Abstract
The substrate specificity of aspartate aminotransferase was successfully modified by directed molecular evolution using a combination of DNA shuffling and selection in an auxotrophic Escherichia coli strain. After five rounds of selection, one of the evolved mutants showed a 105-fold increase in the catalytic efficiency (kcat/Km) for β-branched amino and 2-oxo acids and a 30-fold decrease in that for the native substrates compared with the wild-type enzyme. The mutant had 13 amino acid substitutions, 6 of which contributed 80–90% to the total effect. Five of these six substitutions were conserved among the five mutants that showed the highest activity for β-branched substrates. Interestingly, only one of the six functionally important residues is located within a distance of direct interaction with the substrate, supporting the idea that rational design of the substrate specificity of an enzyme is very difficult. The present results show that directed molecular evolution is a powerful technique for enzyme redesign if an adequate selection system is applied.
To alter the substrate specificity of an existing enzyme is worth trying because it provides not only efficient catalysts with designed substrate specificity but also valuable information on the mechanism of substrate recognition. Despite numerous studies to engineer enzymes over the past decade, only a few of them have reported ∼103-fold enhancements in the catalytic efficiency for the nonnative substrates (1–3). Recently, strategies based on directed molecular evolution have created macromolecules with novel properties (4–10). A method called DNA shuffling has proved promising as a technique to introduce random mutations and recombinations among a population of mutant genes (11–14). A major obstacle that restricts the utility of these methods in redesigning enzymes is the technical difficulty of establishing a rapid and sensitive system to screen >104 mutant enzymes for the desired activity. Chromogenic substrates were successfully used for a screening system utilizing 96-well plates and a plate reader (6) or a visual screening of the colored colonies (14). These substrates are, however, available only for a limited range of reactions. An ideal screening system should be a convenient and reliable one, of which the sensitivity can be adjusted depending on the progress of the selection steps. One possible system is to select mutant enzymes that can reverse the phenotype of a bacterial strain that is deficient in an enzyme with the desired activity. The sensitivity or stringency of the selection can be adjusted by supplementing the medium with an adequate amount of the substrate or product.
Aminotransferases catalyze amino group transfer between amino acids and 2-oxo acids and play central roles in amino acid metabolism in organisms ranging from bacteria to mammals. The structure (15, 16) and reaction mechanism (17) of aspartate 2-oxoglutarate aminotransferase (AspAT) have been studied extensively. Among the natural amino acids, AspAT from Escherichia coli shows the highest activity for acidic substrates and moderate activity for aromatic substrates (18). The activity for β-branched amino acids is barely detectable and is even lower than that for basic amino acids. Initial attempts to increase the activity for basic or aromatic substrates by site-directed mutagenesis resulted in limited success (19, 20). Recently, the catalytic efficiency for aromatic substrates was increased 1500-fold by six residue substitutions, which were chosen based on the crystal structure of AspAT and the sequence comparison between AspAT and tyrosine aminotransferase (3).
In the present study, we established a selection system where the growth of an auxotrophic E. coli strain is assisted by mutant AspATs with the activity for branched-chain substrates. AspAT was then evolved toward higher activity for β-branched substrates through “Darwinian” selection of the plasmid-encoded mutant E. coli AspATs.
MATERIALS AND METHODS
Construction of a ΔilvE∷kan Strain.
