Abstract
Posttranslational modifications of histone proteins regulate gene expression via complex protein–protein and protein–DNA interactions with chromatin. One such modification, the methylation of lysine, has been shown to induce binding to chromodomains in an aromatic cage [Nielsen PR, et al. (2002) Nature 416:103–107]. The binding generally is attributed to the presence of cation–π interactions between the methylated lysine and the aromatic pocket. However, whether the cationic component of the interaction is necessary for binding in the aromatic cage has not been addressed. In this article, the interaction of trimethyllysine with tryptophan is compared with that of its neutral analog, tert-butylnorleucine (2-amino-7,7-dimethyloctanoic acid), within the context of a β-hairpin peptide model system. These two side chains have near-identical size, shape, and polarizabilities but differ in their charges. Comparison of the two peptides reveals that the neutral side chain has no preference for interacting with tryptophan, unlike trimethyllysine, which interacts strongly in a defined geometry. In vitro binding studies of the histone 3A peptide containing trimethyllysine or tert-butylnorleucine to HP1 chromodomain indicate that the cationic moiety is critical for binding in the aromatic cage. This difference in binding affinities demonstrates the necessity of the cation–π interaction to binding with the chromodomain and its role in providing specificity. This article presents an excellent example of synergy between model systems and in vitro studies that allows for the investigation of the key forces that control biomolecular recognition.
Keywords: cation–pi interactions, histone code, lysine methylation, posttranslational modifications, protein–protein interactions
With rapid advancements in genomics, epigenetics has become the next major challenge in understanding how genetic information is controlled (1). It is becoming clear that posttranslational modifications of proteins are a key component in controlling gene expression. These modifications include a number of subtle structural changes, including Lys and Arg methylation, Lys acylation, and Ser/Thr/Tyr phosphorylation, which act as chemical switches to induce or repress protein–protein interactions. Among all histone modifications, lysine methylation is especially important for chromatin function because of its stability and direct contribution to heritable patterns of gene expression (for review, see ref. 2). To understand how such modest structural modifications can control biomolecular recognition events, it is critical to understand the underlying noncovalent interactions involved.
Methylation of Lys induces a protein–protein interaction through the binding of methyl lysine (KMen, n = 1–3) in an aromatic cage. This interaction first was described for the binding of methylated histone 3 (H3) tail to the HP1 chromodomain (Fig. 1) (3, 4). HP1 and methylated H3 interact specifically whether lysine 9 is mono-, di-, or trimethylated. However, the binding is most effective when lysine is trimethylated (5). In addition, more recent findings have shown that phosphorylation of serine 10 prevents interaction of HP1 with methylated H3 (for review, see ref. 6). Therefore, a binary switch mechanism has been proposed for the recognition of methyllysine-containing peptides by chromodomains. Interestingly, binding of a methylated lysine in an aromatic cage is not exclusive to chromodomains. Plant homeobox domain (PHD) fingers and Tudor domains also assemble three aromatic residues around methyllysine of the H3 tail (for review, see refs. 7 and 8).
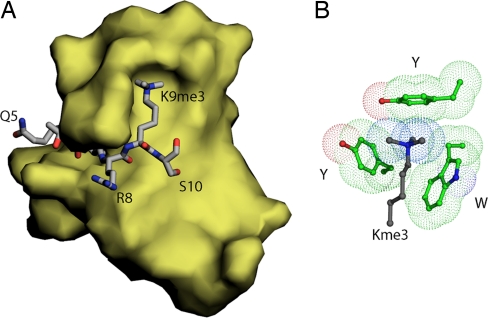
Fig. 1.
Structure of the histone 3A-HP1 chromodomain interaction. (A) Crystal structure of the HP1 chromodomain (yellow surface) in complex with lysine 9-trimethylated H3 tail residues 5 through 10 (gray stick). (B) Aromatic cage (green) formed by two tyrosines and one tryptophan captures the methyllysine (gray).
