Abstract
The Ets family transcription factor PU.1, encoded by the gene Sfpi1, is essential for normal hematopoiesis. A number of studies have suggested that changes in PU.1 concentration play a role in directing cell fate decisions during hematopoiesis. However, the stages of hematopoietic development at which changes in PU.1 concentration are important have not been defined until recently. Experiments using conditional null alleles, reporter alleles, and hypomorphic alleles of the Sfpi1 gene in mice demonstrate that PU.1 concentration is uniformly high during early stages of hematopoietic development. However, reduction of PU.1 concentration is required for normal development of megakaryocyte-erythroid progenitors, B cell progenitors, and T cell progenitors. PU.1 concentration increases in granulocyte-macrophage progenitors. Furthermore, experimental reduction of PU.1 concentration in the myeloid lineages leads to failed differentiation, abnormal proliferation, and leukemia. In this review, we synthesize recent studies to develop a new model of PU.1 function in hematopoiesis.
Introduction
The transcription factor PU.1 was first described in 1988 as a gene dysregulated by proviral insertion of the spleen focus forming virus (SFFV) in murine erythroleukemia [1]. This gene, called Sfpi1 in mice and Spi-1 in humans, was found to encode a transcription factor of the Ets family [2]. PU.1 is highly expressed in the immune system, and regulates many genes in cell types such as macrophages and B cells. The importance of PU.1 in hematopoiesis is underscored by the observation that null mutation of the Sfpi1 gene in mice results in fetal or perinatal lethality and multiple defects in blood cell development [3,4]. While PU.1 is not essential for erythrocyte or megakaryocyte development, mice homozygous for a null allele of Sfpi1 fail to generate B lymphocytes, macrophages, or neutrophils. Transplantation and organ culture experiments demonstrate that PU.1 is also required for the normal development of T lymphocytes and natural killer cells [5,6]. Abnormal levels of PU.1 expression are associated with acute myeloid leukemia in human patients [7] as well as in mouse models [8,9,10]. These studies suggest that appropriate concentration of PU.1 protein is critically important for normal hematopoiesis.
Concentration-dependent function of PU.1 in hematopoietic cell fate determination
Soon after its discovery, PU.1 was recognized to be expressed at significantly higher levels in myeloid lineage cells than in B cells [11]. Subsequently, experiments using retroviral transduction of PU.1 cDNA into Sfpi1 null hematopoietic progenitors resulted in several striking observations. First, B cell progenitors (pro-B cells) generated in culture after infection of Sfpi1−/− fetal liver progenitors with PU.1 cDNA expressed exclusively low levels of PU.1 transcripts and protein. Second, macrophages generated in such cultures expressed exclusively high levels of PU.1 transcripts and protein. Finally, infection of Sfpi1+/− fetal liver progenitors with PU.1 cDNA efficiently blocked the generation of pro-B cells and instead appeared to divert differentiation of infected cells into macrophages [12]. These observations led to the hypothesis that low concentration of PU.1 might direct differentiation of hematopoietic stem cells (HSCs) to the lymphoid lineages, whereas high concentration of PU.1 might direct differentiation of HSCs to the myeloid lineages [12]. This hypothesis has recently been tested directly, using mice with inducible null alleles, reporter alleles, or hypomorphic alleles of the Sfpi1 gene. The results of these studies are described below.
Function of PU.1 during hematopoiesis revealed by studies using conditional null alleles
Two alleles of Sfpi1 have recently been described in which the PU.1 DNA binding domain can be conditionally deleted using Cre-mediated excision of loxP elements [13,14]. These model systems allow analysis of the consequences of deletion of Sfpi1 in purified progenitor cells by infection with viral vectors encoding Cre recombinase; deletion in committed B cells (by crossing to CD19-Cre knock-in mice), or deletion in HSCs (by crossing to Mx1-Cre transgenic mice). Experiments in which Sfpi1 is deleted in common lymphoid progenitors (CLPs) or in B cell progenitors (pro-B cells) suggest that PU.1 is not essential for the generation of B cells from committed CLPs. In fact, B cells lacking PU.1 can secrete immunoglobulin and participate in immune responses [14,15,16]. Experiments in which Sfpi1 is deleted in purified common myeloid progenitors (CMPs) or granulocyte-macrophage progenitors (GMPs) demonstrate that PU.1 is essential for the normal differentiation of macrophages and granulocytes at all stages tested [13,14]. Finally, deletion of Sfpi1 in adult HSCs results in the loss of phenotypically recognizable CLPs, CMPs, and B cell development [13,14]. However, in one model, deletion of Sfpi1 in adult HSCs resulted in an unexpected expansion of immature myeloid cells [13]. Taken together, these studies suggest that PU.1 is essential for specification of the earliest lymphoid progenitors, but not essential for the specification of myeloid progenitors from HSCs. PU.1 is not essential for generation of B cells from committed lymphoid progenitors, but is required for generation of macrophages and neutrophils from committed myeloid progenitors. These experiments should not be interpreted as showing that PU.1 has no function in B cells, since gene expression in Sfpi1−/− pro-B cells is profoundly different from wild type B cells [17].
