Abstract
The G protein-coupled receptor (GPCR) superfamily represents the most important class of pharmaceutical targets. Therefore, the characterization of receptor cascades and their ligands is a prerequisite to discovering novel drugs. Quantification of agonist-induced second messengers and downstream-coupled kinase activities is central to characterization of GPCRs or other pathways that converge on GPCR-mediated signaling. Furthermore, there is a need for simple, cell-based assays that would report on direct or indirect actions on GPCR-mediated effectors of signaling. More generally, there is a demand for sensitive assays to quantify alterations of protein complexes in vivo. We describe the development of a Renilla luciferase (Rluc)-based protein fragment complementation assay (PCA) that was designed specifically to investigate dynamic protein complexes. We demonstrate these features for GPCR-induced disassembly of protein kinase A (PKA) regulatory and catalytic subunits, a key effector of GPCR signaling. Taken together, our observations show that the PCA allows for direct and accurate measurements of live changes of absolute values of protein complex assembly and disassembly as well as cellular imaging and dynamic localization of protein complexes. Moreover, the Rluc-PCA has a sufficiently high signal-to-background ratio to identify endogenously expressed Gαs protein-coupled receptors. We provide pharmacological evidence that the phosphodiesterase-4 family selectively down-regulates constitutive β-2 adrenergic- but not vasopressin-2 receptor-mediated PKA activities. Our results show that the sensitivity of the Rluc-PCA simplifies the recording of pharmacological profiles of GPCR-based candidate drugs and could be extended to high-throughput screens to identify novel direct modulators of PKA or upstream components of GPCR signaling cascades.
Keywords: G protein-coupled receptor, complementation assays, protein–protein interactions, protein fragment
G protein-coupled receptors (GPCRs) represent the largest family of cell-surface molecules involved in signal transmission. GPCRs play roles in a broad range of biological processes through regulating the majority of cell-to-cell and cell-to-environment communication, and, consequently, their dysfunction manifests in numerous diseases (1, 2). The GPCR family has enormous pharmacological importance, as demonstrated by the fact that >30% of approved drugs elicit their therapeutic effect by selectively acting on known members of this family (3). The human genome harbors >800 putative GPCRs including a considerable number with unknown physiological function or ligands. GPCR cascades hence remain a major focus of molecular pharmacology (4, 5).
Signal transduction by GPCRs is mediated by activation of protein kinases (4), among which the most intensively studied is the cAMP-dependent protein kinase A (PKA) (6). Various extracellular signals converge on the cAMP/PKA pathway through ligand binding to GPCRs. The adenylyl cyclase then converts ATP to the ubiquitous second messenger cAMP. Intracellular cAMP gradients are shaped through the sole means of degrading cAMP in the cells by phosphodiesterases (PDE), providing a negative feedback system for down-regulating receptor-mediated signaling cascades (7). The release of this second messenger induces the activation of its main effector, PKA, by provoking the dissociation of activated catalytic subunits from the inhibiting regulatory subunits of PKA (Fig. 1) (7), which enables the specific phosphorylation of a plethora of substrates (8, 9).
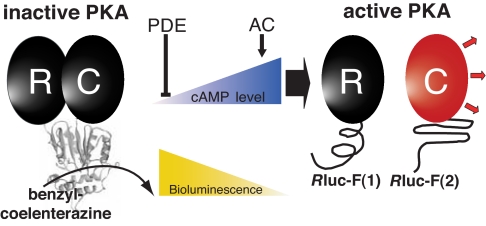
Fig. 1.
Schematic representation of the PCA strategy using Rluc fragments to study the dynamic complex of the PKA heterodimer [regulatory (R) and catalytic (C) PKA subunits] in vivo. Cellular cAMP levels are controlled directly by adenylyl cyclases (AC, production) and PDE (degradation). cAMP elevation and association with the R subunit of PKA induces dissociation of R and C subunits, resulting in decreasing Rluc-PCA activity.
Several cell-based assays have been developed to detect specific activation of PKA, including fluorescence and bioluminescent resonance energy transfer assays for detecting catalytic activity (10) or cAMP-induced PKA subunit dissociation (11–13). These methods have been invaluable to the study of protein complex dynamics and particularly to the integration of compartmentalized GPCR signaling pathways (14–16). However, their range of application is limited because of cellular autofluorescence (for fluorescence resonance energy transfer), limited signal-to-background, and narrow dynamic range. Hence, more general and broadly desired cell-based applications are not easily performed with these assays. Among the most important are high-throughput screenings to discover direct and indirect modulators of protein kinase activities (17).