Recognition of methylated lysine by an aromatic cage appears to be mediated by cation–π interactions between the methylated ammonium group and the side chains of three aromatic residues. The cation–π interaction is defined by the attractive interaction between a positively charged moiety (simple cations, ammonium groups, etc.) and the quadrupole moment of an aromatic ring (9). The magnitude of the cation–π interaction in proteins depends on a number of factors, including the electron density of the aromatic ring (Phe versus Trp, for example), the distribution of positive charge across the cationic moiety, and the degree of solvent exposure of the interaction. Other forces, such as van der Waals interactions and the hydrophobic effect, also contribute to the magnitude of the interaction (9). Numerous examples of functional cation–π interactions exist in structural biology, and they have been demonstrated to be important to protein structure and stability and the functioning of enzymes and ion channels (9).
Because of the potential importance of cation–π interactions in the recognition of the posttranslationally modified amino acids KMen (n = 1–3) and the still-growing body of theoretical and experimental knowledge concerning the various energetic components of the interaction, a number of questions remain that need to be addressed experimentally regarding the interplay among electrostatics, van der Waals interactions, and the hydrophobic effect (10–16). Moreover, in the context of chromodomain, it is not clear to what degree a charge–quadrupole interaction imbues specificity to a biologically significant ligand-receptor interaction or if the interaction is primarily caused by hydrophobic and/or van der Waals interactions between the methyl groups and the aromatic pocket. To this end, we have synthesized the neutral analog of KMe3, tert-butyl norleucine (2-amino-7,7-dimethyloctanoic acid; tBuNle), investigated its interaction with Trp in a β-hairpin model system, and then compared these results to in vitro binding assays with the HP1 chromodomain. This hairpin model system has been used previously to investigate the cation–π interaction between KMe3 and Trp (17). In this model system, we find striking contrasts between the behavior of the two side chains with Trp that provide insight into the role of hydrophobicity and the importance of the charge–quadrupole component of the Trp–KMe3 interaction. Furthermore, in vitro binding studies of the neutral tBuNle analog of the H3 tail peptide demonstrate that the positive charge is required to give a specific interaction between the histone tail and the HP1 protein.
Results
In our previous study of peptides WK (18, 19) and WKMe3 (Fig. 2) (17, 20),¶ we found that methylation of Lys enhanced its interaction with Trp significantly, but that the driving force became more entropically favorable, suggesting an increased hydrophobic component; in contrast, the enthalpic component, attributed to the charge–quadrupole interaction, decreased relative to unmodified Lys (17). This finding led to the question of whether the positive charge in KMe3 indeed is necessary for interaction with an aromatic residue or an aromatic pocket in aqueous solution or whether hydrophobic and van der Waals interactions alone will suffice. We chose to investigate the interaction of Trp with tBuNle because the size and shape of KMe3 and tBuNle are virtually identical; volumes are calculated to be 158.0 Å3 and 160.2 Å3, respectively, as are the polarizabilities (11.3 versus 12.0 Å3) (see Experimental Procedures). However, the charge and the hydrophobicity of the two side chains differ significantly. Differences in hydrophobicity are reflected by the differences in log P (octanol/water partition coefficients) for the two amino acids. tBuNle has a log P value of 1.84, which is consistent with a favorable hydrophobic driving force. In contrast, KMe3 has a log P value of −0.10, signifying little or no hydrophobic driving force (see Experimental Procedures for log P determination). Hence, if hydrophobicity is the primary driving force for interaction with Trp, then the interaction should be significantly more favorable for tBuNle, but if the charge–quadrupole component is important, then it can compensate for the lower hydrophobicity of KMe3.
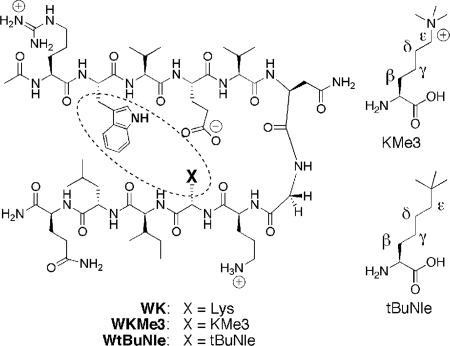
Fig. 2.