Use of reporter alleles to measure levels of PU.1 during hematopoiesis
Changes in PU.1 concentration are important for differentiation of several hematopoietic lineages, but it has been unclear at what stage of development these changes occur. Recently, two laboratories independently reported generation and analysis of mice with reporter alleles of Sfpi1 [18,19]. In both cases, cDNA encoding green fluorescent protein (GFP) was inserted into the Sfpi1 locus by gene targeting, such that the pattern of GFP expression resembles PU.1 expression. These studies suggest that PU.1 is expressed at uniformly high levels in HSCs, CMPs, and CLPs. However, PU.1 expression is substantially upregulated in GMPs and substantially downregulated in megakaryocyte-erythrocyte progenitors (MEPs), B cell progenitors (pro-B cells), and T cell progenitors (pro-T cells). Therefore, these data are not compatible with the idea that changes in PU.1 concentration direct differentiation of HSCs into CMPs or CLPs. Instead, changes in PU.1 concentration are initiated prior to or during specification of MEPs, GMPs, pro-B cells, and pro-T cells.
Function of PU.1 during hematopoiesis revealed by studies using hypomorphic alleles
The most direct test of whether PU.1 concentration plays a role in lymphoid-myeloid cell fate decisions is to experimentally increase or decrease concentration in developing hematopoietic cells. PU.1 concentration can be experimentally increased by retroviral infection of wild type hematopoietic progenitors with PU.1 cDNA. The results from these types of experiments demonstrate that increased expression of PU.1 blocks development of erythrocytes, B cells, and T cells [12,20,21]. Forced expression of PU.1 can also lead to immortalization of erythroid lineage cells and eventually erythroleukemia [22,23]. Therefore, PU.1 concentration must be reduced for the proper development of these cell types.
One method to experimentally decrease PU.1 concentration is to modify the Sfpi1 gene to reduce levels of expression. Two groups have recently generated and analyzed alleles of Sfpi1 with altered levels of PU.1 expression [10,24]. Rosenbauer et al. [10] generated mice with a deletion of an upstream regulatory region (ΔURE) in the Sfpi1 gene. ΔURE mice express reduced levels of PU.1 in most tissues examined, except for T cells, which exhibit an increase in PU.1 levels [10,25]. Reduced PU.1 expression resulted in a block to B cell development, impaired myeloid differentiation, and acute myeloid leukemia [10,25].
More recently our own laboratory has generated a novel hypomorphic allele of Sfpi1 termed BN [24]. This allele was generated by insertion of the bacterial β-lactamase gene into exon 1 of the Sfpi1 gene, replacing two known translation start codons. Analysis of cells homozygous for the BN allele (Sfpi1BN/BN) revealed that the BN allele produces ~20% of wild type levels of PU.1 protein as a consequence of reduced transcription and translation initiation at a third start codon at position 32 of the protein. The BN allele generates a PU.1 protein lacking the N-terminal 31 amino acids. It is unclear if there is a biological function for these first 31 amino acids, since ΔN31 PU.1 protein has activity identical to full-length protein in experiments performed to date [24,26].
