AAG TTA GAA CGG CAA G 3' FseI |
| Reverse: 5' CAT ACC TCC AGG CGC GCC ACC AAG AGA GCT AAT AAT CTG 3' NotI | |
Underlined sequence indicates noted restriction site.
SC Spreads and Conventional Fluorescence Microscopy.
Sporulating cells were spheroplasted and surface spreads produced by using the method of Loidl et al. (39). Conventional fluorescence microscopy was performed on live or surface-spread spheroplasted cells at ×1000 magnification by using either GFP or blue filters (for DAPI-stained DNA) and an ORCA digital camera (Hamamatsu Photonics, Hamamatsu City, Japan) fitted to an Olympus (Melville, NY) microscope. Fluorescence intensity was measured by determining the amount of emitted light at three points on randomly chosen SCs in at least 200 different randomly selected pachytene cells for each sample using WASABI software (version II, Hamamatsu Photonics).
Live Cell Imaging.
Live cells were investigated as described (40) by using a temperature-controlled microscope room equipped with a Zeiss (Jena, Germany) Axiovert epifluorescence microscope with a ×100 plan-neofluor oil-immersion lens (N.A. 1.42) and a 12-bit black-and-white CCD digital camera (PCO; SensiCam, Kelheim, Germany) controlled by TILLvisION ver. 4.0 software (Till Photronics, Munich, Germany). Fluorophors were excited by using a polychrome IV monochromator (TILL Photonics) in combination with a GFP filter set (Chroma, Brattleboro, VT). Cells were first incubated in liquid sporulation medium for 2.5–3 h as described (40). Six hundred microliters of cell suspension was then pipetted onto Con A-coated coverslips (25) sitting in a custom-made well slide (H.S., unpublished work). Cells were immediately transferred to the microscope, and images of live cells were recorded every 0.5 sec for 7 min or every 5 sec for 44 min with an exposure time of 300 msec per frame by using a preset focal plane at the nuclear equator to allow for extended viewing. To record longer periods, videos were generated by using 200-ms-per-frame exposure time and 8-min image intervals. 2D images were chosen to limit bleaching over extended periods.
To examine movement in nuclei in virtual 3D, stacks of 19 images were taken by using 250-nm steps along the z axis with the TILLvision-driven Axiovert equipped with a PIFOC z-SCAN (Physike Instruments, Walbronn, Germany) (30). Stacks were recorded every 30 or 60 sec, converted by the maximum projection algorithm of TILLvision, and displayed sequentially.
Chromosome movements were quantitated by tracing the displacement of single SC ends relative to the immobile ones in the same optical plane using either TILLvision (TILL Photonics) or VirtualDub 1.7 (Free Software Foundation, Cambridge, MA). Nuclear-shape deformations were quantified by using the same software by measuring changes over time in the ratio of two arbitrarily chosen perpendicular axes that were defined by the perimeter of the nucleus made apparent by enhancing diffuse Zip1-GFP fluorescence (Δ[l/w]). For each sample, 10 cells were analyzed by determining the l/w quotient in 11 consecutive images captured at 5.0- or 0.7-sec intervals. Quotients were then normalized to the initial value [l/w0] by determining the absolute value of the difference, dividing it by the initial value and averaging the values for the entire time course  . The average value measured for formaldehyde treated cells (Δ[l/w] = 0.090 ± 0.005) was then subtracted to account for nonbiological movement.
. The average value measured for formaldehyde treated cells (Δ[l/w] = 0.090 ± 0.005) was then subtracted to account for nonbiological movement.
LatB (Sigma, St. Louis, MO), benomyl (Dupont, Wilmington, DE), nocodazole (Sigma), or DMSO (Sigma) was added directly to live cells during recording. Final concentrations were LatB, 30 μM; benomyl, 25 and 125 μg/ml; nocodazole, 15 μg/ml; and DMSO, 0.3% (vol/vol) (19, 40). To examine LatB reversibility, drug was added to a 3.5-h sporulating cell suspension and an aliquot examined to ensure cessation of movement. Cells in the remaining suspension were incubated for 30 min, washed three times, resuspended with fresh sporulation medium, and then examined and recorded by fluorescence microscopy. Acid-free formaldehyde was added to cells at a final concentration of 3.7% (vol/vol) and the cells mounted and imaged immediately thereafter.
Supplementary Material
Acknowledgments
We thank Arnold Barton for help throughout and C. Menzel for technical support. In addition, we are grateful to H. H. Ropers (Max Planck Institute for Molecular Genetics, Berlin, Germany) for continued support. This work was supported in the United States by grants from the United States National Science Foundation and the United States Public Health Service (USPHS; D.B.K.) and from the USPHS (W.Z.C.) and in Germany by the Max Planck Institute.