β-Hairpin peptide containing KMe3 or tBuNle. In the text, peptides are referred to by the residues at positions 2 (Trp) and 9 (X).
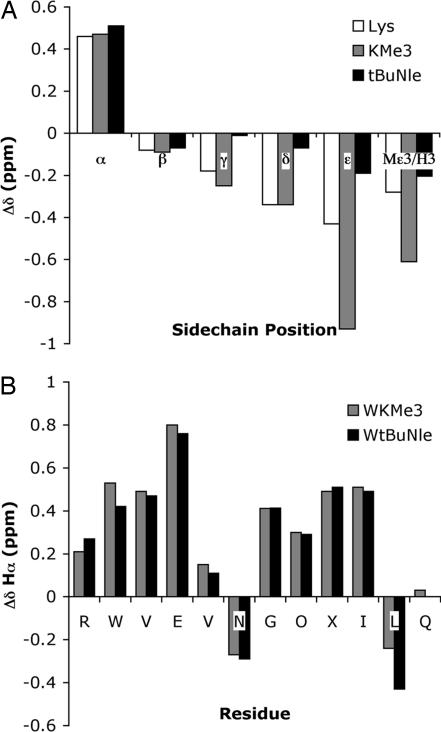
The unnatural amino acid, tBuNle, was synthesized from pseudoephedrine glycinamide and 1-bromo-5,5-dimethylhexane (21, 22) via the method of Myers et al. (23) and incorporated into the β-hairpin peptide via standard solid-phase peptide synthesis to give the peptide WtBuNle (see Experimental Procedures). The side-chain–side-chain interaction was investigated by NMR and compared with WKMe3. The interaction between residue 9 (X) and Trp can be characterized by the extent of upfield shifting of the X side chain (Fig. 3). Greater upfield shifting indicates greater proximity to the face of the Trp indole ring (24); for examples in β-hairpin peptides, see refs. 18, 19, and 25. We previously have shown that KMe3 exhibits enhanced interaction with Trp relative to the unmethylated Lys, particularly at the ε-CH2 and methyl positions (Fig. 3A) (17). Surprisingly, tBuNle exhibits little upfield shifting at any position along the side chain, which indicates that tBuNle has no preference for interaction with the face of Trp, despite its large hydrophobic surface area. This result clearly demonstrates the importance of the electrostatic component of the cation–π interaction. Without polarization of the methyl groups, there is no specific interaction between the side chain and the face of the aromatic ring. These results recall prescient studies by Dougherty et al. (26), who showed binding discrimination between interaction of an aromatic host with a trimethyl ammonium group and a tert-butyl group in a small-molecule model system in borate buffer.
Fig. 3.
NMR chemical shift data for WKMe3 and WtBuNle. (A) tBuNle and KMe3 side-chain upfield shifts relative to random coil values. (B) Hα shifts relative to random coil values. Glycine shifts reflect the splitting.
Despite the lack of interaction between Trp and tBuNle, WtBuNle is as well folded as WKMe3, as determined from the similarity of the downfield shifting of the Hα protons of the two peptides relative to random coil values (Fig. 3B). Quantification of the fraction folded indicates that WtBuNle is a very well folded hairpin (96 ± 1% based on Gly splitting; 90 ± 7% based on Hα chemical shifts), as is WKMe3 (93 ± 1% based on Gly splitting; 91 ± 15% based on Hα chemical shifts). (For NMR methods for determining fraction folded, see Experimental Procedures and refs. 18, 19, and 27.) Thus, tBu must provide stability to the hairpin through means other than specific interaction with the face of the indole ring of Trp, likely because of nonspecific hydrophobic interactions between tBuNle and other sites on the face of the β-hairpin, as has been observed with norleucine (Nle) (19).