We have examined hematopoiesis in fetal and neonatal Sfpi1BN/BN mice to determine the consequences of reduced PU.1 concentration in vivo. Sfpi1BN/BN mice are runted and osteopetrotic, and all die by one month of age, suggesting a macrophage deficiency similar to that observed in one line of Sfpi1−/− mice [27]. B cells were not detectable in Sfpi1BN/BN fetal liver, bone marrow, or spleen at any day of development. Furthermore, Sfpi1BN/BN fetal liver progenitors could not reconstitute B cell development when transplanted into immunodeficient recipient mice. These data suggest that reduced concentration of PU.1 results in a failure to specify B cell progenitors in vivo. Interestingly, Sfpi1BN/BN mice have a thymus, although it is reduced in size and contains T cells with altered frequencies of CD4 and CD8-expressing cells (Figure 1). The development of T cells while B cell development is blocked raises interesting questions about what progenitor cell has seeded the thymus in these mice, and why development of this progenitor is not blocked by the Sfpi1BN/BN mutation.
Fig. 1.
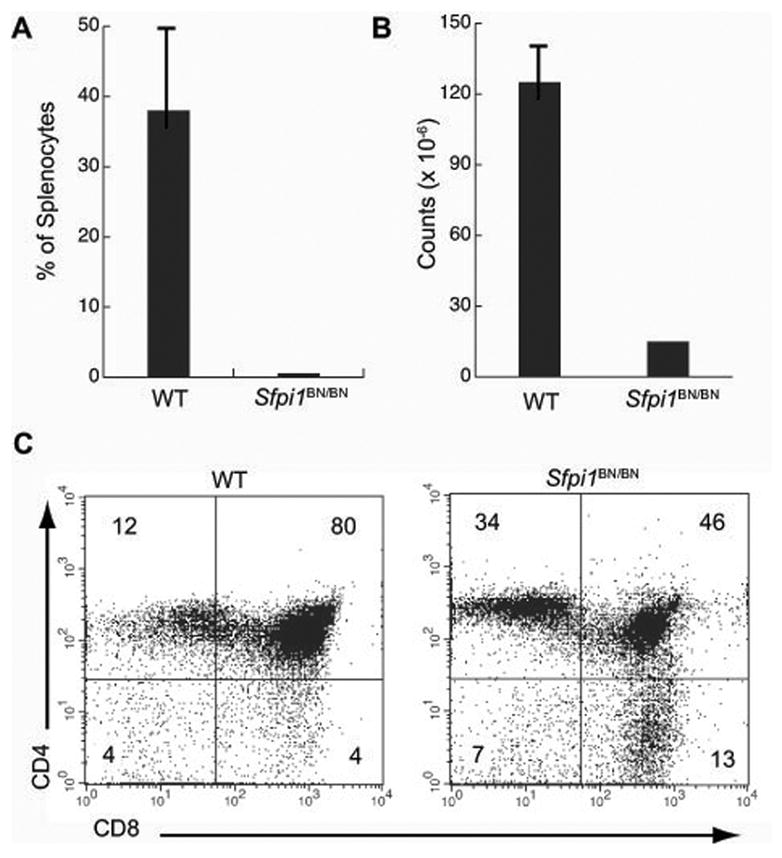
Lymphocyte development in mice homozygous for a hypomorphic allele of Sfpi1. A) Engraftment of wild type (WT) or Sfpi1BN/BN fetal liver progenitors into RAG2−/−il2rγ−/− immunodeficient mice. The percentage of B220+ CD19+ B cells in the spleen of recipient mice (n = 5) was measured by flow cytometry six weeks after i.v. injection of 2 × 106 fetal liver cells. B) Total cellularity of the thymus of wild type (WT) or Sfpi1BN/BN mice. C) Flow cytometric analysis of single-cell suspensions from thymus of either wild type (WT) or Sfpi1BN/BN mice at 10 days of age. Cells were gated for proper size and granularity and incubated with an antibody to the indicated cell surface markers. Numbers represent percentages of gated cells in each quadrant.
We also analyzed myeloid development in fetal and neonatal Sfpi1BN/BN mice. Cells expressing the myeloid-specific markers Gr-1, CD11b or FcγRII/III are undetectable in E14.5 Sfpi1BN/BN fetal liver. The frequency of myeloid colonies that can be generated from Sfpi1BN/BN fetal liver progenitors in response to cytokines such as granulocyte-macrophage colony-stimulating factor (GM-CSF), M-CSF, and G-CSF is dramatically reduced compared to wild type fetal liver progenitors. Unexpectedly, cells with myeloid cell surface markers appear in the bone marrow and spleen of Sfpi1BN/BN mice after birth. The number of these cells increases rapidly in the first week after birth, and by two weeks of age the spleens of Sfpi1BN/BN mice essentially contain only immature myeloid cells. These cells have an abnormal phenotype in which the immature cell marker c-Kit is co-expressed with myeloid cell-specific markers such as Gr-1 and CD11b. From these data, several major conclusions can be drawn. First, high PU.1 concentration is essential for the generation of B cells in vivo. This requirement is probably at the level of the generation of CLPs from HSCs [14]. Second, high PU.1 concentration is not essential for the generation of myeloid lineage cells in vivo, although such cells fail to differentiate normally. Third, the function of PU.1 in generation of myeloid lineage cells appears different between fetal and adult hematopoiesis. During fetal hematopoiesis, PU.1 seems to be essential for the generation of myeloid cells. However, after birth, PU.1 is not required for the generation of myeloid cells, and low concentration of PU.1 appears to stimulate proliferation of immature myeloid cells.