We reasoned that the desired features of a cell-based assay would be met by one that could capture the dynamics of PKA subunit assembly and reassembly in cell populations and would be based on a reporter system that could be easily implemented with simple, off-the-shelf technology. On one hand we tried to develop a reporter that addresses limitations of high-throughput screening studies like sensitivity and signal stability, and on the other hand the same sensor should offer the possibility to be used at different stages of pharmacological drug evaluation (e.g., single cells, cell populations, and animal models). We report here a protein-fragment complementation assay (PCA) based on the reporter enzyme Renilla reniformis luciferase (Rluc) that meets these requirements. The PCA strategy allows the detection of protein complex formation by fusing each of the proteins of interest to two fragments of a “reporter” protein that has been rationally dissected into two fragments by using protein engineering strategies (18–21). Binding of the two proteins of interest brings the unfolded fragments into proximity, allowing for folding and reconstitution of measurable activity of the reporter protein, which can be of different types (19, 22–29). We chose to develop the PKA sensor with the Rluc-PCA because of the inherent sensitivity and lack of any cellular background of a luminescent reporter. Although assays based on this enzyme have been reported (25), we chose to redesign the assay for improved signal based on structural constraints obtained by structural homology modeling (30) (described below) and to demonstrate its reversibility. Reversibility is absolutely necessary for an effective PKA reporter because it is the cAMP-mediated dissociation of the catalytic and regulatory subunit complex that results in activation of PKA. Commonly used PCAs based on GFP and its variants are irreversible and therefore act as complex traps, preventing detection of protein complex dissociation as we show here for the case of PKA. We previously proposed and have demonstrated that PCAs could be designed to be reversible (19, 29, 31, 32). Here we not only rationally design the Rluc-PCA to be reversible but we also show that it can accurately report known dissociation–association kinetics of PKA subunits and known pharmacological responses in a natural biological system. It is the first demonstration of a reversible PCA upon natural dissociation of a protein complex due to allosteric changes or posttranslational modifications. We also show that the Rluc-PCA fragments unfold and separate after dissociation of the PKA complex, demonstrating that the detected disruption of the PKA complex represents true molecular dissociation of the PKA subunit–PCA fragment fusion proteins. We illustrate pharmacological applications of this tool to (i) identify endogenously expressed GPCRs and (ii) to uncover a selective role of PDE-4 family in down-regulating constitutive β-2 adrenergic receptor (β2AR)- but not vasopressin-2 receptor (V2R)-mediated PKA activities. These examples illustrate the simplicity and versatility of the Rluc-PCA to study the dynamics of protein complexes that are key to the understanding of pharmacology in vivo.
Results
Design of the Rluc-PCA.
The purpose of our study was to create a PCA for quantification of dynamic protein complexes. We chose to generate a PCA based on the Rluc, which is, because of its simplicity and sensitivity, a widely used bioluminescence reporter. In contrast to the reported PCA version of Rluc (25), we used a homology-modeled structure of Rluc to test different dissection sites. Points of reporter protein dissection into two PCA fragments are generally chosen based on the following criteria: (i) the cut sites are as far as possible from the catalytic site, (ii) the fragments represent recognizable subdomains, (iii) the reporter protein structure can be accessibly folded from fragments fused to interacting proteins, and (iv) the cut sites are in nonstructured regions (19, 21, 22, 33). The structure of Rluc, isolated from the marine “sea pansy” R. reniformis, has not been solved; however, a simple Blast search (34) immediately identified several sequences with >40% sequence identity to the bacterial haloalkane dehalogenases [supporting information (SI) Fig. 6 and SI Methods]. Based on this observation we submitted the Rluc sequence to the Modeller database (35) and retrieved three high-scoring structures of which, predictably, the highest scoring was a haloalkane dehalogenase from the proteobacterium Xanthobacter autotrophicus (Protein Data Bank ID code 1EDE).
Based on the predicted structure of Rluc and the PCA design criteria we identified and tested several potential sites to dissect the protein into separate complementary fragments (SI Fig. 7A). Among these, fragments that dissect the protein between amino acids 110 and 111 proved to provide reconstitution of the highest bioluminescence activity in overexpression experiments in Saccharomyces cerevisiae (SI Fig. 7B and SI Methods).
Design of Rluc-PCA Sensor of PKA Subunit Complex Dynamics.
The general scheme for construction and detection of the Rluc-PCA PKA sensor consists of fusing complementary fragments of Rluc to the regulatory (Reg) and catalytic (Cat) PKA subunits of PKA (Figs. 1 and 2A). In our first tests with HEK293T cells, quantification of the first 10 seconds of bioluminescence gave the highest luciferase activities for coexpressed PKA combinations of Reg-F(1):Cat-F(2) and Reg-F(1):Reg-F(2) after addition of the Rluc substrate benzyl-coelenterazine. Neither expression of individual nor coexpression of PCA fusion proteins Cat and the Ras binding domain of serine threonine kinase Raf (RafRBD), which should not interact directly [Cat-F(1):Cat-F(2) and RafRBD-F (1):Cat-F(2)], gave significant bioluminescence signals, confirming the specificity of the assay (Fig. 2B). These results indicate that a direct protein–protein interaction is necessary to promote reconstitution of Rluc enzyme activity.
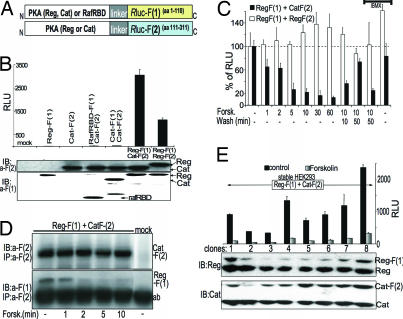
Fig. 2.