Double-mutant cycles were performed in which the interacting residues at positions 2 and 9 were mutated to noninteracting residues Ser and Val to determine the magnitude of the isolated side-chain–side-chain interaction (17–19). These experiments give a value of −1.0 (± 0.1) kcal/mol for the Trp–KMe3 interaction and a value of −0.6 (± 0.1) kcal/mol for the Trp–tBuNle interaction. The magnitude of the Trp–KMe3 interaction is consistent with what has been seen for the interaction of a tetraalkylammonium group with an aromatic ring in a range of different systems (9). An analysis of the factors that contribute to this overall interaction energy provides insight into the driving force of each interaction. Based on the similar surface areas and polarizabilities of KMe3 and tBuNle, the van der Waals interactions of these two side chains with Trp will be of similar magnitude. Comparison of the log P values for KMe3 and tBuNle provides information about the relative contribution of the hydrophobic effect to interaction with Trp. The interaction between Trp and tBuNle comprises a favorable desolvation energy, as indicated by the positive log P value of 1.84, which is consistent with a favorable hydrophobic driving force. In contrast, the interaction of Trp and KMe3 consists of negligible desolvation energy, as indicated by the log P value of −0.10, signifying little or no hydrophobic driving force. Lastly, the Trp–KMe3 interaction also consists of a cation–π component, which is not possible between Trp and tBuNle. Hence, the fact that the Trp–KMe3 interaction is stronger than the Trp–tBuNle interaction thus indicates that the cation–π interaction more than compensates for the loss of hydrophobicity, even though the interaction is solvent exposed.
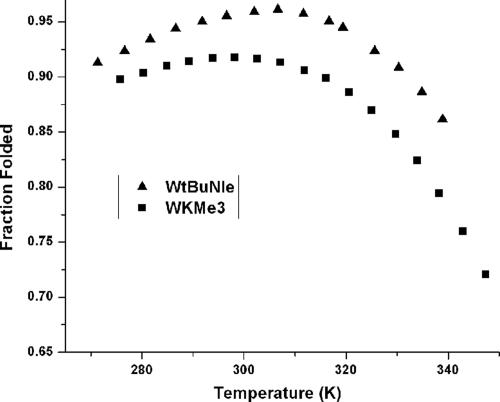
Both WKMe3 and WtBuNle also exhibit high thermal stability, as determined by NMR (Fig. 4). However, WtBuNle exhibits greater cold denaturation, consistent with a greater hydrophobic driving force for folding. Fitting of the thermal denaturation data with a modified Van't Hoff equation (28, 29) reveals that WKMe3 exhibits a favorable entropy of folding and a negligible enthalpy of folding, indicative of multiple sites of favorable interaction with the Trp ring and a favorable hydrophobic component to folding (Table 1). By comparison, the WtBuNle shows a much greater favorable folding entropy and a significantly unfavorable enthalpy of folding, as well as a larger ΔCp value. The considerable increase in folding entropy on going from the KMe3 to the tBuNle side chain, and the corresponding decrease in enthalpic favorability, is consistent with a greater hydrophobic driving force for folding of WtBuNle as well as elimination of the favorable electrostatic interaction between Trp and KMe3. Comparison of the thermodynamic parameters for WKMe3, WtBuNle, and WK indicates that the attraction between KMe3 and Trp has a significant hydrophobic component, but the specificity of the interaction is not attributable to a simple hydrophobic effect, as is the case for WtBuNle.
Fig. 4.
Thermal denaturation profiles of WKMe3 and WtBuNle peptides as determined by NMR. The fraction folded was determined from the Gly splitting. Error is ± 0.5 K in temperature and ± 1% in fraction folded. Conditions: 50 mM NaOAc-d4 buffer (pD 4.0, uncorrected).
Table 1.
Thermodynamic parameters for WKMe3 and WtbutylNle at 298 K (19)
| Peptide | ΔH°, kcal/mol | ΔS°, cal/mol·K | ΔCp°, cal/mol·K |
|---|---|---|---|
| WK | −2.6 (0.1) | −6.2 (0.2) | −180 (30) |
| WKMe3L | −0.1 (0.1) | +4.5 (0.3) | −240 (40) |
| WtBuNle | +2.0 (0.4) | +12.9 (1.4) | −330 (50) |
Parameters were determined from the temperature dependence of the Gly chemical shift from 0°C to 80°C. Errors (in parentheses) are determined from the fit. The error for ΔCp° values is estimated at 15%.