The gene for branched-chain amino acid aminotransferase, ilvE (21), was knocked out as follows. A genomic region of the ilv operon (22) was amplified from the chromosomal DNA of JM103 by LA PCR (TaKaRa Biomedicals) with two primers: 5′-TACGGGATCCTATTGAAATTATTAAACGCATCATAAAAATCGGCC-3′ and 5′-CGTAGGATCCGGAGCACCGGACAGGGGTTGCGAGTCAGCCAT-3′. The amplified 5-kb fragment contained the ilv LGED genes, and BamHI sites were incorporated at the 5′ and 3′ ends. After the fragment was cloned into an AccI site-free pUC18 at the BamHI site, the plasmid was digested with AccI to remove 86% of the ilvE coding region including the NH2 terminus. The larger fragment was agarose gel purified and ligated with the AccI-digested kanamycin resistance (kan) gene (Pharmacia). The resultant plasmid was BamHI digested, and the fragment comprising the kan gene flanked by ilvLG and ilvD was gel purified. The fragment was used to transform a hyper-recombination strain JC7623 (recB, recC, sbcB) by homologous recombination, and the transformants were screened on an LB plate containing 50 μg/ml kanamycin. Kanamycin-resistant colonies were picked up, and the genotype was verified by restriction analysis of the ilv operon amplified by PCR from the chromosomal DNA. The strain, a derivative of the hyper-recombination strain, cannot be used for the present purpose. Thus, the ilvE-deficient ilv operon was transferred to JM103 by phage P1-mediated transduction. The resultant strain, YJ103, was kanamycin resistant and showed the expected auxotrophy except for proline requirement. Proline had to be supplemented to the minimal medium in addition to branched-chain amino acids. This is probably because the F′ plasmid was lost during the construction. YJ103 is recA+, but the plasmids used for this study were stable in this strain.
Screening for Higher Transamination Activity for 2-Oxovaline.
DNA shuffling was done as described by Stemmer (11, 12). 100–300-bp fragments were recovered after DNaseI digestion and reassembled by PCR without primers; the rate of point mutation per one DNA shuffling was 0.8%. For the first round of selection, the 1.9-kb HindIII–EcoRI fragment (23) containing the wild-type aspC gene and its cognate promoter was shuffled to introduce point mutations. The shuffled fragment was ligated with the HindIII- and EcoRI-digested pBR322, and YJ103 was transformed with the ligated DNA by electroporation. A library of transformants (>90% ligation efficiency and an effective library size of 1.0 × 106 colonies) was spread on eight 15-cm plates. The medium contained the standard Davis salts supplemented with 5 μg/ml each of thiamin and pyridoxine, 10 μg/ml each of five nucleic acid bases, and 20 μg/ml each of proline, isoleucine, leucine, and 2-ketoisovalerate (2-oxovaline) (ILkV plate). After a 40-h incubation period at 37°C, 94 colonies were picked up and spotted on an LB plate containing 50 μg/ml ampicillin. After an overnight incubation, the cells were scraped from the plate with sterile water, and a mixture of the plasmid (pAV1 mix) was prepared. For the next round, the same 1.9-kb region was PCR amplified from pAV1 mix. The fragments were shuffled to recombine the genes among the mutants concomitantly introducing point mutations and ligated with pBR322 as described above. The second round of selection was done as follows: 4.2 × 106 colonies, ILkV plates, and a 27-h incubation period. Ninety-six colonies were picked up, pAV2 mix was prepared, and the 1.9-kb region was shuffled. The third round of selection was as follows: 4.5 × 106 colonies, 2-oxovaline was omitted from the medium (IL plate), and a 28-h incubation period. pAV3 mix was prepared from 99 colonies, and a 1.3-kb region, which contains only the coding region, was PCR amplified with 5′ and 3′ primers, which have BamHI and SalI sites, respectively. The promoter-less genes were shuffled and ligated with the BamHI–SalI fragment of pBR322, which puts the gene under the promoter of the tetracycline resistance (Tetr) gene of pBR322 and decreases the expression level of the aspC gene by about 20-fold. The fourth round of selection was 3.1 × 107 colonies, IL plates, and a 28-hr incubation period. pAV4 mix was prepared from 92 colonies, and the 1.3-kb region was PCR amplified and shuffled. The fifth round of selection was done in two different ways, AV5A and AV5B selections. The AV5A selection was the same plasmid construction as that used for the fourth round, 3.5 × 107 colonies, and IL plates. The colonies, however, did not grow much faster than those of the fourth round of selection. After a 28-hr incubation period, 98 large colonies were picked up anyway, and pAV5A mix was prepared. For AV5B selection, the −10 region of the Tetr promoter of pBR322 was mutated, TTTAAT to TTGAAT, and −10 region of the anti-Tetr promoter (24) was also mutated, TAAACT to CGAACT. The expression of the aspC gene put downstream of this mutant Tetr promoter was decreased by 10-fold compared with that from the native Tetr promoter. The AV5B selection was 2.4 × 107 colonies, IL plates, and a 42-hr incubation period. pAV5B mix was prepared from 112 colonies. The primers used for PCR in DNA shuffling were 5′-AACGCATTAAAGCTTGCATGCCTGCAG-3′ and 5′-TCGTCTTCAAGAATTCCCCTGATAAGC-3′ for the first to third rounds of selection; 5′-GACTGTACGGGATCCGTAACCATAATGGAACCTCGTCATG-3′ and 5′-GACTGTACGGTCGACCCCTGATAAGCGTAGCCATCAGGCA-3′ were used for the fourth and fifth rounds of selection.