A dynamic PKA activity sensor based on Rluc-PCA. (A) Schematic representation for the design of a PCA-based PKA reporter. The regulatory (Reg) and catalytic (Cat) PKA subunits were fused to fragment 1 [F(1)] or fragment 2 [F(2)] of Rluc. (B) The Rluc-PCA was detected from transiently transfected HEK293T cells in suspension aliquoted to 96-well microtiter plates. Immunoblot analysis verifies expression of PCA-tagged proteins (representative experiment ± SD of triplicates). RLU, relative luminescence units. (C) Effect of forskolin (100 μM) and 3-isobutyl-1-methylxanthine (100 μM) treatment on Reg-F(1):Cat-F(2) and Reg-F(1):Reg-F(2). The Rluc-PCA was detected from transiently transfected HEK293T cells in suspension and aliquoted to 96-well microtiter plates (mean ± SD from three independent experiments). (D) HEK293T cells coexpressing Reg-F(1):Cat-F(2) were treated for the indicated times with forskolin (100 μM) and were subjected to immunoprecipitation and Western blotting using anti-Rluc antibodies. (E) The Rluc-PCA was detected from untreated and forskolin-treated (100 μM, 15 min) HEK293T cells stably expressing Reg-F(1):Cat-F(2) in suspension (± SD from three independent samples). Immunoblot analysis shows the expression of endogenously expressed and overexpressed PKA subunits.
Use of the PKA Sensor to Monitor Protein Complex Dynamics in Vivo.
To show that the Rluc-PCA could be used for kinetic studies of protein complexes, we analyzed the effect of cAMP elevation on Reg-F(1):Cat-F(2). We treated transiently transfected HEK293T cells with the cAMP-elevating agent forskolin. Forskolin induced disruption of the Reg-F(1):Cat-F(2) within 1 min of treatment, and after 10 min almost 80% of the PCA fragment-tagged PKA subunits were dissociated (Fig. 2C). In agreement with in vitro data, an increase in cAMP concentration causes partial dissociation of the type II PKA holoenzyme with ≈20% of the complex remaining associated (36). We measured reassociation of the PKA subunit complex upon removal of forskolin (Fig. 2C). Also consistent with this observation, pretreatment of cells with the general nonselective PDE inhibitor 3-isobutyl-1-methylxanthine and subsequent forskolin treatment for 10 min followed by its removal maintained cAMP levels elevated and prevented apparent reassociation of the PKA subunits. In contrast to the PKA heterodimer we detected no dissociation of Reg-F(1):Reg-F(2) in response to forskolin (Fig. 2C).
Results of the PKA Rluc-PCA are consistent with the known mechanism of dissociation and reassociation of PKA subunits. In terms of the Rluc-PCA reporter, these results could be interpreted in two ways: either the cAMP-promoted dissociation of the PKA subunits leads to a complete disassembly of the Rluc reporter that dissociates as free fragments or it promotes a conformational change within the assembled Rluc that results in a loss of Rluc catalytic activity. To distinguish between these two possibilities, we examined the association and dissociation of Reg-F(1):Cat-F(2) in response to forskolin directly by immunoprecipitation. We confirmed that the Reg-F(1):Cat-F(2) complex is formed under basal conditions. Dissociation of the PCA–PKA complex could be detected within the first minutes after forskolin treatment (Fig. 2D). The time course of PKA dissociation appears consistent with the observation we have obtained with the bioluminescent readout. To demonstrate the high sensitivity of the Rluc-PCA in measuring dynamic protein complexes we generated stable HEK293T cell lines expressing the Rluc-PCA PKA sensor. In response to forskolin we recorded the dissociation of Reg-F(1):Cat-F(2) in individual clones with expression levels of the biosensor similar to or far below endogenously expressed PKA subunits (Fig. 2E). The reversibility of the Rluc-PCA PKA illustrated here stands in contrast with what was observed with other popular PCAs based on the fragmentation of GFP. Indeed we could not detect disruption of the PKA subunit interaction using the Venus YFP-based PCA reporter in response to cAMP elevation (SI Fig. 8 and SI Methods). Thus, although GFP-based PCAs may have broad applications in visualization, localization, and translocation of protein complexes, existing GFP and mutant variant PCAs cannot be applied to study dynamic protein complex assembly and disassembly.
Imaging of the Dynamic PKA Sensor in Living Cells.
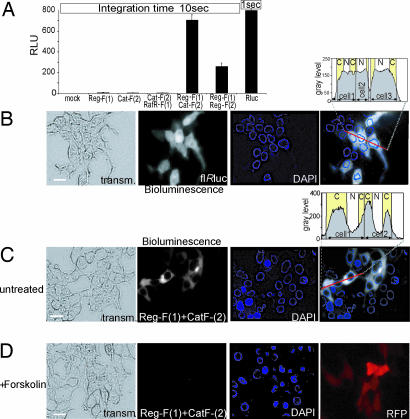
To test whether our reporter could be used for localization studies, we performed live cell imaging of transiently transfected HEK293T cells. Fig. 3A shows the comparison of bioluminescence intensities measured by luminometry of full-length and complemented Rluc activities of adherent cells. We have obtained ≈10% of the full-length Rluc bioluminescence with the coexpressed PCA–PKA combination Reg-F(1):Cat-F(2). As predicted, we observed bioluminescence signal throughout the cell upon expression of the full-length Rluc (Fig. 3B). In contrast, the signal generated by reconstituted Reg-F(1):Cat-F(2) was selectively localized to the cytoplasm with a clear exclusion from the nucleus (Fig. 3C). Upon pretreatment with forskolin for 30 min the bioluminescence signal was no longer detectable (Fig. 3D). These data indicate that imaging of bioluminescence is not only feasible for localization studies but can also be used to detect alterations of protein complexes.