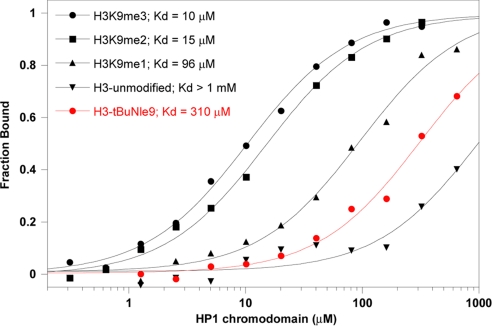
Although there is no specific interaction between tBuNle and Trp in our β-hairpin model system, it was not clear that the same effect would be observed in the binding of the H3 peptide to the HP1 chromodomain: the aromatic pocket made up of a Trp and two Tyr residues is designed to perfectly accommodate a group with the same size and shape as either a trimethylammonium or a tert-butyl group (Fig. 1B). Given the greater hydrophobicity of the tBuNle side chain relative to KMe3 and the similar polarizability of the two residues, as well as the exposure of the aromatic pocket to solvent on the surface of the protein, it seemed likely that a mutant H3 peptide containing the tBuNle side chain at position 9 would bind to chromodomain strongly, despite the lack of a positive charge. Hence, we synthesized the modified H3 peptide (NH2-ARTKQTAR(tBuNle)STGGKAY-COOH, H3-tBuNle9) and compared it to the native sequence containing KMe3 (NH2-ARTKQTAR(KMe3)STGGKAY-COOH, H3-K9Me3) as a positive control, the partially methylated variants KMe2 (NH2-ARTKQTAR(KMe2)STGGKAY-COOH, H3-K9Me2) and KMe (NH2-ARTKQTAR(KMe)STGGKAY-COOH, H3-K9Me), as well as Lys (NH2-ARTKQTARKSTGGKAY-COOH, H3-K9) as a negative control. At pH 7.5, specific binding was found for the trimethylated peptide, with a Kd of 10 μM, with concomitantly weaker binding for H3-K9Me2 and H3-K9Me and virtually no binding for H3-K9 (Fig. 5). We then tested the binding of the neutral analog, H3-tBuNle9, and found that it is nearly as poor as the unmethylated peptide H3-K9 for binding to the HP1 chromodomain (Fig. 5). This observation was surprising given the similarity in size and shape, as well as the greater hydrophobicity of the tert-butyl group relative to the trimethylammonium group of KMe3 but is consistent with results from the model system. Indeed, H3-K9Me and H3-K9Me2 bind more strongly than does H3-tBuNle9, despite the fact that they do not fill the binding pocket of HP1 chromodomain and must sequester a water molecule to do so, as shown in the crystal structures of the histone-chromodomain complexes (5). This finding clearly demonstrates the essential nature of the cation–π component to binding of the lysine-methylated H3 tail to chromodomains.
Fig. 5.
Binding of H3-K9Me3, H3-K9Me2, H3-K9Me, H3-tBuNle9, and H3-K9 to the HP1 chromodomain as determined by fluorescence polarization assay. Drosophila HP1 chromodomain was purified and used in binding studies with fluoresceinated peptides as described in ref. 5. Binding assays were performed at 15°C in phosphate buffer (pH 7.5) and containing 25 mM NaCl and 1 mM DTT.
