Abstract
The dissociation constant for an ionizable ligand binding to a receptor is dependent on its charge and therefore on its environmentally-influenced pKa value. The pKa values of sphingosine 1-phosphate (S1P) were studied computationally in the context of the wild type S1P1 receptor and the following mutants: E3.29Q, E3.29A, and K5.38A. Calculated pKa values indicate that S1P binds to S1P1 and its site mutants with a total charge of −1, including a +1 charge on the ammonium group and a −2 charge on the phosphate group. The dissociation constant of S1P binding to these receptors was studied as well. The models of wild type and mutant proteins originated from an active receptor model that was developed previously. We used ab initio RHF/6–31+G(d) to optimize our models in aqueous solution, where the solvation energy derivatives are represented by conductor-like polarizable continuum model (C-PCM) and integral equation formalism polarizable continuum model (IEF-PCM). Calculation of the dissociation constant for each mutant was determined by reference to the experimental dissociation constant of the wild type receptor. The computed dissociation constants of the E3.29Q and E3.29A mutants are 3–5 orders of magnitude higher than those for the wild type receptor and K5.38A mutant, indicating vital contacts between the S1P phosphate group and the carboxylate group of E3.29. Computational dissociation constants for K5.38A, E3.29A and E3.29Q mutants were compared with experimentally determined binding and activation data. No measurable binding of S1P to the E3.29A and E3.29Q mutants was observed, supporting the critical contacts observed computationally. These results validate the quantitative accuracy of the model.
INTRODUCTION
Sphingosine 1-phosphate (S1P) is a bioactive lipid with broad biological effects. In the last decade, S1P was found to act as an agonist of a G protein-coupled receptor (GPCR), EDG-1/S1P1.1 This led to the discovery and classification of additional S1P-responsive GPCR in the endothelial differentiation gene (EDG) family, EDG-3/S1P3,2 EDG-5/S1P2,2,3 EDG-6/S1P44,5 and EDG-8/S1P56,7 with 40–50% sequence identity.8 S1P receptors regulate endothelial cell migration both positively (S1P1 and S1P3) and negatively (S1P2).9,10 S1P receptors are important for enhancement of cell survival, cell proliferation, regulation of the actin-based cytoskeleton affecting cell shape, adherence, chemotaxis, and the activation of Cl− and Ca2+ ion conductances.11–13 The S1P1 receptor is the target of a novel immunosuppressive agent in phase III clinical trials to treat transplant rejection14 and is the focus of ongoing efforts in multiple laboratories to identify novel agonists with similar therapeutic promise.15–24
GPCR exhibit conformational equilibrium between active and inactive conformations.25,26 In the simplest model of ligand influence on GPCR equilibria, ligands can bind to and stabilize the active conformation (agonist), the inactive conformation (inverse agonist) or can bind to both conformations without preference (neutral antagonist). We have previously reported models of active (S1P1, S1P4, LPA1–3) and inactive (LPA1–3) conformations of EDG family members in complex with both agonists and antagonists.27–33 These previous studies have largely focused on validating qualitative structure-based predictions regarding relative binding affinities and roles of amino acids in binding. The present study focuses on the validation of the active conformation of the S1P1 receptor as a quantitatively accurate tool to examine agonist binding. However, the charge on the S1P phosphate group in the receptor binding site is ambiguous due to the overlap of the second pKa value with the biological pH range. As binding affinity depends strongly on the charge of the S1P phosphate group, the environmental dependence of the phosphate group pKa must be computed before binding affinities can be addressed. Accurate pKa and binding affinity computation requires a model that includes coulombic interactions, hydrophobic interactions, and hydrogen bond interactions between the ligand and the receptor as well as intramolecular interactions of these types within the ligand.
The pKa of receptor-bound S1P was determined using the method Li and Jensen34 applied to determine amino acid sidechain pKa values. This method extends from initial theoretical models by Tanford and Kirkwood that treated all ionizable sidechains as points on an impenetrable spherical protein surface,35 by Shire, Hanania and Gurd who incorporated static solvent accessibility terms to compensate for the assumption of a smooth boundary between the exterior and interior of the protein,36 by Warshel who described the importance of electrostatic solvation differences due to both permanent and induced protein dipoles,37 and by Bashford and Karplus who eliminated the need to estimate intrinsic pKa corrections.38 Since our protein structure is a computational model, we validated its structure by calculating dissociation constants for a series of receptor mutants and compared the computed binding affinities to experimental results. Accurate binding affinity results validate both the computed pKa values and the use of homology models of EDG receptors for quantitative studies of agonist binding. In this paper we present the dissociation constant calculation approach, and the pKa values and binding constants of S1P in the wild type S1P1 receptor and its mutants.
METHODOLOGY
THEORETICAL BASIS
pKa calculation
Following the method developed by Li and Jensen34 for carboxyl pKa values, the pKa of the phosphate group in S1P when it is bound to a receptor, R HS1P, is related to the standard free energy change, ΔG, of the proton exchange reaction with a reference molecule, Ref.:
| (1) |
By the equation
| (2) |
The value 5.66 is the experimental pKa value of the reference molecule O-phosphoethanolamine,39 at 298K. This pKa value describes the transition between ionization states of 0 and −1. The value 1.36 is 2.303RT at the same temperature in kilocalories per mole. The free energy of a molecule X, G(X), is given by the sum of ground state electronic energy Eele and the solvation energy Gsol.
| (3) |
Free energies were calculated using structures optimized at the RHF/6–31(+)G* level including solvation energy using a new geometry optimization algorithm, developed by Li and Jensen.34 Solvation energy and energy derivatives at each optimization step were calculated using the conductor-like polarizable continuum model (C-PCM)40,41 and integral equation formalism polarizable continuum model (IEF-PCM),42 using GAMESS43 parameters: IEF=−10, and RET=100, to prevent use of additional spheres. The iterative solvent was water. Diffuse functions (L shell) were added to the heavy atoms on the negatively charged groups (one carbon, four oxygen, and one phosphorus atom of the phosphate group and two carbon and two oxygen atoms of the glutamic acid side chain) under consideration. The United Atom Hartree Fock (UAHF)44 radii determined with Gaussian9845 were used to define the molecular cavities. The energies were calculated using the quantum mechanics program GAMESS.43 The solvation energy contribution to the free energy, Gsol,, was obtained during geometry optimization. Density functional theory (DFT) with the B3LYP functional46,47 at the [DFT/6–31(+)G(2d, p)] level was used to calculate the electronic contribution to the free energy, Eelec, for the optimized geometry described previously using GAMESS.43
Kd Calculation
Binding constants, in the form of Kd values, are calculated relative to a reference molecule. The procedure is based on the thermodynamic cycle shown in Figure 1. R is the reference receptor with known experimental Kd value. M is a mutant receptor or a receptor from the same family (an S1P receptor in the current case). As free energy is a state function, all paths which start and end at the same points in the cycle are equivalent, so:
Figure 1.
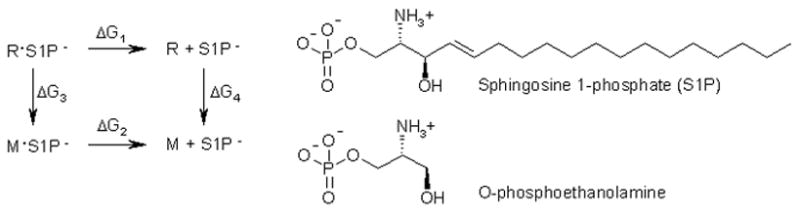
Thermodynamic cycle for binding affinity of S1P− to a mutant (M) relative to the reference receptor (R).
| (4) |
Reaction free energy is related to binding affinity by the following relation:
| (5) |
In equation 5, R is the gas constant and T is the temperature. This relation and the known Kd of 38.7 nM for S1P in S1P1 allow substitution of known values for ΔG1.28 Additionally, since ΔG = Gproduct − Greactant, we can write:
| (6) |
All the free energy terms can be calculated using ab-initio quantum mechanical methods as explained above.