Fig. 3.
Cellular imaging of bioluminescence of transiently transfected HEK293T cells expressing full-length Rluc and Rluc-PCA. (A) The Rluc-PCA was detected from HEK293T cells expressing indicated PCA fusion proteins (10 seconds) or full-length Rluc (1-second integration time) grown on 96-well microtiter plates (mean ± SD from triplicates). (B and C) Visualization of Rluc bioluminescence of HEK293T cells. By using a CCD camera and integration time of 30 seconds we imaged the bioluminescence (shown in gray scale) of full-length Rluc (B) and localized bioluminescence of HEK293T cells expressing Reg-F(1):Cat-F(2) (120 seconds) in PBS supplemented with 5 μM benzyl-coelenterazine (C). (D) Effect of 30 min of forskolin (100 μM) pretreatment on the bioluminescence of Reg-F(1):Cat-F(2) (120 seconds). Cotransfection of the red fluorescent protein (RFP) serves as control for transfection. C, cytoplasm; N, nucleus. (Scale bars: 5 μm.)
Monitoring GPCR Activity Using the Rluc-PCA PKA Reporter.
We next examined whether the Rluc-PCA PKA reporter could be used to study the dynamics of PKA activation under the control of GPCR-mediated signal transduction pathways. We used physiological stimuli to activate two Gαs-coupled receptors, β2AR and V2R. Treatment of stable receptor cell lines [HEK293-β2AR (37) or HEK293-V2R (38)] expressing the PCA-tagged PKA sensor with the agonist isoproterenol (Fig. 4A) or the antidiuretic hormone arginine–vasopressin (AVP) (Fig. 4B) induced the dissociation of up to 80% of Reg-F(1):Cat-F(2) within the first 15 min. The specificity of the receptor-mediated response was confirmed by the observation that the β-adrenergic antagonist alprenolol and the V2R antagonist SR121363B blocked the activation of the PKA reporter by isoproterenol and AVP, respectively (Fig. 4 A and B).
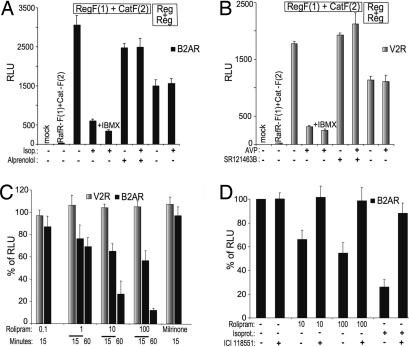
Fig. 4.
Effect of GPCR agonists, antagonists, and PDE inhibition on PKA activities. The Rluc-PCA was detected from transiently transfected HEK293T cells grown on white-walled 96-well microtiter plates. (A) Effect of combinations of alprenolol (10 μM, 60 min) pretreatment and isoproterenol (10 μM, 15 min) treatment of stable HEK293 clones expressing the β2AR on Reg-F(1):Cat-F(2) or Reg-F(1):Reg-F(2) (mean ± SD from triplicates). (B) Effect of SR121463B (10 μM, 60 min) pretreatment and AVP (100 nM, 15 min) treatment of stable HEK293 clones expressing the V2R on Reg-F(1):Cat-F(2) or Reg-F(1):Reg-F(2) (mean ± SD triplicates). (C) Stable V2R-HEK293 and β2AR-HEK293 cells were pretreated for indicated times with milrinone (10 μM) and increasing concentrations of rolipram (μM) followed by plate reader analysis of the effect on the Reg-F(1):Cat-F(2) (mean ± SD from three independent experiments). (D) Stable β2AR-HEK293 cells were pretreated for 30 min with the selective β2AR-antagonist ICI118,551 (1 μM) and isoproterenol (1 μM, 15 min) or increasing concentrations of rolipram (μM, 15 min) followed by Rluc-PCA analysis of Reg-F(1):Cat-F(2) (mean ± SD from three independent experiments).
These results indicate that discrete GPCR activation induces the dissociation of the cAMP/PKA reporter Reg-F(1):Cat-F(2). In contrast, isoproterenol or AVP treatment did not induce the dissociation of the regulatory PKA homodimer.
PDE4 Regulates Basal Activities of the β2AR.
The accumulation of cAMP is regulated through the activation of adenylyl cyclase by Gαs and its degradation by PDE activity. To test the role of PDE in the regulation of constitutive basal GPCR activities we quantified PKA activities in HEK293 cells treated with specific inhibitors for those PDEs that have the highest activity in HEK293 cells, PDE3 and PDE4 (39). In HEK293 cells that stably express the Gαs-coupled receptors V2R or β2AR, we transiently transfected Reg-F(1):Cat-F(2) and treated the cells with the PDE3 or PDE4 selective inhibitors milrinone or rolipram, respectively. Milrinone had no effect on PKA activity, but rolipram caused concentration- and time-dependent activation of PKA in stable β2AR-HEK293 cells in the absence of any ligand as well as in the presence of the neutral β2AR antagonist alprenolol (SI Fig. 9). No response could be detected in V2R-HEK293 cells (Fig. 4C) or HEK293 cells (data not shown). Pretreatment of β2AR-HEK293 cells with the selective β2AR antagonist ICI118,551 (inverse agonist) inhibited the rolipram effect on PKA activation as well as the isoproterenol-mediated response (Fig. 4D). This finding indicates that inhibition of PDE4 increases the β2AR-mediated accumulation of cAMP and therefore PKA activity. Binding of inverse agonists ICI118,511 and propranolol (data not shown) to the β2AR prevented the receptor-mediated PKA response evoked by PDE4 inhibition under basal conditions. These data indicate that the β2AR spontaneous activity is a major contributor to the basal cAMP levels that can be regulated by PDE4 in these cells.