Activity Measurements with Crude Lysates.
The lysates of YJ103 cells carrying the plasmids were added to the reaction mixtures containing 50 mM aspartate, 10 mM 2-oxovaline, 0.5 units of malate dehydrogenase (Boehringer Mannheim), 0.1 mM NADH, and 10 μM pyridoxal 5′-phosphate. The buffer system was 50 mM Hepes, pH 8.0, containing 0.1 M KCl and 10 μM EDTA. Aspartate was chosen as the amino group donor in this assay because it was easy to monitor the reactions by coupling malate dehydrogenase. The reactions were monitored at 25°C by following the decrease in absorption at 340 nm. Steady-state transamination activity between aspartate and 2-oxovaline, undetectable with the wild-type AspAT, was observed in the lysates containing evolved mutants.
Expression and Purification of Mutant AspATs.
The coding region of the mutant AspATs was subcloned into pUC vectors. The mutant enzymes were expressed in E. coli TY103 (25), which is defective in the AspAT gene, and purified as described (26).
Activity Measurements with Purified AspATs.
Activity for each substrate was measured at 25°C by spectrophotometrically monitoring the single-turnover reaction using an Applied Photophysics stopped-flow apparatus (model SX.17MV) as described (18). The dead time of this system was 2.0 ms under a pressure of 500 kPa. The reactions of the wild-type AspAT with valine, isoleucine, and their 2-oxo acids were slow and thus followed by a conventional spectrophotometer. The buffer system was 50 mM Hepes, pH 8.0, containing 0.1 M KCl and 10 μM EDTA.
RESULTS AND DISCUSSION
Strategy for the Selection.
Our selection system is based on the auxotrophy of an E. coli strain that is deficient in the branched-chain amino acid aminotransferase gene, ilvE. Branched-chain amino acid aminotransferase catalyzes the last step of branched-chain amino acid biosynthesis, and thus the ilvE-deficient strain cannot grow on a minimal plate without the supplement of valine, isoleucine, and leucine. If the efficiency with which a plasmid-encoded mutant AspAT synthesizes branched-chain amino acids is increased, the deficient strain carrying the plasmid should be able to grow on a minimal plate. To establish a reliable selection system, the NH2-terminal 86% of the chromosomal ilvE gene of E. coli JM103 was replaced with the kanamycin resistance gene. The resultant strain, YJ103 (ΔilvE∷kan), showed the expected auxotrophy and grew on a minimal plate when transformed with a plasmid containing the ilvE gene.
Evolution of Transamination Activity for 2-Oxovaline.
The stringency of the selection was increased during the successive rounds of selection; 2-oxovaline was omitted from the medium, the incubation time was shortened, and the expression level of mutant AspATs was decreased by manipulating the construction of the plasmid. Isoleucine and leucine were supplemented throughout the selections because they were expected to serve as amino group donors to 2-oxovaline. At each step, a library of 106–107 transformants was spread on plates, 90–100 of the largest colonies were picked, and a mixture of the plasmids from these colonies was prepared for subsequent DNA shuffling. This selection system directs the evolution of AspAT to higher transamination activity for 2-oxovaline. After the fifth round of selection, YJ103 was retransformed by pAV5A mix and pAV5B mix and spread on IL plates. Twelve large colonies were picked up from each plate, cultured in LB + ampicillin and the plasmids, pAV5A-1–12 and pAV5B-1–12, were prepared. The transamination activity between aspartate and 2-oxovaline was measured with the lysate from each culture, and AV5A-1, AV5A-7, AV5B-1, AV5B-4, and AV5B-5, which showed the highest activity among the 24 clones, were picked up for further analysis.