Discussion
This study provides substantial insight into why lysine methylation is successful in making critical interactions that are required for controlling gene expression. Using a peptide model system, we have demonstrated the essential nature of the cation–π component to the interaction of the H3 tail with HP1 chromodomain by providing affinity and specificity. Moreover, these data enhance our fundamental understanding of how the methylation of lysine functions cooperatively within the broad spectrum of posttranslational modifications. The cation–π component for docking of a chromodomain to a lysine-methylated histone tail is especially useful for controlling epigenetic signaling via a phosphorylation switch. Recent studies have shown that phosphorylation of the residue adjacent to the methyllysine reduces the affinity of the chromodomain for the histone tail by 100-fold, which effectively blocks recognition of the methyllysine signal (30, 31). The presence of a phosphate group immediately adjacent to the methyllysine dramatically reduces the stability of the cation–π bond between the methyllysine and the aromatic cage. Among well known posttranslational modifications that contribute to biomolecular signaling, lysine methylation of histone tails is a stable modification during the cell cycle that is inherited during cell division (2). Although recent studies have identified bona fide nucleosome-specific lysine-demethylases, it appears that these contribute to the resetting of a fraction of methyllysine signals by mechanisms that are poorly understood. Therefore, the reversible phosphorylation of the residue adjacent to the methyllysine is a novel biomolecular feature for on–off switching of the lysine methylation signal. Phosphorylation blocks the docking of proteins to methyllysines, thus chromatin can undergo maximal compaction in preparation of the metaphase chromosomes (30, 32). Subsequent events that reverse the phosphorylation allow reestablishing functional chromatin boundaries by recruiting specific methyllysine-docking factors. This finding is consistent with a histone code that suggests distinct histone modifications act sequentially or in combination to bring about important events for eukaryotic gene regulation (33).
In conclusion, valuable mechanistic information regarding the nature of molecular recognition between the H3A tail and the HP1 chromodomain readily was obtained from the peptide model system, which is not directly available from the protein–peptide interaction. We now have a better understanding of the forces underlying the preference for KMe3 recognition that firmly establishes the importance of the charge–quadrupole interaction to binding and specificity. In a broader context, this study provides insight into the subtle features of noncovalent interactions that contribute to biomolecular recognition and indicates that the simple separation of interactions into polar and hydrophobic can be too simplistic to fully understand or control biomolecular recognition.
Experimental Procedures
Peptide Synthesis.
The synthesis of all peptides was performed on an Applied Biosystems (Foster City, CA) Pioneer peptide synthesizer by using Applied Biosystems PEG-PAL resin (PAL, aminomethyl-3,5-dimethoxyphanoxy pentanoic acid). Peptides were synthesized on a 0.1- or 0.07-mmol scale. All amino acids with functionality were protected during synthesis as follows: Arg(Pbf), Asn(trt), Lys(Boc), Orn(Boc), Gln(trt), Trp(Boc), and Glu(tBu). Coupling reagents were HBTU/HOBt. The N terminus was acylated for all peptides with a solution of 5% acetic anhydride and 6% 2,6-lutidine in dimethylformamide. Cleavage conditions removed all side-chain protection with a mixture of 90% TFA/5% triisopropylsilane/5% H2O. Peptides were purified by reverse-phase HPLC on a C18 column. Peptides were purified with a gradient of A (95% H2O/5% acetonitrile with 0.1% TFA) and B (95% acetonitrile/5% H2O with 0.1% TFA). Once purified, peptides were lyophilized to powder and characterized by MALDI mass spectroscopy and NMR.
NMR Measurements.
NMR samples were made to concentrations of 0.3–1 mM and analyzed on a Varian (Palo Alto, CA) Inova 600-MHz instrument. Samples were dissolved in D2O/acetate-d3 buffer and referenced to 2,2-dimethyl-2-silapentane-5-sulfonate (DSS; pD (log [D+]) 4.0, uncorrected). By using a 1- to 1.5-s presaturation or solvent suppression, 1D NMR spectra were collected with between 8 and 32 scans. All 2D NMR experiments used pulse sequences from the chempack software, including total correlation spectroscopy (TOCSY), double quantum-filtered correlation spectroscopy (DQCOSY), gradient-correlated spectroscopy (gCOSY), and NOESY. Typical NOESY mixing times were 0.5 s. The 2D NMR scans were taken with 16 or 32 scans in the first dimension and 128–256 in the second dimension. All spectra were analyzed by using standard window functions (sinebell and Gaussian with shifting). Assignments were made by using standard methods (34). Thermal denaturations were run in duplicate, with standard deviations of <0.005 ppm. Samples were allowed to equilibrate for 7 min at each temperature before the measurement was taken. The temperature was calibrated with methanol and ethylene glycol standards.