Detecting Endogenously Expressed GPCRs Using the Rluc-PCA.
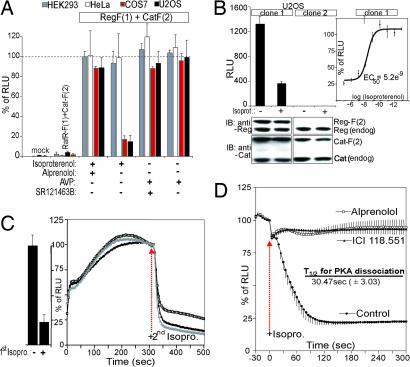
We next applied the Rluc-PCA PKA reporter to assess the presence of endogenously expressed GPCRs in selected cell lines. We treated HEK293T, HeLa, COS7, and U2OS cells, transiently transfected with Reg-F(1):Cat-F(2), with combinations of V2R and β2AR agonists and antagonists. After agonist treatment we observed PKA activation only in COS7 and U2OS cells in response to the nonselective β-adrenergic agonist isoproterenol. In HEK293T and HeLa cells we could not detect any isoproterenol-mediated response, suggesting little or no activity of β-adrenergic receptors. Pretreatment with the nonselective β-adrenergic antagonist alprenolol inhibited the isoproterenol-induced activation of the PKA reporter. No expression of functional V2R can be detected in any tested cell line. (Fig. 5A). These results confirm that COS7 and U2OS cells express at least one subtype of β-adrenergic receptors endogenously.
Fig. 5.
Identification of endogenously expressed GPCR cascades using the Rluc-PCA PKA reporter. (A) The Rluc-PCA was detected from attached HEK293T, HeLa, COS7, or U2OS cells grown on white-walled 96-well microtiter plates. Shown is the effect of combinations of alprenolol (10 μM, 60 min) pretreatment and isoproterenol (10 μM, 15 min) treatment and combinations of SR121463B (1 μM, 60 min) pretreatment and AVP (100 nM, 15 min) treatment on Reg-F(1):Cat-F(2) (mean ± SD from three independent experiments). (B) The Rluc-PCA was detected from U2OS cells stably expressing Reg-F(1):Cat-F(2) in the presence and absence of isoproterenol (1 μM) (Left). Immunoblot analysis verifies the expression of endogenously expressed and overexpressed regulatory and catalytic PKA subunits. EC50 values for isoproterenol-mediated β-adrenergic activation were obtained by using increasing concentrations of the ligand measured as changes of Reg-F(1):Cat-F(2) (Right; mean ± SD from three independent experiments). (C) Real-time kinetics (normalized on the control experiment) in response to isoproterenol (1 μM) of Reg-F(1):Cat-F(2) recorded with attached U2OS cells (stable clone 1; three independent samples, representative experiment). Washing steps were performed four times with PBS after 10 min of first isoproterenol treatment. (D) Real-time kinetics (normalized on the control experiment) of changes of Reg-F(1):Cat-F(2) recorded with attached U2OS cells (stable clone 1) when treated with combinations of ICI118,551, alprenolol (both 10 μM, pretreatment 60 min), and isoproterenol (1 μM). Half-times of PKA dissociation were calculated from three independent experiments (mean ± SD). Calculation of EC50 and t1/2 values and curve fittings were done with Prism 3.01 (GraphPad) (mean ± SD from at least three independent samples).
Kinetics of β2AR-Mediated PKA Dissociation and Reassociation.
We generated a stable U2OS cell line expressing the Rluc-PCA PKA sensor to analyze responses of endogenously expressed GPCRs(SI Methods). We performed immunoblotting analysis and detected expression levels of Reg-F(1):Cat-F(2) at similar levels of endogenously expressed PKA subunits in clone one (Fig. 5B Lower). In response to isoproterenol treatment, we detected the dissociation of Reg-F(1):Cat-F(2) (Fig. 5B Upper). The half-maximal effective concentrations of isoproterenol required to induce PKA activation (EC50 value) by endogenously expressed GPCR was determined to be 5.2 ± 3.5 nM (Fig. 5B Right), in the range of values obtained by in vitro cAMP measurements of isoproterenol-treated β2AR-expressing HEK293 cells (13.9 ± 3.9 nM) (40).