Sequence Analysis of Mutant AspATs.
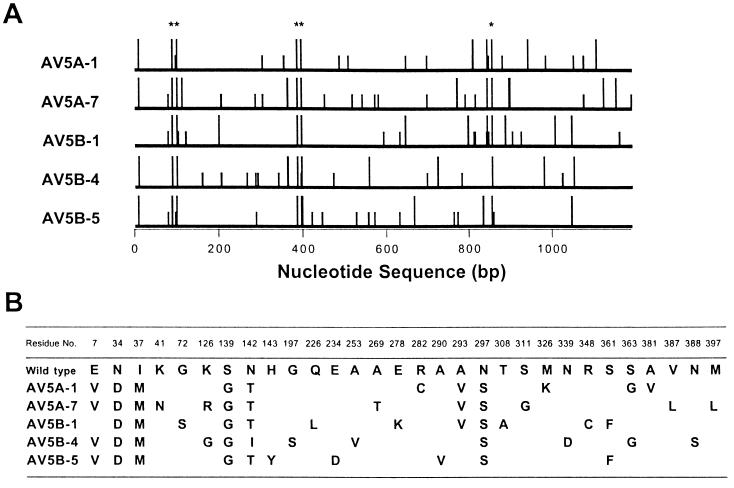
The genes encoding the five clones exhibiting the highest activity for 2-oxovaline were sequenced. Each mutant contained 23–28 point mutations evenly distributed over the coding region, 10–13 of which were non-silent (Fig. 1A). No insertion or deletion mutations were found. Among 30 amino acid substitutions, only five substitutions were “conserved” in all five mutant AspATs, although Asn142 was replaced by isoleucine in one mutant and by threonine in the other four mutants (Fig. 1B). Fifty-three silent mutations were found in total: 46 mutations were unique to each mutant AspAT, 6 mutations were common in two of the five mutants, and one mutation was found in three mutants, the G to A mutation at base position 696 in AV5A-1, AV5A-7, and AV5B-4. These findings indicate that the five mutants are not immediate descendants of the small population of elite survivors of the last round of selection. Molecular evolution theory predicts in such a case that the conserved five substitutions must be functionally important, although the present results based on a small number of mutants may not be statistically reliable enough to apply the theory.
Figure 1.
Sequence analysis of the evolved AspATs that have high activity for 2-oxovaline. (A) Lines show the entire coding region of the aspC gene. Longer bars indicate the nucleotide positions of missense mutations, which caused amino acid substitutions, and shorter bars indicate those of silent mutations. Asterisks are the missense mutations conserved in all five mutant AspATs. (B) Amino acid substitutions. Residues were numbered according to the sequence of cytosolic AspAT from pig as described (27).
Characterization of a Mutant AspAT.
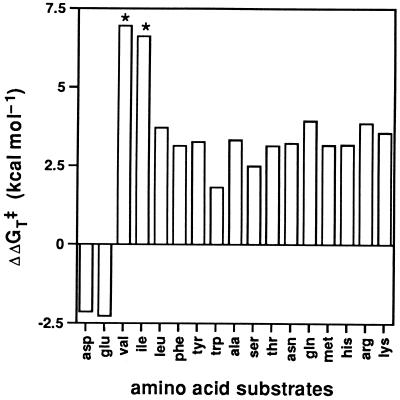
The coding region of AV5A-7, which showed the highest activity among the five mutants, was subcloned to a high copy plasmid, and the overproduced AV5A-7 was purified. Compared with the wild-type enzyme, AV5A-7 showed a 1.3 × 105-fold increase in the kcat/Km value for 2-oxovaline, a 7.3 × 104-fold increase for 2-oxoisoleucine, and a 20-fold decrease for acidic 2-oxo acids (Table 1). Similarly, the activity of AV5A-7 for valine increased >1.1 × 105-fold, that for isoleucine increased >6.6 × 103-fold, and that for acidic amino acids decreased 40-fold. As summarized in Fig. 2, AV5A-7 has specifically evolved high catalytic efficiency for β-branched substrates, valine and isoleucine. Interestingly, 20–800-fold increases in kcat/Km values were observed for the other amino acid substrates examined. Tyrosine (kcat/Km = 52000 s−1 M−1) was the best amino acid substrate for AV5A-7 (the corresponding value of the wild-type AspAT is 400 s−1 M−1). Because AV5A-7 is essential for valine biosynthesis in YJ103, this enzyme may be named tyrosine 2-oxovaline aminotransferase. It is, however, difficult to tell which amino acid primarily serves as an amino group donor to 2-oxovaline in YJ103 cells under the selection conditions.