Electrostatic Potential Maps and Side-Chain Volumes.
Electrostatic potential maps and side-chain volumes of lysine side-chain analogues were calculated at the HF/6–31g* level by using MacSpartan Pro version 1.0.4 (Wavefunction, Irvine, CA).
Polarizabilities.
Side-chain polarizabilities were calculated with Gaussian 03 (HF/6–31g**; keyword: Polar) and reported as polarizability volumes. Structures used in polarizability calculations are the side-chain mimics shown in Fig. 3.
CLog P values.
The octanol/water partition coefficients (CLog P) were calculated for the N- and C-capped amino acids AcNHCH(R)CONH2 by using Chemdraw Ultra version 10.0 (CambridgeSoft, Cambridge, MA).
Quantification of Folding.
To determine the chemical shifts of the fully folded state, 14-residue disulfide-linked analogs of peptides were synthesized with the sequence of Ac-CRWVEVNGOXILQC-NH2, where X = KMe3 or tbutylNle. The disulfide bond between Cys-1 and Cys-14 constrains the peptide to a β-hairpin. To determine the unfolded chemical shifts, 7-mer were synthesized with sequences Ac-RWVEVNG-NH2 and Ac-NGOXILQ-NH2, where X = KMe3 or tBuNle. The chemical shifts for residues in the strand and one turn residue were obtained from each 7-mer peptide. The fraction folded was determined from Eq. 1.
Characterization of Structure.
Methods used to indicate the formation of β-hairpin structure include the analysis of Hα shifting relative to random coil (Fig. 3B), backbone amide shifts relative to random coil [see supporting information (SI)], and the identification of cross-strand NOEs (see SI), as described previously (17–19, 28, 35).
Thermodynamic Analysis.
Peptides were analyzed by assuming two-state folding. The equilibrium constant was determined from the fraction folded (f) by K = f/(1 − f). The free energy then was calculated from ΔG° = −RT lnK (where R is the ideal gas constant and T is temperature). To determine the thermodynamic parameters, ΔH°, ΔS°, and ΔCp°, the temperature dependence of the Gly chemical-shift difference was fit to Eq. 3 (27):
where x = T(ΔS°298 + a ln(T/298) + b (T − 298) − (c/2) (1/T2 − 1/2982)) − (ΔH°298 + a (T − 298) + (b/2) (T2 − 2982) − c (1/T − 1/298)).
Supplementary Material
Acknowledgments
Work in S.K.'s laboratory was supported by National Institutes of Health Grants R01 GM064786 and T32 GM08136. Work in M.L.W.'s laboratory was supported by National Institutes of Health Grant GM071589. R.M.H. gratefully acknowledges support from a Burroughs Wellcome Foundation fellowship and an American Chemical Society Division of Organic Chemistry fellowship.
Abbreviations
- H3
histone 3
- tBuNle
tert-butyl norleucine (2-amino-7,7-dimethyloctanoic acid).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.A.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610850104/DC1.
We previously have reported the use of a β-hairpin model system to study of cation–π interactions between Trp and Lys or KMe3, which were placed in a diagonal relationship to each other on the same face of the β-hairpin. Leu was placed laterally cross-strand from the Trp residue to provide a hydrophobic interaction with Trp, and Glu was placed cross-strand from Lys or Nle to increase water solubility. Previous studies, including a pH study and NMR structure of WK and WKMe3, indicate that the Leu and Glu residues do not interfere with the diagonal Trp–Lys and Trp–KMe3 interactions of interest. See refs. 17–20.