Finally, we studied real-time continuous kinetics of PKA activation by recording live cell bioluminescence of the whole cell population. Subsequent to cAMP-triggered dissociation (first isoproterenol) (Fig. 5C Left), reformation of the PCA-tagged PKA complex could be detected immediately after removal of isoproterenol within the first few seconds, reaching the maximum amount of PKA holoenzyme in a biphasic process within 2–3 min. Repeated isoproterenol treatment induced dissociation of the Rluc-PCA PKA sensor, demonstrating that the PCA detects PKA subunit reassociation (Fig. 5C Right). Immediately after isoproterenol treatment we observed the dissociation of the PKA reporter (t1/2 = 30.47 ± 3.03 seconds). A similar pattern and timeframe of PKA activation has been observed with fluorescence resonance energy transfer-based PKA activity reporters in different cell types (10, 11). Pretreatment of U2OS cells with the nonselective β-adrenergic antagonist alprenolol and with the selective β-2 adrenergic antagonist ICI118,551 prevented isoproterenol-mediated PKA activation to the same extent (Fig. 5D), indicating that the response was mediated by the β-2 adrenergic subtype.
Discussion
Our results demonstrate that the Rluc-PCA sensor meets several requirements to study cell biological aspects of signal transmission in vivo. First we show that folding of rationally designed Rluc fragments is reversible, which is a prerequisite to study signaling events by the dynamics of protein complex assembly and disassembly. The simplicity of this technology allows the detection of interacting proteins even at expression levels far below the endogenously expressed proteins, which causes fewer perturbations of the cellular physiology. Second, the very high signal-to-background ratio due to folding of the Rluc fragments in living cells permits more sensitive detection of protein complex dynamics by imaging single cells or more simply by spectroscopic monitoring of whole cell populations as would be desirable for high-throughput screening applications.
Another advantage of the Rluc-PCA is the ability to record and quantify repeatedly live changes of protein complexes in cell populations. This approach provides the basis for accurate determination of dose dependence of pharmacologically induced alterations of protein complexes with temporal resolution in the timeframe of seconds. In contrast to fluorescence and bioluminescent resonance energy transfer approaches, Rluc-PCA is a readout for absolute values of protein complexes, which allows for the precise quantification of even modest perturbations. Kinetics of protein complexes can be tracked via a simple and undemanding protocol in high-throughput screening format using 1,536-well plates (SI Fig. 10 and SI Methods). Reconstitution of Rluc enzyme activity can be continuously measured for >60 min (SI Fig. 11). Overall we demonstrate that Rluc-PCA can be used as a reliable real-time sensor for detecting dynamic protein complexes in living cells featured by its sensitivity and simplicity.
We could not observe reversibility, a precondition to study dynamic protein complexes, with the “barrel”-structured GFP-based PCA (SI Fig. 8). We have tested every potential dissection site in GFP and several variants and found none that are reversible (data not shown). A key criterion for choosing a PCA reporter has been to select those that have recognizable subdomains (regions with more contact surface among a group of residues than with the rest of the structure) and to choose dissection sites between these domains (21). The GFP β-barrel structure has no such subdomains. It is possible that, given the fewer contacts between subdomains, the dissociation of fragments observed with the Rluc-PCA occurs first through dissociation of its subdomains. The mechanistic details of PCA dissociation are under investigation.
To assess pharmacological effects of cAMP modulation in living cells we set out to use the Rluc-PCA in a biological system to study protein complex dynamics of a key player in GPCR cascades, PKA. Through the analysis of signaling cascades of two prototypical Gαs-coupled receptors we evaluated the extent to which PDEs modulate constitutive basal GPCR activities (41). We demonstrated that PDE4 inhibition selectively augments β2AR-mediated PKA activation in the absence of any ligand (Fig. 4C) as well as in the presence of the neutral β2AR antagonist alprenolol (SI Fig. 9) while having no effect on another Gαs-coupled receptor (V2R). These findings better define the role of the PDE4 family, whose involvement in agonist-mediated β2AR signaling had been described (42, 43) and whose inhibitors are therapeutic targets for a variety of diseases (44). The existence of PDE4 and β2AR in the same complex under basal conditions could explain these results (45, 46). Interestingly, the effect of PDE4 inhibition could not be observed after binding of the inverse agonists ICI118,551 (Fig. 4D) or propranolol (data not shown) to β2AR. Inverse agonists stabilize the receptor in its inactive conformation, thereby maximally inhibiting interaction of the receptor with Gs. The neutral antagonist leaves the equilibrium between active and inactive receptor more or less unchanged (47). Given the large numbers of inverse agonists used clinically one could predict unexpected synergistic effects of simultaneous treatment with PDE4 inhibitors.
Our study also confirms that the Rluc-PCA PKA sensor is sensitive enough to report rapid alterations of protein complexes originating from endogenously expressed receptors and effectors. This is a prerequisite to reproduce known dissociation–association kinetics and known pharmacological responses with fidelity in response to GPCR stimulation. We further show that it is possible to identify and characterize functional, endogenously expressed receptors in a parallel fashion.
Given that the GPCR family is the most important class of drug target, the Rluc-based PKA sensor represents a widely applicable assay to study various aspects of GPCR signaling in the context of drug discovery. Our findings demonstrate that using combinations of selective agonists and antagonists it is possible to assess key features of GPCRs such as (i) identifying regulatory components and mechanism of constitutive GPCR activities, (ii) identifying subtypes of endogenously expressed receptors in distinct cell lines, (iii) estimating dose–response curves for ligands, and (iv) recording quantitatively the real-time kinetics of PKA activation and inactivation.