Table 1.
Kinetic parameters of AspATs for branched and acidic substrates, pH 8.0, 25°C
| Substrate | WT*
|
AV5A-7
|
AV5A-76
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) | |
| l-Valine | —† | —† | <1 × 10−3 | 46 (6) | 420 (70) | 110 | —† | —† | 14 (0.2) |
| l-Isoleucine | —† | —† | <1 × 10−3 | —† | —† | 6.6 (0.09) | —† | —† | 0.5 (0.005) |
| l-Leucine | —† | —† | 2.4 | 160 (90) | 120 (10) | 1.3 × 103 | —† | —† | 160 (1) |
| 2-Oxovaline‡ | 5.7 (0.1) × 10−3 | 100 (3) | 0.057 | 28 (1) | 3.8 (0.3) | 7.4 × 103 | 60 (4) | 33 (4) | 1.8 × 103 |
| 2-Oxoisoleucine‡ | 3.3 (0.3) × 10−3 | 52 (9) | 0.063 | 220 (20) | 48 (4) | 4.6 × 103 | 230 (20) | 170 (20) | 1.4 × 103 |
| 2-Oxoleucine‡ | 1.2 (0.03) | 43 (1) | 28 | 140 (6) | 3.4 (0.3) | 4.1 × 104 | 290 (10) | 19 (2) | 1.5 × 104 |
| l-Aspartate | 550 | 4.5 | 1.2 × 105 | 220 (10) | 68 (8) | 3.2 × 103 | 70 (2) | 11 (0.9) | 6.4 × 103 |
| l-Glutamate | 700 | 38 | 1.8 × 104 | —† | —† | 380 (6) | 120 (9) | 220 (20) | 550 |
| Oxalacetate | 800 | 0.035 | 2.3 × 107 | 650 (70) | 0.52 (0.08) | 1.3 × 106 | 1.4 (0.1) × 103 | 0.46 (0.06) | 3.0 × 106 |
| 2-Oxoglutarate | 600 | 1.3 | 4.6 × 105 | 120 (4) | 3.9 (0.3) | 3.1 × 104 | 610 (30) | 4.5 (0.4) | 1.4 × 105 |
Numbers in parentheses are the standard deviations.
Data from ref. 18, except for those for branched-chain 2-oxo acids.
Reactions did not show saturation kinetics in the substrate concentrations examined.
Abbreviations: 2-oxovaline, 2-ketoisovaleric acid; 2-oxoisoleucine, dl-2-keto-3-methyl-n-valeric acid; 2-oxoleucine, 2-ketoisocaproic acid.
Figure 2.
Stabilization of the activation free energy for various amino acid substrates by AV5A-7, relative to the wild-type AspAT. ΔΔGT‡ = RTln{(kcat/Km)AV5A-7/(kcat/Km)WT}. Substrate amino acids are shown by three-letter abbreviations. Positive bars indicate that the catalytic efficiency of AV5A-7 is higher than that of the wild-type AspAT, and negative bars indicate vice versa. Exact values could not be determined for valine and isoleucine; therefore, ΔΔGT‡ values for their 2-oxo acids are shown for these amino acids (asterisks).