THEORETICAL CALCULATIONS
Receptor and Receptor Complex Structures
Protein structures used to derive S1P microenvironment models for quantum mechanical calculations were derived from a previously published, experimentally validated model of S1P in the S1P1 receptor.48–50 This model of the wild type receptor was used to generate mutant receptor models by amino acid sidechain replacement. Each mutant was modeled with and without bound S1P. The bound S1P was modeled with a total charge of zero or negative one. Each model (fifteen complexes in all) was refined using force-field based molecular dynamics, followed by geometry optimization. All force field molecular dynamics and force field energy minimizations were done using the software package MOE (version 2005.06, Chemical Computing Group, Montreal, Canada), with the MMFF94 force field.51 Molecular dynamics simulations of 5 ns for the native S1P1 complex with both acid and base forms of S1P showed that temperature and geometry both converged within the first 100 ps. Root mean square deviations of atom positions contributing to the model were less than 1.5 Å between the structure obtained after 100 ps and that at 5 ns, a typical geometric fluctuation for sidechain atoms due to the kinetic energy in the system during molecular dynamics simulations. Therefore, molecular dynamics simulations were performed for 100 picoseconds, at 300K, followed by geometry optimization with termination criteria of 100 steps or a gradient of 0.1 kcal/mol*Å. After the force field refinement completed, S1P microenvironment models were extracted and subjected to constrained geometry optimization with the same quantum mechanical approach described above.
S1P Microenvironment Models
The ability to calculate the binding constant for GPCR agonists would assist in the development of agonists with lower binding constants as candidate leads for drug development. In order to achieve that goal, we assess whether our method can predict reasonable relative binding constants for known agonists. To reliably calculate binding constants, we need to know the charge (and therefore the pKa) of the S1P phosphate group within the microenvironment of the receptor. Therefore, our microenvironment model includes the phosphate group and its immediate chemical environment. Similar to a previous study,52 we constructed multiple models (Figure 2) to determine the minimal subset of atoms that adequately reflect the environmental influence on pKa. The atoms and bonds that were energetically optimized are denoted using bold fonts in Figure 2. The smallest model (Figure 2A) contains the first three of eighteen S1P carbon atoms bearing the phosphate, ammonium and hydroxyl groups, two arginine (R3.28, R7.34) guanidinium groups, a lysine (K5.38) ammonium and a glutamic acid (E3.29) carboxylate group. These functional groups include all atoms within 2.4 Å of the ionizable groups of S1P. An intermediate model (Figure 2B) considers hydrophobic interactions and cation-pi interactions by introducing the indole ring of W4.64. The intermediate model includes all functional groups approaching within 2.9 Å of the ionizable groups of S1P. A larger model (Figure 2C) considers hydrophobic contributions from the sidechains of M3.32, I5.41, L6.55, A7.35 and F7.38 and dipole contributions from the amide bonds linking M3.32 to F3.33, R3.28 to E3.29, P4.60 to I4.61 and H5.37 to K5.38. The functional groups in the larger model include all atoms within 5.5 Å of the ionizable groups of S1P. No additional charged sidechains are found in the model within a 10 Å radius of the ionizable groups of S1P, and the closest buried ionizable sidechain atom is more than 20 Å away from the S1P phosphate group.
Figure 2.

Models of S1P in the S1P1 receptor. Optimized atoms are shown in bold. A. Small microenvironment model including functional groups within 2.4 Å of S1P phosphate and ammomium groups. B. Intermediate microenvironment model including functional groups within 2.9 Å of S1P phosphate and ammomium groups. C. Larger microenvironment model including functional groups within 5.5 Å of S1P phosphate and ammomium groups.
Coordinates were extracted from the energy minimized model of the receptor that was obtained after molecular dynamics. Hydrogen atoms were added using the program MOE. Several conformations were modeled using a stochastic search algorithm. For the pKa reference molecule, O-phosphoethanolamine all 41 possible conformations were generated manually. The total free energy for each molecule was taken to be the ‘conformational average’ of the free energies of each conformer (Gi):53
| (7) |
Where G0 is the lowest energy conformer, and ΔGi = Gi − G0
For the mutant studies, we used models analogous to the intermediate microenvironment model shown in Figure 2B. The appropriate sidechain functional group, ammonium or formate, was excluded from the mutant model for the alanine mutants, K5.38A, or E3.29A, respectively. For the E3.29Q mutant, formamide was used in place of formate. Optimization of the K5.38A mutant resulted in shift of the phosphate group into a location occupied by a phenylalanine residue, F7.38, in the context of the receptor, thus the phosphorus and attached oxygen atoms were held fixed during optimization of this mutant. In Figure 3 the mutation sites are presented three-dimensionally in the intermediate microenvironment model.
Figure 3.
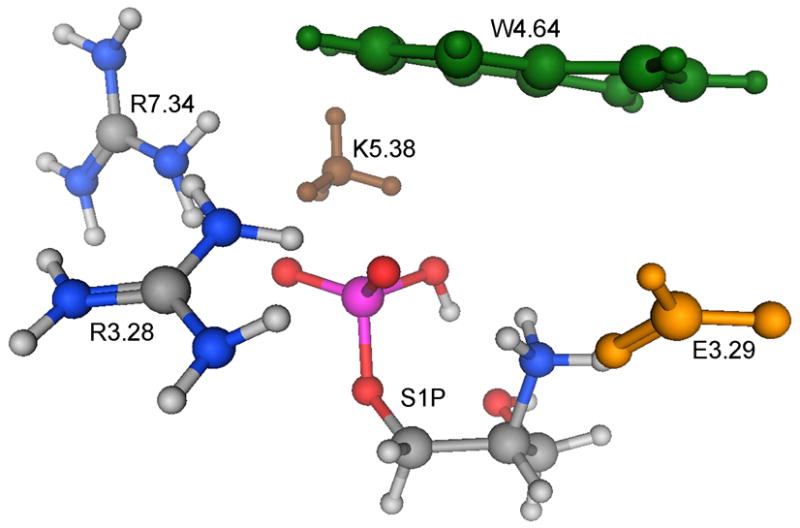
Three-dimensional structure of the intermediate microenvironment model of S1P bound to S1P1. Amino acid functional groups are colored green (W4.64), brown (K5.38) and yellow (E3.29).
EXPERIMENTAL MATERIALS AND METHODS
Reagents
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) and were of analytical purity unless specified otherwise. S1P was obtained from Avanti Polar Lipids (Alabaster, AL).
Site-directed mutagenesis
Mutants were prepared from the N-terminal FLAG epitope-tagged S1P1 receptor construct (GenBank™ accession number AF233365) provided by Dr. Timothy Hla (University of Connecticut). Site-specific mutations were generated using the ExSite™ mutagenesis kit (Stratagene, La Jolla, CA) as described in our previous publications.29,48,49,54,55 S1P1 and the mutants were subcloned into the pcDNA3.1 vector (Invitrogen). Clones were verified by complete insert sequencing.