In conclusion, our data indicate that using the newly developed Rluc-PCA PKA sensor we can quantify and image dynamic protein complex assembly and disassembly, which are modulated by distinct modules of the cAMP machinery. We have identified a mechanism of how constitutive basal GPCR activities are regulated and demonstrate a strategy to identify endogenously expressed Gαs-coupled receptors. Thus, this approach opens the door to the identification and characterization of multiple effectors of PKA. The design of further PCA sensors for distinct Gα-coupled signaling cascades would provide a functional assay platform to establish and analyze interconnected signaling routes of kinases and the GPCR family.
Methods
Construction of Plasmids.
Fragments [F(1):1–110aa; F(2):111–310aa] of the humanized Rluc gene were PCR-amplified (template, phRL-CMV; Promega). Regulatory (rat, type II, Reg) and catalytic (mouse, type α, Cat) subunits of PKA [cDNAs generously provided by M. Zaccolo, Dulbecco Telethon Institute, Padua, Italy (11)] were subcloned into the 5′ end of the 10-aa linker (GGGGS)2 and the Rluc-PCA fragments [Rluc-F(1) or Rluc-F(2); pcDNA3.1].
Cell Culture and Immunoblot Analysis.
Indicated cell lines were plated into 12- or 96-well dishes and grown in DMEM (Invitrogen) supplemented with 10% FBS. Transient transfections were performed with FuGENE-6 reagent (Roche). Cells were treated with forskolin, 3-isobutyl-1-methylxanthine, AVP, isoproterenol, alprenolol, ICI118,551, rolipram, milrinone (Sigma), and SR121463B. The reactions were terminated and immunoblotted with anti-Rluc antibodies [MAB4400 versus Rluc-F(2), MAB4410 versus Rluc-F(1); Chemicon] or anti-Reg and anti-Cat PKA antibodies (BD Transduction Laboratories).
Coimmunoprecipitation.
PCA-tagged PKA complexes were immunoprecipitated from agonist-treated six-well dishes of HEK293T cells. Cell lysate proteins were immunoprecipitated with 0.5 μg of anti-Rluc antibodies (MAB4400; Chemicon) and protein A/G-Sepharose (Calbiochem).
Bioluminescence Assay.
Immediately after treatment, exchange of medium, and addition of 100 μl of PBS to the 96-well white-walled plates (Corning), the bioluminescence analysis using the LMaxII384 luminometer (Molecular Devices) was initiated. Cells grown in 12-well plates were resuspended in 600 μl of PBS. A total of 100 μl of cell suspension (≈105 cells) was transferred to 96-well plates and subjected to bioluminescence analysis. Rluc activities were monitored for the first 10 seconds after addition of the substrate benzyl-coelenterazine (5 μM; Nanolight).
Bioluminescence Imaging.
Transfected HEK293T cells were imaged on a Nikon Eclipse TE2000U inverted microscope connected to a CoolSnap HQ Monochrome CCD camera (Photometrics) with binning 4 and CCD format of 6.45 × 6.45-μm pixels. Bioluminescence images were background-corrected and processed by using Metamorph software (Universal Imaging).
Acknowledgments
We thank M. Vasseur and M. Oueslati for their support and E. Manderson for critically reading the manuscript. This research was supported by Canadian Institutes of Health Research Grant MOP-152556 (to S.W.M.). S.W.M. holds the Canada Research Chair in Integrative Genomics. M.B. holds the Canada Research Chair in Signal Transduction and Molecular Pharmacology.
Abbreviations
- GPCR
G protein-coupled receptor
- PCA
protein-fragment complementation assay
- PKA
protein kinase A
- Rluc
Renilla luciferase
- PDE
phosphodiesterase
- β2AR
β-2 adrenergic receptor
- V2R
vasopressin-2 receptor
- AVP
arginine–vasopressin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704257104/DC1.