To discuss how the mutations affect the substrate specificity, one needs to determine a minimum set of mutations required for the altered activity. To this end, each of the 13 amino acid substitutions of AV5A-7 was mutated back to the wild-type sequence, and the activity of the single-residue revertants for 2-oxovaline was compared with that of AV5A-7 (data not shown). Seven of the substitutions were functionally nonessential, but the following six mutations were clearly important; Asn34Asp, Ile37Met, Ser139Gly, Asn142Thr, Asn297Ser, and Val387Leu. Five of these six substitutions (except for Val387Leu) were conserved in all mutants (Fig. 1). To determine if these mutations constitute a minimum set required for the altered activity, a mutant AspAT with these six substitutions, AV5A-76, was constructed. The purified AV5A-76 enzyme showed a 10- or 3-fold decrease in kcat/Km values for branched-chain amino or 2-oxo acids, respectively, compared with AV5A-7 (Table 1). The six substitutions in AV5A-76, therefore, contributed 80–90% to the stabilization of the activation free energy by AV5A-7 for valine or 2-oxovaline: [RTln{(kcat/Km)AV5A-76/(kcat/Km)WT}]/[RTln{(kcat/Km)AV5A-7/(kcat/Km)WT}]. The other seven substitutions may cooperatively contribute to the full activity of AV5A-7.
These findings provide insight into the nature of the mutations. First, the two mutations at residues 34 and 37 were separated by only 11 bp, and those at residues 139 and 142 were separated by 10 bp. Although it was possible that one of the two mutations of each pair was not necessary but was conserved simply because it was too closely “linked” to the other functionally essential mutation to be separated by recombination during DNA shuffling, all of these mutations proved to be indispensable. Second, the Glu7Val substitution was found in four of five mutant AspATs but did not seem to be functionally important for AV5A-7. It remains to be examined if this mutation is simply a conserved neutral mutation or somehow benefits the growth of YJ103 cells under the selection conditions. Third, Val387Leu is the only substitution that is unique to AV5A-7 and functionally important. Mutant AspATs other than AV5A-7 may have similar unique yet important mutations. For example, AV5A-1 has higher activity for 2-oxovaline than a mutant AspAT with only the five conserved substitutions (data not shown). Finally, Onuffer and Kirsch (3) previously reported a mutant AspAT with six amino acid substitutions that showed a 1500-fold increase in the kcat/Km value for phenylalanine. AV5A-7 also showed a 200-fold increase for phenylalanine. These mutants have two sites substituted in common, Lys41 and Asn297, although residue 41 was Tyr in their mutant but is Asn in AV5A-7.
Possible Effects of the Substitutions on Substrate Recognition.
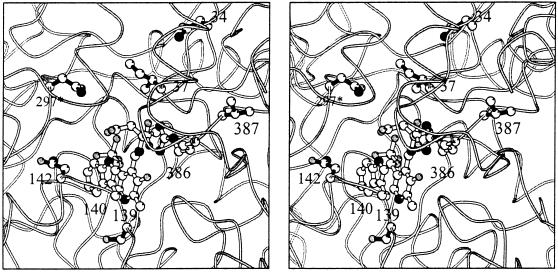
The role of each substitution may be proposed from an analysis of the x-ray crystal structure of E. coli AspAT complexed with a substrate analog, 2-methyl-l-aspartate (Fig. 3 and ref. 16). The γ-methyl group of the side chain of Ile37 is close to the β-methylene of 2-methylaspartate (C-C distance 3.8 Å) and thus might make direct contact with the γ-methyl group of β-branched substrates. The Ile37Met substitution likely removes this steric hindrance. The other residues, Asn34, Ser139, Asn142, Asn297, and Val387, are positioned farther away from the substrate. It is therefore more difficult to speculate on their effects. The side chain of Asn142 directly contacts the side-chain indole ring of Trp140, which, in turn, is stacked against the pyridine ring of the coenzyme pyridoxal 5′-phosphate during the course of catalysis (16). The Asn142Thr and Ser139Gly substitutions may affect the reaction by reorienting Trp140. As for the Asn297Ser substitution, crystallographic analysis previously showed that this mutation moves a water molecule away from the substrate binding site to make space for the binding of aromatic substrates (28). The side chains of β-branched substrates are, however, smaller than those of aromatic substrates, and thus it is not known if the same explanation holds for the reaction with β-branched substrates. Next to Val387 is Arg386, whose side chain directly interacts with the α-carboxylate group of the substrates. The Val387Leu substitution might adjust the orientation of the side chain of Arg386 to bind β-branched substrates. Because Asn34 is located >10 Å distance from the substrate binding site, it is impossible to speculate on the effect of the Asn34Asp substitution.