Cell Culture and Transfection
RH7777 and HEK293 cells (ATCC, Manassas, VA) were cultured in Dulbecco’s modified minimal essential medium (DMEM) containing 10% fetal bovine serum (Hyclone, Logan, UT). Cells (2 × 106) were transfected with 2 μg of plasmid DNA with Effectene (Qiagen, Valencia, CA) according to the manufacturer protocol, for 24 h. Before ligand binding and receptor activation assays, the cells were washed twice with serum-free DMEM and serum-starved in DMEM for at least 6 h.
[32P]S1P Radioligand Binding Assays
The S1P binding assays were done as previously described.28 Transfected RH7777 cells (5 × 105) were incubated at 4°C in 20 mM Tris-HCl (pH 7.5) binding buffer containing 100 mM NaCl, 15 mM NaF, protease inhibitor cocktail (Sigma-Aldrich), and 20% (w/v) essentially fatty-acid free BSA with 1 nM [32P]S1P in 50 nM S1P for 40 min. Cells were centrifuged and washed twice with binding buffer. The pellet was resuspended in 2:1 CHCl3/MeOH and equilibrated in scintillation fluid overnight. Cell-bound radioactivity was measured by liquid scintillation counting using a Beckman LS5000 TA counter (Beckman Coulter, Irvine, CA). Specific binding was defined as the difference between total binding and non-specific binding (in the presence of 2–5 μM cold S1P). Standard errors were computed on the basis of triplicate samples from two simultaneous transfections.
HEK-293 cells were used for the competition assays. Briefly, 4×105 cells were plated in 24-well dishes and allowed to adhere overnight. The cells were then transfected with 0.4 μg of cDNA using Lipofectamine 2000 (Invitrogen) and the transfection proceeded for 48 h. After duplicate washes with ice-cold binding buffer (20 mM Tris-HCl, pH 7.4 and 150 mM NaCl), 0.1 nM [32P]S1P and competing concentrations of cold S1P (1 nM-10 μM), resuspended in binding buffer containing 0.4% (w/v) BSA, were applied to the cells and incubated on ice for 30 min. After duplicate washes cells with ice-cold binding buffer containing 0.04% (w/v) BSA, the pelleted cells were lysed with 0.5% (w/v) SDS and equilibrated in scintillation fluid. Samples were measured in triplicate. Kd values were determined using GraphPad Prism software (San Diego, CA).
S1P1 Receptor Activation Assays
Receptor function was assayed in transiently transfected RH7777 cells by measuring S1P-activated [35S]GTPγS binding as described in our previous publication.28 Cells were harvested by scraping in homogenization buffer (20 mM HEPES, pH 7.4, 50 mM NaCl, and 2 mM EDTA), lysed by sonication for 30 s on ice (Kontes Micro-Ultrasonic Cell Disrupter), and centrifuged at 2000 ×g at 4°C for 5 min. The supernatant was then centrifuged at 100,000 ×g for 1 h at 4°C. The pellet was resuspended in the storage buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM MgCl2, and 2 mM EDTA) and stored at −80°C. The assay was performed in triplicate in a total volume of 100 μl with 5–10 μg of protein. Membranes were incubated in the binding buffer (50 mM HEPES (pH 7.5), 100 mM NaCl, 1mM MgCl2, 10 μM GDP, and 2 mM dithiothreitol) containing 0.1 nM [35S]GTPγS (1000 Ci/mmol; Amersham Pharmacia Biotech, Piscataway, NJ) and 0–10 μM S1P for 30 min at 30°C. Membrane-bound radioactivity was separated by vacuum filtration using a 96-well Brandel Cell Harvester (Gaithersburg, MD) through a Whatman GF/B filter, and washed three times with ice-cold washing buffer (20 mM Tris, 120 mM NaCl, and 25 mM MgCl2). Bound radioactivity was quantitated by liquid scintillation counting. Specific binding was determined by subtracting the counts of unstimulated samples from the samples to which S1P was added.
RESULTS
Impact of Mutations on S1P1 Structure
Molecular dynamics studies were used to refine the S1P1 mutant complexes with both protonated and unprotonated forms of S1P. Significant differences in the relative orientation of the S1P polar headgroup and surrounding sidechains were observed between the wild type receptor and its mutants. In general, mutation to a smaller amino acid resulted in shifts of surrounding amino acids toward the vacated space in the mutant (Figure 4 and data not shown). These shifts rearranged, rather than disrupted, the ionic interactions between the S1P phosphate and cationic amino acids R3.28, K5.38 and R7.34 (Table I). Notably, a significant improvement in ionic interactions between the S1P phosphate and cationic residues R3.28 and R7.34 was noted in the case of the K5.38A mutant (Figure 4 and Table I). This improvement is reflected in an average distance between phosphate oxygen atoms and nitrogen atoms of surrounding cationic residues of 2.51 Ǻ relative to the wild type receptor average of 2.74 Ǻ. One fewer cationic residue allowed the remaining two to adopt bifurcated interactions, either involving a single arginine nitrogen interacting with two phosphate oxygens, or two arginine nitrogen atoms each interacting with a phosphate oxygen atom.
Figure 4.

Comparison of ionic interactions between S1P phosphate group and surrounding residues in the wild type S1P1 complex (left) and the K5.38A complex (right).
Table I.
Distances in Ǻ between S1P phosphate oxygen atoms and nitrogen atoms of surrounding cationic amino acid residues.
| S1P1 | E3.29A | E3.29Q | K5.38A | |
|---|---|---|---|---|
| S1P-O1···R3.28-N1 | 2.64 | 2.60 | 2.57 | 2.57 |
| S1P-O2···R3.28-N1 | * | * | * | 2.75 |
| S1P-O2···R3.28-N2 | * | * | * | * |
| S1P-O2···R7.34-N1 | 2.63 | 2.86 | 2.86 | 2.30 |
| S1P-O2···K5.38-N | 2.99 | * | * | * |
| S1P-O3···R7.34-N2 | * | * | 2.72 | 2.43 |
| S1P-O3···K5.38-N | 2.70 | 2.58 | 2.45 | * |
| Number < 3.00 Ǻ | 4 | 3 | 4 | 4 |
| Average Distance | 2.74 | 2.68 | 2.65 | 2.51 |
Distance greater than 3.00 Ǻ.
S1P Head Group pKa
Three microenvironment models of the S1P head group bound to the S1P1 receptor were developed (Figure 2). Computed pKa values for the second ionization of the phosphate group are 3.5 and 4.8 pH units, using the small and intermediate models, respectively. Computed pKa values using a comparable subset of conformations for the intermediate and larger models differed by less than half a unit, indicating that long-range contributions to the pKa were negligible. All models indicate that the phosphate group is deprotonated when it is bound to the wild type S1P1 receptor.
The difference in pKa value determined with the small and intermediate models indicates that the indole ring of W4.64 influences the pKa value, thus all calculations on mutant receptors were performed using the intermediate microenvironment model. The centroid of the pyrrole ring of W4.64 is located 4.59 Å from the ammonium group of S1P. This cation-pi interaction is responsible for the pKa difference determined using the small and intermediate models. All mutants showed identical or decreased S1P pKa values relative to S1P1. The E3.29Q mutant showed no change in pKa relative to the wild type S1P1 receptor, whereas the K5.38 and E3.29A mutants showed decreases in pKa from 4.8 for the wild type S1P1 receptor to 2.5 and −1.0, respectively. All mutants will therefore bind S1P with a total charge of negative one (−2 on phosphate and +1 on ammonium). All pKa values and binding constants are summarized in Table II.
Table II.