Abbreviations
- SC
synaptonemal complex
- LatB
latrunculin B.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704860104/DC1.
References
- 1.Baker BS, Carpenter AT, Esposito MS, Esposito RE, Sandler L. Annu Rev Genet. 1976;10:53–134. doi: 10.1146/annurev.ge.10.120176.000413. [DOI] [PubMed] [Google Scholar]
- 2.Trelles-Stricken E, Loidl J, Scherthan H. J Cell Sci. 1999;112:651–658. doi: 10.1242/jcs.112.5.651. [DOI] [PubMed] [Google Scholar]
- 3.Zickler D, Kleckner N. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 4.Esponda P, Gimenez-Martin G. Chromosoma. 1972;38:405–417. doi: 10.1007/BF00320159. [DOI] [PubMed] [Google Scholar]
- 5.Byers B, Goetsch L. Proc Natl Acad Sci USA. 1975;72:5056–5060. doi: 10.1073/pnas.72.12.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padmore R, Cao L, Kleckner N. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 7.Loidl J, Nairz K, Klein F. Chromosoma. 1991;100:221–228. doi: 10.1007/BF00344155. [DOI] [PubMed] [Google Scholar]
- 8.White EJ, Cowan C, Cande Z, Kaback DB. Genetics. 2004;167:51–63. doi: 10.1534/genetics.167.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allers T, Lichten M. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 10.Hunter N, Borner GV, Lichten M, Kleckner N. Nat Genet. 2001;27:236–238. doi: 10.1038/85781. [DOI] [PubMed] [Google Scholar]
- 11.Borner GV, Kleckner N, Hunter N. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 12.Henderson KA, Keeney S. BioEssays. 2005;27:995–998. doi: 10.1002/bies.20310. [DOI] [PubMed] [Google Scholar]
- 13.Parvinen M, Soderstrom K. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- 14.Poorman PA, Moses MJ, Russell LB, Cacheiro NL. Chromosoma. 1981;81:507–518. doi: 10.1007/BF00285846. [DOI] [PubMed] [Google Scholar]
- 15.Scherthan H. Biochem Soc Trans. 2006;34:550–553. doi: 10.1042/BST0340550. [DOI] [PubMed] [Google Scholar]
- 16.Sym M, Engebrecht JA, Roeder GS. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 17.Meuwissen R, Offenberg H, Dietrich A, Reisewijk A, Iersel MV, Heyting C. EMBO J. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, Botstein D. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- 19.Hochwagen A, Wrobel G, Cartron M, Demougin P, Niederhauser-Wiederkehr C, Boselli MG, Primig M, Amon A. Mol Cell Biol. 2005;25:4767–4781. doi: 10.1128/MCB.25.11.4767-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trelles-Sticken E, Dresser ME, Scherthan H. J Cell Biol. 2000;151:95–106. doi: 10.1083/jcb.151.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chua PR, Roeder GS. Genes Dev. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- 22.Peoples-Holst TL, Burgess SM. Genes Dev. 2005;19:863–874. doi: 10.1101/gad.1293605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad MN, Dominguez AM, Dresser ME. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 24.Zickler D, Olson LW. Chromosoma. 1975;50:1–23. doi: 10.1007/BF00284959. [DOI] [PubMed] [Google Scholar]
- 25.Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 26.Conrad MN, Lee C-Y, Wilkerson JL, Dresser ME. Proc Natl Acad Sci USA. 2007;104:8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 28.Zickler D, Kleckner N. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 29.Taxis C, Maeder C, Reber S, Rathfelder N, Miura K, Greger K, Stelzer EH, Knop M. Traffic. 2006;7:1628–1642. doi: 10.1111/j.1600-0854.2006.00496.x. [DOI] [PubMed] [Google Scholar]
- 30.Liebe B, Alsheimer M, Hoog C, Benavente R, Scherthan H. Mol Biol Cell. 2004;15:827–837. doi: 10.1091/mbc.E03-07-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pederson T, Aebi U. Mol Biol Cell. 2005;16:5055–5060. doi: 10.1091/mbc.E05-07-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kane SM, Roth R. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2000. [Google Scholar]
- 34.King JS, Mortimer RK. Genetics. 1990;126:1127–1138. doi: 10.1093/genetics/126.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 36.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski RS, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scherer S, Davis RW. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loidl J, Klein F, Scherthan H. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. J Cell Biol. 2005;170:213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.