References
- 1.Marinissen MJ, Gutkind JS. Trends Pharmacol Sci. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 2.Dorsam RT, Gutkind JS. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 3.Hopkins AL, Groom CR. Nat Rev. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 4.Pierce KL, Premont RT, Lefkowitz RJ. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 5.Jacoby E, Bouhelal R, Gerspacher M, Seuwen K. Chem Med Chem. 2006;1:761–782. doi: 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- 6.Taylor SS, Yang J, Wu J, Haste NM, Radzio-Andzelm E, Anand G. Biochim Biophys Acta. 2004;1697:259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 7.Beavo JA, Brunton LL. Nat Rev Mol Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 8.Shabb JB. Chem Rev. 2001;101:2381–2411. doi: 10.1021/cr000236l. [DOI] [PubMed] [Google Scholar]
- 9.Smith FD, Langeberg LK, Scott JD. Trends Biochem Sci. 2006;31:316–323. doi: 10.1016/j.tibs.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Ma Y, Taylor SS, Tsien RY. Proc Natl Acad Sci USA. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaccolo M, De Giorgi F, Cho CY, Feng L, Knapp T, Negulescu PA, Taylor SS, Tsien RY, Pozzan T. Nat Cell Biol. 2000;2:25–29. doi: 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- 12.Rich TC, Karpen JW. Ann Biomed Eng. 2002;30:1088–1099. doi: 10.1114/1.1511242. [DOI] [PubMed] [Google Scholar]
- 13.Prinz A, Diskar M, Erlbruch A, Herberg FW. Cell Signalling. 2006;18:1616–1625. doi: 10.1016/j.cellsig.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 15.Terrin A, Di Benedetto G, Pertegato V, Cheung YF, Baillie G, Lynch MJ, Elvassore N, Prinz A, Herberg FW, Houslay MD, et al. J Cell Biol. 2006;175:441–451. doi: 10.1083/jcb.200605050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodge-Kafka KL, Soughayer J, Pare GC, Carlisle Michel JJ, Langeberg LK, Kapiloff MS, Scott JD. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen P. Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 18.Johnsson N, Varshavsky A. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michnick SW, Remy I, Campbell-Valois FX, Vallee-Belisle A, Pelletier JN. Methods Enzymol. 2000;328:208–230. doi: 10.1016/s0076-6879(00)28399-7. [DOI] [PubMed] [Google Scholar]
- 20.Pelletier JN, Arndt KM, Pluckthun A, Michnick SW. Nat Biotechnol. 1999;17:683–690. doi: 10.1038/10897. [DOI] [PubMed] [Google Scholar]
- 21.Pelletier JN, Campbell-Valois FX, Michnick SW. Proc Natl Acad Sci USA. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Nat Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 23.Kaihara A, Kawai Y, Sato M, Ozawa T, Umezawa Y. Anal Chem. 2003;75:4176–4181. doi: 10.1021/ac0300800. [DOI] [PubMed] [Google Scholar]
- 24.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Proc Natl Acad Sci USA. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulmurugan R, Gambhir SS. Anal Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spotts JM, Dolmetsch RE, Greenberg ME. Proc Natl Acad Sci USA. 2002;99:15142–15147. doi: 10.1073/pnas.232565699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. Proc Natl Acad Sci USA. 2002;99:3469–3474. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu CD, Kerppola TK. Nat Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remy I, Michnick SW. Nat Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 30.Remy I, Campbell-Valois F-X, Ghaddar G, Aquin S, Michnick SW. In: Protein–Protein Interactions: A Molecular Cloning Manual. Golemis EA, Adams PD, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2005. pp. 637–672. [Google Scholar]
- 31.Remy I, Michnick SW. Proc Natl Acad Sci USA. 1999;96:5394–5399. doi: 10.1073/pnas.96.10.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remy I, Wilson IA, Michnick SW. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh I, Hamilton AD, Regan L. J Am Chem Soc. 2000;122:5658–5659. [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sali A, Blundell TL. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 36.Vigil D, Blumenthal DK, Brown S, Taylor SS, Trewhella J. Biochemistry. 2004;43:5629–5636. doi: 10.1021/bi0499157. [DOI] [PubMed] [Google Scholar]
- 37.Lavoie C, Mercier JF, Salahpour A, Umapathy D, Breit A, Villeneuve LR, Zhu WZ, Xiao RP, Lakatta EG, Bouvier M, et al. J Biol Chem. 2002;277:35402–35410. doi: 10.1074/jbc.M204163200. [DOI] [PubMed] [Google Scholar]
- 38.Morello JP, Salahpour A, Laperriere A, Bernier V, Arthus MF, Lonergan M, Petaja-Repo U, Angers S, Morin D, Bichet DG, et al. J Clin Invest. 2000;105:887–895. doi: 10.1172/JCI8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch MJ, Baillie GS, Mohamed A, Li X, Maisonneuve C, Klussmann E, van Heeke G, Houslay MD. J Biol Chem. 2005;280:33178–33189. doi: 10.1074/jbc.M414316200. [DOI] [PubMed] [Google Scholar]
- 40.Breit A, Lagace M, Bouvier M. J Biol Chem. 2004;279:28756–28765. doi: 10.1074/jbc.M313310200. [DOI] [PubMed] [Google Scholar]
- 41.Seifert R, Wenzel-Seifert K. Naunyn-Schmiedeberg's Arch Pharmacol. 2002;366:381–416. doi: 10.1007/s00210-002-0588-0. [DOI] [PubMed] [Google Scholar]
- 42.Perry SJ, Baillie GS, Kohout TA, McPhee I, Magiera MM, Ang KL, Miller WE, McLean AJ, Conti M, Houslay MD, et al. Science. 2002;298:834–836. doi: 10.1126/science.1074683. [DOI] [PubMed] [Google Scholar]
- 43.Xiang Y, Naro F, Zoudilova M, Jin SL, Conti M, Kobilka B. Proc Natl Acad Sci USA. 2005;102:909–914. doi: 10.1073/pnas.0405263102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houslay MD, Schafer P, Zhang KY. Drug Discovery Today. 2005;10:1503–1519. doi: 10.1016/S1359-6446(05)03622-6. [DOI] [PubMed] [Google Scholar]
- 45.Wong W, Scott JD. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 46.Willoughby D, Wong W, Schaack J, Scott JD, Cooper DM. EMBO J. 2006;25:2051–2061. doi: 10.1038/sj.emboj.7601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azzi M, Charest PG, Angers S, Rousseau G, Kohout T, Bouvier M, Pineyro G. Proc Natl Acad Sci USA. 2003;100:11406–11411. doi: 10.1073/pnas.1936664100. [DOI] [PMC free article] [PubMed] [Google Scholar]