Phosphate pKa and S1P binding constant in S1P1 and its mutants. Theoretical pKa values computed using a subset of conformations are shown in parentheses as are standard errors of experimental measurements.
| pKa | Kd (nM) | Receptor Activation | |||
|---|---|---|---|---|---|
| Model | Theoretical | Theoretical | Experimental | EC50(nM) | Emax(%) |
| Small model of S1P1 | 3.5 | ||||
| Intermediate model of S1P1 | 4.8 (4.6) | 38.7 (7.84) | 1.4 | 100 | |
| Large model of S1P1 | ---(4.2) | ||||
| E3.29A | −1.0 | 1.1×104 | >10000* | >10000* | <10* |
| E3.29Q | 4.8 | 1.1×104 | >10000* | >10000* | 17* |
| K5.38A | 2.5 | 40 | 49.8 (13.3) | 32.4 | 97 |
From 48
S1P Receptor Binding Affinities
Single mutations of several residues involved in phosphate recognition in the S1P receptor family completely abolish ligand binding and ligand-induced receptor activation.28,30,33 The polar headgroup therefore contributes significantly to the overall binding free energy. To validate our receptor models, we calculated the relative binding constants (Kd) for the S1P headgroup microenvironment in several mutants. We used a reference molecule approach, where, as explained in the method section, we based our calculation on a known wild type S1P1 binding constant. The K5.38A mutation has a calculated Kd value of 40 nM which is close to the experimental Kd, 49.8 ± 13.3 nM.
The E3.29Q and E3.29A mutations have calculated binding constants five orders of magnitude higher than the wild type receptor, which means that their binding with the active conformation of S1P is very poor. These results indicate that E3.29 makes critical contacts with S1P in the activated receptor. Both of these mutants show 25% or less activation in response to S1P stimulation relative to the wild type S1P1 receptor confirming the modeling prediction that S1P does not bind and stabilize the active conformation of these mutants. Experimental S1P binding assays with these mutants indicate that position 3.29 is critically important for S1P binding, as well as S1P-stimulated receptor activation, as no measurable binding was observed for the E3.29A and E3.29Q mutants.
DISCUSSION
S1P receptors are of interest as therapeutic targets due to their proven role in immune function,21,56–58 angiogenesis,59–62 and cell migration.9,10,63–67 Computationally-guided drug design for these targets requires not only the qualitative understanding of ligand recognition that has been demonstrated in our previous modeling studies,29,48,49,54,68–72 but also a quantitative understanding. A prerequisite for quantitative determination of relative binding affinities is accurate modeling of ionizable groups. While assumptions can be made about the prevalent form of some ionizable groups, the second pKa of phosphate groups in many biologically important molecules is too close to the biological pH range for accurate estimation of ionization state. We therefore examined the ionization state of S1P bound to the S1P1 receptor in order to accurately predict the effect of the surrounding microenvironment73 on the pKa. Our approach differs from previous investigations of titratable functional groups in proteins due to our focus on the bound ligand ionization state under the pH typical of biological assay conditions rather than appropriate ionization behavior of all titratable groups under a range of conditions as previously investigated.35–38
The pKa value of 4.8 computed for S1P bound to wild type S1P1 is lower than the solution pKa value of 5.66 reported for the reference molecule, O-phosphoethanolamine. However, the pKa value computed with a fixed position of the S1P hydroxyl group, 5.5, suggests that the difference is related to the structural difference between S1P and the reference molecule, which lacks this hydroxyl group. Thus electrostatic charge stabilization and hydrophobic solvent exclusion in the headgroup binding site are balanced and contribute to an overall environment similar to aqueous solution. Most mutations disrupted the balance among the hydrophobic and electrostatic contributions to the microenvironment and showed decreased S1P pKa values relative to S1P1. The K5.38A and E3.29A mutants showed decreases in pKa from 4.8 for the wild type S1P1 receptor to 2.5 and −1.0, respectively. Interestingly, the smallest and largest shifts in pKa relative to wild type S1P1 occur for different mutations at a common site. The mutation of E3.29 to alanine caused the S1P phosphate pKa to decrease by 5.8 pH units. The absence of a negatively charged group in the receptor enhances its preference for the anionic form of S1P. In contrast, the pKa did not change relative to the wild type receptor in the E3.29Q mutant, due in part to the partially negative formamide group as well as geometry differences in the mutant microenvironment. Given the large S1P pKa decrease in the E3.29A mutant relative to wild type S1P1, an increase in pKa might be expected for the K5.38A mutant due to the loss of a positive charge capable of stabilizing the anion. However, as Table I and Figure 5 indicate, significant rearrangement of the cationic residues surrounding the phosphate in the K5.38A mutant was observed relative to wild type S1P1. Thus the pKa of S1P was 3.0 units lower in the K5.38A microenvironment than in wild type S1P1. The role of hydrophobic and cation-pi interactions is evident from lack of the W4.64 indole ring in the small model, which causes the pKa to decrease by 1.3 pH units, from 4.8 for the intermediate model to 3.5 in the small model of the wild type S1P1 receptor. The centroid of the pyrrole ring of W4.64 is 4.59 Å from the cationic ammonium group of S1P, indicating a strong cation-pi interaction that is lacking in the W4.64A mutant.
The microenvironment affecting the pKa of S1P includes not only the residues in the receptor, but also functional groups present within the S1P structure itself. The hydroxyl group of S1P stabilizes the unprotonated form of S1P phosphate by hydrogen bonding as can be seen in Figure 4. Conformations lacking this intramolecular hydrogen bond had higher energy, and pKa values computed using such conformations were higher (data not shown). A similar interaction was noted in computational modeling of myo-inositol 2-monophosphate using B3LYP/6–31+G(d), where an intramolecular hydrogen bond interaction was observed in the most stable conformation of each structure studied.74 Experimental studies using magic angle spinning 31P NMR indicate that a hydrogen bond interaction decreases the pKa of LPA relative to phosphatidic acid and dehydroxy-LPA by half of a pH unit, in a phosphatidyl choline bilayer.75 A hydrogen on an electronegative atom in an acidic residue can serve as a hydrogen bond donor, which decreases pKa.73 This internal stabilization of the more negatively charged ionization state provides a framework to understand recent structure activity studies of S1P analogs. Lim et al. reported that an oxidized S1P analog, with a C3 ketone, has a significantly lower binding affinity for S1P1, S1P2 and S1P3.76 In contrast, they later reported that a C3-dehydroxy 4,5-dihydro S1P analog failed to bind to S1P1, but showed significant binding to S1P2 and S1P3.77 These findings can now be reinterpreted to indicate that an intramolecular hydrogen bond donated from the C3 hydroxyl to the phosphate group of S1P is required to stabilize the deprotonated form for high affinity binding to S1P1, but not S1P2 or S1P3. Both S1P2 and S1P3 have a lysine residue at position 7.34, where S1P1 has an arginine residue. The microenvironment in S1P1 is therefore distinct from that of S1P2 and S1P3. Microenvironment differences may influence the ionization state of S1P that is relevant for activation of different receptors in the S1P family.