Figure 3.
Stereo representation of the structure of the wild-type E. coli AspAT complexed with 2-methyl-l-aspartate (16). The side chains of the six functionally important residues mutated in this study are shown by full bonds (Asn34, Ile37, Ser139, Asn142, Asn297, and Val387). The coenzyme pyridoxal 5′-phosphate, the substrate analog, and the side chains of Trp140 and Arg386 are also shown (open bonds). Asn297 belongs to the other subunit of the dimer (asterisk).
Conclusions.
The activity of AspAT for valine and 2-oxovaline was increased by five orders of magnitude. Naturally occurring enzymes have evolved high reaction and substrate specificity. During their evolution, each enzyme has gained a specialized structure–function relationship, for example, in the form of intricate hydrogen-bonding networks. Some have speculated that this is why creating an enzyme with novel properties from an existing enzyme is so difficult (29); attempts to manipulate the enzyme disrupt the finely tuned networks, inactivating the enzyme rather than redesigning it as intended. Our present results demonstrate that directed molecular evolution is a powerful technique to overcome such difficulty when an adequate method of mutagenesis and selection is applied. DNA shuffling is an ideal method to accumulate infrequent and functionally important mutations in a given genetic region (11–14). The selection system used in this study was convenient and reliable, and the stringency could be easily adjusted. This adjustability was important because the desired activity seldom emerges in one step but increases slowly during successive rounds of selection.
Gene duplication and subsequent divergence are believed to be ubiquitous in all organisms and responsible for the existence of many gene families (30). This process, molecular evolution of proteins, cannot be observed directly but only speculated retrospectively. The selection reported here is thought to simulate the initial phase of speciation of proteins with distinctive functions. Thus, this selection system may provide an opportunity to observe the process and to study theories of molecular evolution of enzymes.
Acknowledgments
We thank V.W. Cornish and D.R. Liu for careful reading and helpful comments on the manuscript and A. Okamoto for help in drawing Fig. 3. This work was supported by the Ministry of Education, Science, Sports, and Culture of Japan (to T.Y.) and the Japan Society for the Promotion of Science (Research for the Future Program) (to H.K.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AspAT, aspartate aminotransferase; 2-oxovaline, 2-ketoisovalerate.
References
- 1.Wilks H M, Hart K W, Feeney R, Dunn C R, Muirhead H, Chia W N, Barstow D A, Atkinson T, Clarke A R, Holbrook J J. Science. 1988;242:1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]
- 2.Hedstrom L, Szilagyi L, Rutter W J. Science. 1992;255:1249–1253. doi: 10.1126/science.1546324. [DOI] [PubMed] [Google Scholar]
- 3.Onuffer J J, Kirsch J F. Protein Sci. 1995;4:1750–1757. doi: 10.1002/pro.5560040910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson C, Szostak J W. Nature (London) 1995;374:777–782. doi: 10.1038/374777a0. [DOI] [PubMed] [Google Scholar]
- 5.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 6.Moore J C, Arnold F H. Nat Biotechnol. 1996;14:458–467. doi: 10.1038/nbt0496-458. [DOI] [PubMed] [Google Scholar]