Role of reduced PU.1 concentration in myeloid leukemia
The results of reporter allele studies suggest that PU.1 concentration is uniformly high during differentiation of the myeloid lineages, and increases further during terminal differentiation [18,19]. Therefore, reductions in PU.1 concentration do not normally occur during myeloid development. If PU.1 concentration is experimentally reduced in myeloid cells, the consequences include abnormal proliferation and myeloid leukemia [10,24,28]. Interestingly, PU.1 expression and/or activity is frequently suppressed in myeloid leukemia by a variety of different mechanisms [reviewed in 29,30]. It will be important to determine whether there is a causal relationship between abnormal myeloid proliferation caused by low PU.1 concentration and neoplastic transformation, which presumably requires additional somatic mutations. Identification of the target genes responsible for abnormal myeloid proliferation when PU.1 concentration is reduced will be an important future area of investigation.
Conclusions: a revised model of PU.1 function in hematopoiesis
The results of studies using conditional null, reporter, and hypomorphic alleles of Sfpi1 provide important insights into the function of PU.1 in hematopoiesis. These studies show that changes in PU.1 concentration probably do not occur during differentiation of HSCs into CMPs or CLPs. However, PU.1 concentration is reduced during the generation of committed progenitor cells for erythrocytes, megakaryocytes, B cells, and T cells (Fig. 2). Reduction of PU.1 concentration from initially high levels is a requirement for megakaryocyte/erythroid and lymphoid differentiation. We speculate that an important function of PU.1 during hematopoiesis might be to repress erythroid and lymphoid cell fates. Identification of the target genes that mediate this cell fate repression will be an important future research direction.
Fig. 2.

A revised model for concentration-dependent function of PU.1 in hematopoiesis. The model of hematopoiesis shown is based on a revised model of hematopoiesis proposed by Adolfsson et al. [31]. PU.1 concentration is indicated by a gradient from red (high) to blue (low) in the top to bottom direction. The position of each cell type within this gradient indicates the relative concentration of PU.1. Mature megakaryocytes, erythrocytes, and T cells do not express PU.1. Abbreviations are as follows: HSC, hematopoietic stem cell; MPP, multipotential progenitor; LMPP, lymphoid-primed multipotential progenitor; CLP, common lymphoid progenitor; pro-B, progenitor B cell; B, mature B cell; pro-NK, progenitor natural killer cell; NK, mature natural killer cell; pro-T, progenitor T cell; T, mature T cell; MEP, megakaryocyte-erythroid progenitor; Meg, megakaryocyte; E, erythrocyte; GMP, granulocyte-macrophage progenitor; Mac, macrophage; Neut, neutrophil.