Relative binding affinities of the deprotonated form of S1P were calculated for S1P1 receptor mutants relative to the binding affinity of wild type S1P1. Two mutants, E3.29A and E3.29Q, showed negligible affinity for S1P based on theoretical Kd values of 1.1×104 nM. GPCR, such as S1P1, exist in an equilibrium involving several receptor conformations, with conformational extremes that either activate (active conformations) or fail to activate (inactive conformations) the coupled G protein.25 Therefore, theoretical calculations involving the active conformation suggest that these three mutants should show negligible activation. Experimental determination of GTPγS binding indicates that the E3.29A and E3.29Q mutants cannot be activated by S1P in the nanomolar concentration range.48 Experimental measurement of S1P binding demonstrates that these mutants fail to bind S1P, indicating that E3.29 is critical for S1P recognition, without which there can be no S1P-induced receptor activation. One mutant, K5.38A, showed maximal activation of 97%, which was nearly equivalent to that of the wild type receptor. This finding is consistent with the theoretical prediction of a Kd value very near that of the wild type receptor. The potency, reflected in the concentration giving half-maximal activation, was 32.4 nM for K5.38A compared with 1.4 nM for the wild type receptor. The calculated binding affinity for K5.38A was 40 nM, quite comparable to the experimentally determined Kd of 49.8 nM. The structural changes induced by the K5.38A mutation (Figure 4 and Table I) are consistent with the finding that K5.38 does not play an irreplaceable role in S1P recognition in the S1P1 receptor. This finding highlights a key difference between the S1P1 and S1P4 receptors. The K5.38A mutation in S1P4 completely abolished S1P-induced GTPγS binding and cell migration.49 This difference is likely due to the difference in amino acid residues appearing at position 7.34 in the two receptors. Arginine occurs at position 7.34 in S1P1 whereas glycine appears at position 7.34 in S1P4. Glycine lacks a positive charge, and cannot compensate for the elimination of lysine in the manner observed for R7.34 of S1P1 in this study.
CONCLUSIONS
In conclusion, these studies demonstrate that S1P binds to S1P1 with a total charge of −1. Mutations to residues surrounding the S1P headgroup tend to shift the receptor-bound pKa to lower values. Examination of mutant binding affinities allowed determination of the relative importance of contacts between S1P and various residues present in the binding pocket. In particular, contact between the carboxylate group of E3.29 and the phosphate group of S1P was identified computationally as critical for binding. Experimental binding assays confirm that no measurable binding of S1P occurs to either the E3.29A or E3.29Q mutant. In contrast, K5.38 is not critical as mutation of this residue to alanine allowed rearrangement of other ion pairing groups giving equivalent binding affinity to the wild type receptor, both computationally and experimentally. These studies validate the S1P1 receptor model not only as a qualitatively accurate tool to examine the role of receptor amino acids in agonist recognition, but also as a quantitatively accurate tool to examine contributions of specific functional groups to binding and activation.
Acknowledgments
MMN and ALP thank The Chemical Computing Group for the MOE software package. We appreciate financial support from NIH (HL084007-01 to AP; CA92160-01 and HL61469 to GT) and The Department of Chemistry and Computational Research on Materials Institute at the University of Memphis.
Financial support for this work provided in part by NIH (HL084007-01 to AP; CA92160-01 and HL61469 to GT).
ABBREVIATIONS
- S1P
Sphingosine 1-phosphate
- IEF-PCM, C-PCM
conductor-like polarization continuum model
- GPCR
G protein-coupled receptor
- EDG
endothelial differentiation gene
- DFT
density functional theory
- DMEM
Dulbecco’s modified minimal essential medium
- RH7777
rat hepatoma 7777
- HEK
human embryonic kidney
- BSA
bovine serum albumin
- FBS
fetal bovine serum
- SDS
sodium dodecyl sulfate
- EDTA
ethylene diamine tetraacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee MJ, Van Brocklyn JR, Thangada S, Liu CH, Hand AR, Menzeleev R, Spiegel S, Hla T. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998;279(5356):1552–1555. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- 2.An S, Bleu T, Huang W, Hallmark OG, Coughlin SR, Goetzl EJ. Identification of cDNAs encoding two G protein-coupled receptors for lysosphingolipids. FEBS Lett. 1997;417(3):279–282. doi: 10.1016/s0014-5793(97)01301-x. [DOI] [PubMed] [Google Scholar]
- 3.MacLennan AJ, Browe CS, Gaskin AA, Lado DC, Shaw G. Cloning and characterization of a putative G-protein coupled receptor potentially involved in development. Mol Cell Neurosci. 1994;5(3):201–209. doi: 10.1006/mcne.1994.1024. [DOI] [PubMed] [Google Scholar]
- 4.Van Brocklyn JR, Gräler MH, Bernhardt G, Hobson JP, Spiegel S. Sphingosine-1-phosphate is a Ligand for the G Protein-Coupled Receptor EDG-6. Blood. 2000;95(8):2624–2629. [PubMed] [Google Scholar]
- 5.Yamazaki Y, Kon J, Sato K, Tomura H, Sato M, Yoneya T, Okazaki H, Okajima F, Ohta H. Edg-6 as a Putative Sphingosine 1-Phosphate Receptor Coupling to Ca2+ Signaling Pathway. Biochem Biophys Res Commun. 2000;268(2):583–589. doi: 10.1006/bbrc.2000.2162. [DOI] [PubMed] [Google Scholar]
- 6.Glickman M, Malek RL, Kwitek-Black AE, Jacob HJ, Lee NH. Molecular Cloning, Tissue-Specific Expression, and Chromosomal Localization of a Novel Nerve Growth Factor-Regulated G-Protein- Coupled Receptor, nrg-1. Mol Cell Neurosci. 1999;14(2):141–152. doi: 10.1006/mcne.1999.0776. [DOI] [PubMed] [Google Scholar]
- 7.Malek RL, Toman RE, Edsall LC, Wong S, Chiu J, Letterle CA, Van Brocklyn JR, Milstien S, Spiegel S, Lee NH. Nrg-1 belongs to the endothelial differentiation gene family of G protein-coupled sphingosine-1-phosphate receptors. J Biol Chem. 2001;276(8):5692–5699. doi: 10.1074/jbc.M003964200. [DOI] [PubMed] [Google Scholar]