- 7.Shao Z, Arnold F H. Curr Opin Struct Biol. 1996;6:513–518. doi: 10.1016/s0959-440x(96)80117-5. [DOI] [PubMed] [Google Scholar]
- 8.Wrighton N C, Farrell F X, Chang R, Kashyap A K, Barbone F P, Mulcahy L S, Johnson D L, Barrett R W, Jolliffe L K, Dower W J. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 9.Cwirla S E, Balasubramanian P, Duffin D J, Wagstrom C R, Gates C M, Singer S C, Davis A M, Tansik R L, Mattheakis L C, Boytos C M, Schatz P J, Baccanari D P, Wrighton N C, Barrett R W, Dower W J. Science. 1997;276:1696–1699. doi: 10.1126/science.276.5319.1696. [DOI] [PubMed] [Google Scholar]
- 10.Wright M C, Joyce G F. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 11.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 12.Stemmer W P C. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crameri A, Dawes G, Rodriguez E, Jr, Silver S, Stemmer W P C. Nature Biotechnol. 1997;15:436–438. doi: 10.1038/nbt0597-436. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J-H, Dawes G, Stemmer W P C. Proc Natl Acad Sci USA. 1997;94:4504–4509. doi: 10.1073/pnas.94.9.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPhalen C A, Vincent M G, Jansonius J N. J Mol Biol. 1992;225:495–517. doi: 10.1016/0022-2836(92)90935-d. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto A, Higuchi T, Hirotsu K, Kuramitsu S, Kagamiyama H. J Biochem. 1994;116:95–107. doi: 10.1093/oxfordjournals.jbchem.a124509. [DOI] [PubMed] [Google Scholar]
- 17.Kirsch J F, Eichele G, Ford G C, Vincent M G, Jansonius J N, Gehring H, Christen P. J Mol Biol. 1984;174:497–525. doi: 10.1016/0022-2836(84)90333-4. [DOI] [PubMed] [Google Scholar]
- 18.Kuramitsu S, Hiromi K, Hayashi H, Morino Y, Kagamiyama H. Biochemistry. 1990;29:5469–5476. doi: 10.1021/bi00475a010. [DOI] [PubMed] [Google Scholar]
- 19.Cronin C N, Kirsch J F. Biochemistry. 1988;27:4572–4579. doi: 10.1021/bi00412a052. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi H, Kuramitsu S, Inoue Y, Morino Y, Kagamiyama H. Biochem Biophys Res Commun. 1989;159:337–342. doi: 10.1016/0006-291x(89)92443-1. [DOI] [PubMed] [Google Scholar]
- 21.Kuramitsu S, Ogawa T, Ogawa H, Kagamiyama H. J Biochem. 1985;97:993–999. doi: 10.1093/oxfordjournals.jbchem.a135176. [DOI] [PubMed] [Google Scholar]
- 22.Cox J L, Cox B J, Fidanza V, Calhoun D H. Gene. 1987;56:185–198. doi: 10.1016/0378-1119(87)90136-3. [DOI] [PubMed] [Google Scholar]
- 23.Kuramitsu S, Okuno S, Ogawa T, Ogawa H, Kagamiyama H. J Biochem. 1985;97:1259–1262. doi: 10.1093/oxfordjournals.jbchem.a135173. [DOI] [PubMed] [Google Scholar]
- 24.Brosius J, Cate R L, Perlmutter A P. J Biol Chem. 1982;257:9205–9210. [PubMed] [Google Scholar]
- 25.Yano T, Kuramitsu S, Tanase S, Morino Y, Hiromi K, Kagamiyama H. J Biol Chem. 1991;266:6079–6085. [PubMed] [Google Scholar]
- 26.Inoue Y, Kuramitsu S, Inoue K, Kagamiyama H, Hiromi K, Tanase S, Morino Y. J Biol Chem. 1989;264:9673–9681. [PubMed] [Google Scholar]
- 27.Mehta P K, Hale T I, Christen P. Eur J Biochem. 1989;186:249–253. doi: 10.1111/j.1432-1033.1989.tb15202.x. [DOI] [PubMed] [Google Scholar]
- 28.Malashkevich V N, Onuffer J J, Kirsch J F, Jansonius J N. Nat Struct Biol. 1995;2:548–553. doi: 10.1038/nsb0795-548. [DOI] [PubMed] [Google Scholar]
- 29.Hwang J-K, Warshel A. Nature (London) 1988;334:270–272. doi: 10.1038/334270a0. [DOI] [PubMed] [Google Scholar]
- 30.Henikoff S, Greene E A, Pietrokovski S, Bork P, Attwood T K, Hood L. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]