References
- 1.Khorasanizadeh S. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 2.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs SA, Khorasanizadeh S. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 5.Fischle W, Wang YM, Jacobs SA, Kim YC, Allis CD, Khorasanizadeh S. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eissenberg JC, Elgin SCR. Nature. 2005;438:1090–1091. doi: 10.1038/4381090a. [DOI] [PubMed] [Google Scholar]
- 7.Mellor J. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Sims RJ, III, Reinberg D. Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 9.Ma JC, Dougherty DA. Chem Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson MAL, Morgantini PY, Kollman PA. J Phys Chem B. 1999;103:4474–4480. [Google Scholar]
- 11.Hunter CA, Low CMR, Rotger C, Vinter JG, Zonta C. Proc Natl Acad Sci USA. 2002;99:4873–4876. doi: 10.1073/pnas.072647899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mo YR, Subramanian G, Gao JL, Ferguson DM. J Am Chem Soc. 2002;124:4832–4837. doi: 10.1021/ja0174433. [DOI] [PubMed] [Google Scholar]
- 13.Ruan CH, Rodgers MT. J Am Chem Soc. 2004;126:14600–14610. doi: 10.1021/ja048297e. [DOI] [PubMed] [Google Scholar]
- 14.Costanzo F, Della Valle RG, Barone V. J Phys Chem B. 2005;109:23016–23023. doi: 10.1021/jp055271g. [DOI] [PubMed] [Google Scholar]
- 15.Reddy AS, Sastry GN. J Phys Chem A. 2005;109:8893–8903. doi: 10.1021/jp0525179. [DOI] [PubMed] [Google Scholar]
- 16.Xu YC, Shen JH, Zhu WL, Luo XM, Chen KX, Jiang HL. J Phys Chem B. 2005;109:5945–5949. doi: 10.1021/jp044568w. [DOI] [PubMed] [Google Scholar]
- 17.Hughes RM, Waters ML. J Am Chem Soc. 2005;127:6518–6519. doi: 10.1021/ja0507259. [DOI] [PubMed] [Google Scholar]
- 18.Tatko CD, Waters ML. Protein Sci. 2003;12:2443–2452. doi: 10.1110/ps.03284003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatko CD, Waters ML. J Am Chem Soc. 2004;126:2028–2034. doi: 10.1021/ja038258n. [DOI] [PubMed] [Google Scholar]
- 20.Hughes RM, Benshoff ML, Waters ML. Chem Eur J. 2007 Apr 12; doi: 10.1002/chem.200700223. [DOI] [Google Scholar]
- 21.Shirahata A. Tetrahedron Lett. 1989;30:6393–6394. [Google Scholar]
- 22.Rybczynski PJ, Zeck RE, Dudash J, Jr, Combs DW, Burris TP, Yang M, Osborne MC, Chen X, Demarest KT. J Med Chem. 2004;47:196–209. doi: 10.1021/jm0301888. [DOI] [PubMed] [Google Scholar]
- 23.Myers AG, Schnider P, Kwon S, Kung DW. J Org Chem. 1999;64:3322–3327. doi: 10.1021/jo990341z. [DOI] [PubMed] [Google Scholar]
- 24.Pople JA. J Chem Phys. 1956;24:1111. [Google Scholar]
- 25.Honda S, Kobayashi N, Munekata E. J Mol Biol. 2000;295:269–278. doi: 10.1006/jmbi.1999.3346. [DOI] [PubMed] [Google Scholar]
- 26.Petti MA, Shepodd TJ, Barrans JRE, Dougherty DA. J Am Chem Soc. 1988;110:6825–6840. [Google Scholar]
- 27.Maynard AJ, Sharman GJ, Searle MS. J Am Chem Soc. 1998;120:1996–2007. [Google Scholar]
- 28.Griffith-Jones SR, Maynard AJ, Searle MS. J Mol Biol. 1999;292:1051–1069. doi: 10.1006/jmbi.1999.3119. [DOI] [PubMed] [Google Scholar]
- 29.Hughes RM, Waters ML. J Am Chem Soc. 2006;128:12735–12742. doi: 10.1021/ja061656g. [DOI] [PubMed] [Google Scholar]
- 30.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 31.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim YC, Minor W, Rastinejad F, Khorasanizadeh S. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 32.Hirota T, Lipp JJ, Toh BH, Peters JM. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 33.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 34.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley-Interscience; 1986. [Google Scholar]
- 35.Maynard AJ, Sharman GJ, Searle MS. J Am Chem Soc. 1998;120:1996–2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.