Acknowledgments
This article is based on a presentation at the Seventh International Workshop on Molecular Aspects of Myeloid Stem Cell Development and Leukemia in Annapolis, Maryland on May 13–16, 2007, sponsored by The Leukemia & Lymphoma Society. This work was supported by NIH grant AI052175.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–80. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 2.Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–24. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 3.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–7. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 4.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DG, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. The EMBO Journal. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 5.Spain LM, Guerriero A, Kunjibettu S, Scott EW. T cell development in PU.1-deficient mice. The Journal of Immunology. 1999;163:2681–2687. [PubMed] [Google Scholar]
- 6.Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97:2625–32. doi: 10.1182/blood.v97.9.2625. [DOI] [PubMed] [Google Scholar]
- 7.Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD, Behre G, Hiddemann W, Ito Y, Tenen DG. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100:998–1007. doi: 10.1182/blood.v100.3.998. [DOI] [PubMed] [Google Scholar]
- 8.Cook WD, McCaw BJ, Herring C, John DL, Foote SJ, Nutt SL, Adams JM. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood. 2004;104:3437–44. doi: 10.1182/blood-2004-06-2234. [DOI] [PubMed] [Google Scholar]
- 9.Walter MJ, Park JS, Ries RE, Lau SK, McLellan M, Jaeger S, Wilson RK, Mardis ER, Ley TJ. Reduced PU.1 expression causes myeloid progenitor expansion and increased leukemia penetrance in mice expressing PML-RARalpha. Proc Natl Acad Sci U S A. 2005;102:12513–8. doi: 10.1073/pnas.0504247102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–30. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 11.Ross IL, Dunn TL, Yue X, Roy S, Barnett CJK, Hume DA. Comparison of the expression and function of the transcription factor PU.1 (SPI-1 proto-oncogene) between murine macrophages and B lymphocytes. Oncogene. 1994;9:121–132. [PubMed] [Google Scholar]
- 12.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–41. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 13.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML, Elf S, Chan S, Kastner P, Huettner CS, Murray R, Tenen DG, Akashi K. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye M, Ermakova O, Graf T. PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J Exp Med. 2005;202:1411–22. doi: 10.1084/jem.20051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polli M, Dakic A, Light A, Wu L, Tarlinton DM, Nutt SL. The development of functional B lymphocytes in conditional PU.1 knock-out mice. Blood. 2005;106:2083–90. doi: 10.1182/blood-2005-01-0283. [DOI] [PubMed] [Google Scholar]
- 17.Schweitzer BL, DeKoter RP. Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J Immunol. 2004;172:144–54. doi: 10.4049/jimmunol.172.1.144. [DOI] [PubMed] [Google Scholar]
- 18.Back J, Allman D, Chan S, Kastner P. Visualizing PU.1 activity during hematopoiesis. Exp Hematol. 2005;33:395–402. doi: 10.1016/j.exphem.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–31. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuetze S, Stenberg PE, Kabat D. The Ets-related transcription factor PU.1 immortalizes erythroblasts. Molecular and Cellular Biology. 1993;13:5670–5678. doi: 10.1128/mcb.13.9.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–96. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 22.Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeaux M, Grimber G, Briand P, Vainchenker W, Tavitian A. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Molecular and Cellular Biology. 1996;16:2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houston IB, Huang KJ, Jennings SR, DeKoter RP. PU.1 immortalizes hematopoietic progenitors in a GM-CSF-dependent manner. Exp Hematol. 2007;35:374–384. doi: 10.1016/j.exphem.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Houston IB, Kamath MB, Schweitzer BL, Chlon TM, DeKoter RP. Reduction in PU.1 activity results in a block to B cell development, abnormal myeloid proliferation, and neonatal lethality. Experimental Hematology. 2007 doi: 10.1016/j.exphem.2007.04.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, Liu C, Hackanson B, Akashi K, Leutz A, Rothstein TL, Plass C, Tenen DG. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 26.Fisher RC, Olson MC, Pongubala JM, Perkel JM, Atchison ML, Scott EW, Simon MC. Normal myeloid development requires both the glutamine-rich transactivation domain and the PEST region of transcription factor PU.1 but not the potent acidic transactivation domain. Mol Cell Biol. 1998;18:4347–57. doi: 10.1128/mcb.18.7.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tondravi MM, McKercher SR, Anderson K, Erdmann JM, Quiroz M, Maki R, Teitelbaum SL. Osteopetrosis in mice lacking hematopoietic transcription factor PU.1. Nature. 1997;386:81–84. doi: 10.1038/386081a0. [DOI] [PubMed] [Google Scholar]
- 28.Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, Klippel S, Steidl C, Bruns I, Costa DB, Wagner K, Aivado M, Kobbe G, Valk PJ, Passegue E, Libermann TA, Delwel R, Tenen DG. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet. 2006;38:1269–77. doi: 10.1038/ng1898. [DOI] [PubMed] [Google Scholar]
- 29.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–17. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 30.Dahl R, Simon MC. The importance of PU.1 concentration in hematopoietic lineage commitment and maturation. Blood Cells Mol Dis. 2003;31:229–33. doi: 10.1016/s1079-9796(03)00152-9. [DOI] [PubMed] [Google Scholar]
- 31.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]