- 8.Parrill AL, Sardar VM, Yuan H. Sphingosine 1-phosphate and lysophosphatidic acid receptors: agonist and antagonist binding and progress toward development of receptor-specific ligands. Semin Cell Dev Biol. 2004;15(5):467–476. doi: 10.1016/j.semcdb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Takuwa Y. Subtype-specific differential regulation of Rho family G proteins and cell migration by the Edg family sphingosine-1-phosphate receptors. Biochim Biophys Acta. 2002;1582(1–3):112–120. doi: 10.1016/s1388-1981(02)00145-2. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi H, Kitayama J, Takuwa N, Arikawa K, Inoki I, Takehara K, Nagawa H, Takuwa Y. Sphingosine-1-phosphate receptor subtype-specific positive and negative regulation of Rac and haematogenous metastasis of melanoma cells. Biochem J. 2003;374(Pt 3):715–722. doi: 10.1042/BJ20030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tigyi G, Fischer DJ, Baker D, Wang DA, Yue J, Nusser N, Virag T, Zsiros V, Liliom K, Miller D, Parrill A. Pharmacological characterization of phospholipid growth-factor receptors. Ann N Y Acad Sci. 2000;905:34–53. doi: 10.1111/j.1749-6632.2000.tb06536.x. [DOI] [PubMed] [Google Scholar]
- 12.Van Brocklyn J, Letterle C, Snyder P, Prior T. Sphingosine-1-phosphate stimulates human glioma cell proliferation through Gi-coupled receptors: role of ERK MAP kinase and phosphatidylinositol 3-kinase beta. Cancer Lett. 2002;181(2):195–204. doi: 10.1016/s0304-3835(02)00050-2. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Love LM, Singh I, Zhang QX, Dewald J, Wang DA, Fischer DJ, Tigyi G, Berthiaume LG, Waggoner DW, Brindley DN. Lipid phosphate phosphatase-1 and Ca2+ control lysophosphatidate signaling through EDG-2 receptors. J Biol Chem. 2000;275(36):27520–27530. doi: 10.1074/jbc.M003211200. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmann V, Cyster JG, Hla T. FTY720: sphingosine 1-phosphate receptor-1 in the control of lymphocyte egress and endothelial barrier function. Am J Transplant. 2004;4(7):1019-1–1025. doi: 10.1111/j.1600-6143.2004.00476.x. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Chen W, Hale JJ, Lynch CL, Mills SG, Hajdu R, Keohane CA, Rosenbach MJ, Milligan JA, Shei GJ, Chrebet G, Parent SA, Bergstrom J, Card D, Forrest M, Quackenbush EJ, Wickham LA, Vargas H, Evans RM, Rosen H, Mandala S. Discovery of potent 3,5-diphenyl-1,2,4-oxadiazole sphingosine-1-phosphate-1 (S1P1) receptor agonists with exceptional selectivity against S1P2 and S1P3. J Med Chem. 2005;48(20):6169–6173. doi: 10.1021/jm0503244. [DOI] [PubMed] [Google Scholar]
- 16.Hale JJ, Lynch CL, Neway W, Mills SG, Hajdu R, Keohane CA, Rosenbach MJ, Milligan JA, Shei GJ, Parent SA, Chrebet G, Bergstrom J, Card D, Ferrer M, Hodder P, Strulovici B, Rosen H, Mandala S. A rational utilization of high-throughput screening affords selective, orally bioavailable 1-benzyl-3-carboxyazetidine sphingosine-1-phosphate-1 receptor agonists. J Med Chem. 2004;47(27):6662–6665. doi: 10.1021/jm0492507. [DOI] [PubMed] [Google Scholar]
- 17.Yan L, Hale JJ, Lynch CL, Budhu R, Gentry A, Mills SG, Hajdu R, Keohane CA, Rosenbach MJ, Milligan JA, Shei GJ, Chrebet G, Bergstrom J, Card D, Rosen H, Mandala SM. Design and synthesis of conformationally constrained 3-(N-alkylamino)propylphosphonic acids as potent agonists of sphingosine-1-phosphate (S1P) receptors. Bioorg Med Chem Lett. 2004;14(19):4861–4866. doi: 10.1016/j.bmcl.2004.07.049. [DOI] [PubMed] [Google Scholar]
- 18.Hale JJ, Yan L, Neway WE, Hajdu R, Bergstrom JD, Milligan JA, Shei GJ, Chrebet GL, Thornton RA, Card D, Rosenbach M, HughRosen Mandala S. Synthesis, stereochemical determination and biochemical characterization of the enantiomeric phosphate esters of the novel immunosuppressive agent FTY720. Bioorg Med Chem. 2004;12(18):4803–4807. doi: 10.1016/j.bmc.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Hale JJ, Doherty G, Toth L, Mills SG, Hajdu R, Keohane CA, Rosenbach M, Milligan J, Shei GJ, Chrebet G, Bergstrom J, Card D, Forrest M, Sun SY, West S, Xie H, Nomura N, Rosen H, Mandala S. Selecting against S1P3 enhances the acute cardiovascular tolerability of 3-(N-benzyl)aminopropylphosphonic acid S1P receptor agonists. Bioorg Med Chem Lett. 2004;14(13):3501–3505. doi: 10.1016/j.bmcl.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 20.Hale JJ, Doherty G, Toth L, Li Z, Mills SG, Hajdu R, Ann Keohane C, Rosenbach M, Milligan J, Shei GJ, Chrebet G, Bergstrom J, Card D, Rosen H, Mandala S. The discovery of 3-(N-alkyl)aminopropylphosphonic acids as potent S1P receptor agonists. Bioorg Med Chem Lett. 2004;14(13):3495–3499. doi: 10.1016/j.bmcl.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 21.Hale JJ, Neway W, Mills SG, Hajdu R, Ann Keohane C, Rosenbach M, Milligan J, Shei GJ, Chrebet G, Bergstrom J, Card D, Koo GC, Koprak SL, Jackson JJ, Rosen H, Mandala S. Potent S1P receptor agonists replicate the pharmacologic actions of the novel immune modulator FTY720. Bioorg Med Chem Lett. 2004;14(12):3351–3355. doi: 10.1016/j.bmcl.2004.02.106. [DOI] [PubMed] [Google Scholar]
- 22.Foss FW, Jr, Clemens JJ, Davis MD, Snyder AH, Zigler MA, Lynch KR, Macdonald TL. Synthesis, stability, and implications of phosphothioate agonists of sphingosine-1-phosphate receptors. Bioorg Med Chem Lett. 2005;15(20):4470–4474. doi: 10.1016/j.bmcl.2005.07.057. [DOI] [PubMed] [Google Scholar]
- 23.Clemens JJ, Davis MD, Lynch KR, Macdonald TL. Synthesis of 4(5)-phenylimidazole-based analogues of sphingosine-1-phosphate and FTY720: discovery of potent S1P1 receptor agonists. Bioorg Med Chem Lett. 2005;15(15):3568–3572. doi: 10.1016/j.bmcl.2005.05.097. [DOI] [PubMed] [Google Scholar]
- 24.Clemens JJ, Davis MD, Lynch KR, Macdonald TL. Synthesis of para-alkyl aryl amide analogues of sphingosine-1-phosphate: discovery of potent S1P receptor agonists. Bioorg Med Chem Lett. 2003;13(20):3401–3404. doi: 10.1016/s0960-894x(03)00812-6. [DOI] [PubMed] [Google Scholar]
- 25.Lefkowitz RJ, Cotecchia S, Samama P, Costa T. Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol Sci. 1993;14(8):303–307. doi: 10.1016/0165-6147(93)90048-O. [DOI] [PubMed] [Google Scholar]
- 26.Samama P, Pei G, Costa T, Cotecchia S, Lefkowitz RJ. Negative antagonists promote an inactive conformation of the beta 2-adrenergic receptor. Mol Pharmacol. 1994;45(3):390–394. [PubMed] [Google Scholar]
- 27.Jo E, Sanna MG, Gonzalez-Cabrera PJ, Thangada S, Tigyi G, Osborne DA, Hla T, Parrill AL, Rosen H. S1P1-selective in vivo-active agonists from high-throughput screening: off-the-shelf chemical probes of receptor interactions, signaling, and fate. Chem Biol. 2005;12(6):703–715. doi: 10.1016/j.chembiol.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 28.Wang DA, Lorincz Z, Bautista DL, Liliom K, Tigyi G, Parrill AL. A single amino acid determines lysophospholipid specificity of the S1P1 (EDG1) and LPA1 (EDG2) phospholipid growth factor receptors. J Biol Chem. 2001;276(52):49213–49220. doi: 10.1074/jbc.M107301200. [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara Y, Sardar V, Tokumura A, Baker D, Murakami-Murofushi K, Parrill A, Tigyi G. Identification of residues responsible for ligand recognition and regioisomeric selectivity of lysophosphatidic acid receptors expressed in mammalian cells. J Biol Chem. 2005;280(41):35038–35050. doi: 10.1074/jbc.M504351200. [DOI] [PubMed] [Google Scholar]
- 30.Inagaki Y, Pham TT, Fujiwara Y, Kohno T, Osborne DA, Igarashi Y, Tigyi G, Parrill AL. Sphingosine 1-phosphate analogue recognition and selectivity at S1P4 within the endothelial differentiation gene family of receptors. Biochem J. 2005;389(Pt 1):187–195. doi: 10.1042/BJ20050046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Virag T, Elrod DB, Liliom K, Sardar VM, Parrill AL, Yokoyama K, Durgam G, Deng W, Miller DD, Tigyi G. Fatty alcohol phosphates are subtype-selective agonists and antagonists of lysophosphatidic acid receptors. Mol Pharmacol. 2003;63(5):1032–1042. doi: 10.1124/mol.63.5.1032. [DOI] [PubMed] [Google Scholar]
- 32.Sardar VM, Bautista DL, Fischer DJ, Yokoyama K, Nusser N, Virag T, Wang DA, Baker DL, Tigyi G, Parrill AL. Molecular basis for lysophosphatidic acid receptor antagonist selectivity. Biochim Biophys Acta. 2002;1582(1–3):309–317. doi: 10.1016/s1388-1981(02)00185-3. [DOI] [PubMed] [Google Scholar]
- 33.Parrill AL, Wang D, Bautista DL, Van Brocklyn JR, Lorincz Z, Fischer DJ, Baker DL, Liliom K, Spiegel S, Tigyi G. Identification of Edg1 receptor residues that recognize sphingosine 1-phosphate. J Biol Chem. 2000;275(50):39379–39384. doi: 10.1074/jbc.M007680200. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Jensen JH. Improving the Efficiency and Convergence of Geometry Optimization with the Polarizable Continuum Model: New Energy Gradients and Molecular Surface Tessellation. Journal of Computational Chemistry. 2004;25(12):1449–1462. doi: 10.1002/jcc.20072. [DOI] [PubMed] [Google Scholar]
- 35.Tanford C, Kirkwood JG. Theory of Protein Titration Curves. I. General Equations for Impenentrable Spheres. J Am Chem Soc. 1957;79(20):5333–5339. [Google Scholar]
- 36.Shire SJ, Hanania GIH, Gurd FRN. Electrostatic Effects in Myoglobin. Hydrogen Ion Equilibria in Sperm Whale Ferrimyoglobin. Biochemistry. 1974;13(14):2967–2974. doi: 10.1021/bi00711a028. [DOI] [PubMed] [Google Scholar]
- 37.Warshel A. Calculations of Enzymatic Reactions: Calculations of pKa, Proton Transfer Reactions, and General Acid Catalysis Reactions in Enzymes. Biochemistry. 1981;20(11):3167–3177. doi: 10.1021/bi00514a028. [DOI] [PubMed] [Google Scholar]
- 38.Bashford D, Karplus M. pKa’s of Ionizable Groups in Proteins: Atomic Details from a Continuum Electrostatic Model. Biochemistry. 1990;29(44):10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 39.Robitaille PL, Robitaille PA, Brown GGJ, Brown GG. An analysis of the pH-dependent chemical-shift behavior of phosphorus-contaning metabolites. Journal of Magnetic Resonance. 1991;92:73–84. [Google Scholar]
- 40.Barone V, Cossi M. Quantum Calculation of Molecular Energies and Enrgy Gradients in Solution by a Conductor Solvent Model. Journal of Physical Chemistry A. 1998;102(11):1995–2001. [Google Scholar]
- 41.Cossi M, Rega N, Scalmani G, Barone V. Energies, Structures, and Electronic Properties of Molecules in Solution With the C-PCM. Journal of Computational Chemistry. 2003;24(6):669–681. doi: 10.1002/jcc.10189. [DOI] [PubMed] [Google Scholar]
- 42.Cances E, Mennucci B, Tomasi J. A new integral equation formalism for the polarizable continuum model: Theoretical background and applications to isotropic and anisotropic dielectrics. Journal of Chemical Physics. 1997;107(8):3032–3041. [Google Scholar]
- 43.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su SJ, Windus TL, Dupuis M, Montgomery JA. General Atomic and Molecular Electronic-Structure System. Journal of Computational Chemistry. 1993;14(11):1347–1363. [Google Scholar]
- 44.Barone V, Cossi M, Tomasi J. A new definition of cavities for the computation of solvation free energies by the polarizable continuum model. Journal of Chemical Physics. 1997;107(8):3210–3221. [Google Scholar]
- 45.Gaussian 98 RAMJ, Frisch GWT, Schlegel HB, Scuseria GE, Robb MA, Zakrzewski JRCVG, Montgomery JA, Jr, Stratmann RE, Dapprich JCBS, Millam JM, Daniels AD, Strain KNKMC, Farkas O, Tomasi J, Barone V, Cammi MCR, Mennucci B, Pomelli C, Adamo C, Clifford S, Petersson JOGA, Ayala PY, Cui Q, Morokuma K, Rabuck DKMAD, Raghavachari K, Foresman JB, Ortiz JCJV, Stefanov BB, Liu G, Liashenko A, Komaromi PPI, Gomperts R, Martin RL, Fox DJ, Al-Laham TKMA, Peng CY, Nanayakkara A, Gonzalez C, Gill MCPMW, Johnson B, Chen W, Wong MW, Gonzalez JLAC, Head-Gordon M, Replogle ES. aJAP. Gaussian I; Pittsburgh PA: 1998. [Google Scholar]
- 46.Becke A. Density-Functional Thermochemistry.3. The Role of Exact Exchange. Journal of Chemical Physics. 1993;98(7):5648–5652. [Google Scholar]
- 47.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Physical Review B Condensed Matter. 1988;37(2):785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 48.Parrill AL, Wang D-A, Bautista DL, Van Brocklyn JR, Lorincz Z, Fischer DJ, Baker DL, Liliom K, Spiegel S, Tigyi G. Identification of Edg1 Receptor Residues that Recognize Sphingosine 1- Phosphate. J Biol Chem. 2000;275(50):39379–39384. doi: 10.1074/jbc.M007680200. [DOI] [PubMed] [Google Scholar]
- 49.Inagaki Y, Pham TT, Fujiwara Y, Kohno T, Osborne DA, Igarashi Y, Tigyi G, Parrill AL. Sphingosine-1-Phosphate Analog Recognition and Selectivity at S1P4 within the Endothelial Differentiation Gene Family of Receptors. Biochem J. 2005;389:187–195. doi: 10.1042/BJ20050046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujiwara Y, Osborne DA, Walker MD, Wang DA, Bautista DA, Liliom K, Van Brocklyn JR, Parrill AL, Tigyi G. Identification of the hydrophobic ligand binding pocket of the S1P1 receptor. J Biol Chem. 2006 doi: 10.1074/jbc.M609648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halgren TA. Merch Molecular Force Field. I. Basis, Form, Scope, Parameterization, and Preformance of MMFF94. Journal of Computational Chemistry. 1996;17(5 6):490–519. [Google Scholar]
- 52.Naor MM, Jensen JH. Determinants of cysteine pKa values in creatine kinase and alpha1-antitrypsin. Proteins. 2004;57(4):799–803. doi: 10.1002/prot.20261. [DOI] [PubMed] [Google Scholar]
- 53.McQuarrie D. Statistical Thermodynamics. Mill Valley, CA: University Science Books; 1973. [Google Scholar]
- 54.Wang D, Lorincz Z, Bautista DL, Liliom K, Tigyi G, Parrill AL. A Single Amino Acid Determines Ligand Specificity of the S1P1 (EDG1) and LPA1 (EDG2) Phospholipid Growth Factor Receptors. J Biol Chem. 2001;276(52):49213–49220. doi: 10.1074/jbc.M107301200. [DOI] [PubMed] [Google Scholar]
- 55.Tsukahara T, Tsukahara R, Yasuda S, Makarova N, Valentine WJ, Allison P, Yuan H, Baker DL, Li Z, Bittman R, Parrill A, Tigyi G. Different residues mediate recognition of 1-O-oleyl-lysophosphatidic acid and rosiglitazone in the ligand binding domain of PPAR1. J Biol Chem. 2005 doi: 10.1074/jbc.M510843200. [DOI] [PubMed] [Google Scholar]
- 56.Brinkmann V, Davis MD, Heise CE, Albert R, Cottens S, Hof R, Bruns C, Prieschl E, Baumruker T, Hiestand P, Foster CA, Zollinger M, Lynch KR. The Immune modulator, FTY720, Targets Sphingosine 1-Phosphate Receptors. J Biol Chem. 2002;277(24):21453–21457. doi: 10.1074/jbc.C200176200. [DOI] [PubMed] [Google Scholar]
- 57.Hla T. Signaling and biological actions of sphingosine 1-phosphate. Pharmacol Res. 2003;47(5):401–407. doi: 10.1016/s1043-6618(03)00046-x. [DOI] [PubMed] [Google Scholar]
- 58.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427(6972):355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 59.English D, Kovala AT, Welch Z, Harvey KA, Siddiqui RA, Brindley DN, Garcia JGN. Induction of Endothelial Cell Chemotaxis by Sphingosine 1-Phosphate and Stabilization of Endothelial Monolayer Barrier Function by Lysophosphatidic Acid, Potential Mediators of Hematopoietic Angiogenesis. J Hematother Stem Cell Res. 1999;8:627–634. doi: 10.1089/152581699319795. [DOI] [PubMed] [Google Scholar]
- 60.Licht T, Tsirulnikov L, Reuveni H, Yarnitzky T, Ben-Sasson SA. Induction of Pro-angiogenic Signaling by a Synthetic Peptide Derived from the 2nd Intracellular Loop of S1P3 (EDG3) Blood. 2003;102(6):2099–2107. doi: 10.1182/blood-2002-12-3634. [DOI] [PubMed] [Google Scholar]
- 61.Chae SS, Paik JH, Furneaux H, Hla T. Requirement for sphingosine 1-phosphate receptor-1 in tumor angiogenesis demonstrated by in vivo RNA interference. J Clin Invest. 2004;114(8):1082–1089. doi: 10.1172/JCI22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osborne N, Stainier DY. Lipid receptors in cardiovascular development. Annu Rev Physiol. 2003;65:23–43. doi: 10.1146/annurev.physiol.65.092101.142235. [DOI] [PubMed] [Google Scholar]
- 63.Kohno T, Matsuyuki H, Inagaki Y, Igarashi Y. Sphingosine 1-phosphate Promotes Cell Migration Through the Activation of Cdc42 in Edg-6/S1P4-expressing Cells. Genes to Cells. 2003;8:685–697. doi: 10.1046/j.1365-2443.2003.00667.x. [DOI] [PubMed] [Google Scholar]
- 64.Graler MH, Grosse R, Kusch A, Kremmer E, Gudermann T, Lipp M. The sphingosine 1-phosphate receptor S1P4 regulates cell shape and motility via coupling to Gi and G12/13. J Cell Biochem. 2003;89(3):507–519. doi: 10.1002/jcb.10537. [DOI] [PubMed] [Google Scholar]
- 65.Osada M, Yatomi Y, Ohmori T, Ikeda H, Ozaki Y. Enhancement of sphingosine 1-phosphate-induced migration of vascular endothelial cells and smooth muscle cells by an EDG-5 antagonist. Biochem Biophys Res Commun. 2002;299(3):483–487. doi: 10.1016/s0006-291x(02)02671-2. [DOI] [PubMed] [Google Scholar]
- 66.Arikawa K, Takuwa N, Yamaguchi H, Sugimoto N, Kitayama J, Nagawa H, Takehara K, Takuwa Y. Ligand-dependent inhibition of B16 melanoma cell migration and invasion via endogenous S1P2 G protein-coupled receptor. Requirement of inhibition of cellular RAC activity. J Biol Chem. 2003;278(35):32841–32851. doi: 10.1074/jbc.M305024200. [DOI] [PubMed] [Google Scholar]
- 67.Rosenfeldt HM, Hobson JP, Macyka M, Olivera A, Nava VE, Milstien S, Spiegel S. EDG-1 Links the PDGF Receptor to Src and Focal Adnesion Kinase Activation Leading to Lamellipodia Formation and Cell Migration. FASEB J. 2001;15:2649–2659. doi: 10.1096/fj.01-0523com. [DOI] [PubMed] [Google Scholar]
- 68.Durgam GG, Tsukahara R, Makarova N, Walker MD, Fujiwara Y, Pigg KR, Baker DL, Sardar VM, Parrill AL, Tigyi G, Miller DD. Synthesis and Pharmacological Evaluation of Second-Generation Phosphatidic Acid Derivatives as Lysophosphatidic Acid Receptor Ligands. Bioorg Med Chem Lett. 2006;16(3):633–640. doi: 10.1016/j.bmcl.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 69.Jo E, Sanna MG, Gonzalez-Cabrera PJ, Thangada S, Tigyi G, Osborne DA, Hla T, Parrill AL, Rosen H. S1P1-selective in vivo-active agonists from high throughput screening: Off-the-shelf chemical probes of receptor interactions, signaling and fate. Chemistry & Biology. 2005;12:703–715. doi: 10.1016/j.chembiol.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 70.Holdsworth G, Osborne DA, Pham TT, Fells JI, Hutchinson G, Milligan G, Parrill AL. A single amino acid determines preference between phospholipids and reveals length restriction for activation of the S1P4 receptor. BMC Biochem. 2004;5(1):12. doi: 10.1186/1471-2091-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Virag T, Elrod DB, Liliom K, Sardar VM, Parrill AL, Yokoyama K, Durgam G, Deng W, Miller DD, Tigyi G. Fatty Alcohol Phosphates are Subtype-Selective Agonists and Antagonists of LPA Receptors. Mol Pharmacol. 2003;63(5):1032–1042. doi: 10.1124/mol.63.5.1032. [DOI] [PubMed] [Google Scholar]
- 72.Sardar VM, Bautista DL, Fischer DJ, Yokoyama K, Nusser N, Virag T, Wang D, Baker DL, Tigyi G, Parrill AL. Molecular Basis for Lysophosphatidic Acid Receptor Antagonist Selectivity. Biochim Biophys Acta. 2002;1582(1–3):309–317. doi: 10.1016/s1388-1981(02)00185-3. [DOI] [PubMed] [Google Scholar]
- 73.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268(5214):1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 74.Yang P, Murthy PP, Brown RE. Synergy of intramolecular hydrogen-bonding network in myo-inositol 2-monophosphate: theoretical investigations into the electronic structure, proton transfer, and pKa. J Am Chem Soc. 2005;127(45):15848–15861. doi: 10.1021/ja053371u. [DOI] [PubMed] [Google Scholar]
- 75.Kooijman EE, Carter KM, van Laar EG, Chupin V, Burger KN, de Kruijff B. What Makes the Bioactive Lipids Phosphatidic Acid and Lysophosphatidic Acid So Special? Biochemistry. 2005;44(51):17007–17015. doi: 10.1021/bi0518794. [DOI] [PubMed] [Google Scholar]
- 76.Lim HS, Oh YS, Suh PG, Chung SK. Syntheses of sphingosine-1-phosphate stereoisomers and analogues and their interaction with EDG receptors. Bioorg Med Chem Lett. 2003;13(2):237–240. doi: 10.1016/s0960-894x(02)00893-4. [DOI] [PubMed] [Google Scholar]
- 77.Lim HS, Park JJ, Ko K, Lee MH, Chung SK. Syntheses of sphingosine-1-phosphate analogues and their interaction with EDG/S1P receptors. Bioorg Med Chem Lett. 2004;14(10):2499–2503. doi: 10.1016/j.bmcl.2004.03.001. [DOI] [PubMed] [Google Scholar]


