Abstract
Controlling protein function through posttranslational manipulations has emerged as an attractive complementary technology to existing genetic systems. Often these methods involve developing pharmacological agents to probe protein function without the need to generate a unique compound for each protein family. One common strategy uses small molecules that act as chemical inducers of dimerization by mediating the interaction of two proteins. Herein we report the use of a chemical inducer of dimerization for the development of a posttranslational technology for the manipulation of protein function. This system, split ubiquitin for the rescue of function (SURF), places the complementation of genetically split ubiquitin under the control of rapamycin-induced dimerization of FK506-binding protein and FKBP12-rapamycin-binding protein. Before complementation a “degron” dooms a protein of interest for destruction by the proteasome. Addition of rapamycin results in a proteolytic shunt away from degradation by inducing ubiquitin complementation and cleavage of the protein of interest from the degron. Importantly, the native protein is rescued. We characterized this system with firefly luciferase and went on to apply it to members of three important classes of proteins: proteases (caspase-3), kinases (v-Src), and transcription factors (Smad3). This general strategy should allow for inducible rescue of a variety of proteins in such a way that their native structure and function are maintained.
Keywords: degradation, rescue of function
Genetic techniques that alter protein function have generated a remarkable wealth of information over the past century. Mutational screens (forward genetics) can identify unknown members of a pathway responsible for generating a phenotype. Targeted techniques (reverse genetics), such as tet/dox and Cre/lox (1), have been applied to many systems, allowing the function of a particular gene of interest to be interrogated. More recently, RNA interference has emerged as an attractive method for controlling function through posttranscriptional gene silencing (2). Despite the obvious power of these techniques, experimental limitations exist. Embryonic lethal phenotypes are not uncommon, arising from the disruption of a gene that is essential for development, limiting the effectiveness of this approach for the study of essential genes in an adult organism. Alternatively, the mutation may result in no observable phenotype, usually because of the presence of redundant or alternative pathways, making definitive interpretation of the protein's significance difficult.
Cell-permeable small-molecule modulators of protein function can overcome some of the drawbacks of genetic techniques and have therefore become a valuable partnering strategy in the investigation of protein function (3). Small molecules are useful because they can be used in a rapid, dose-dependant, and reversible manner. However, they often lack the specificity inherent in reverse genetic techniques. Indeed, many small molecules target one, if not multiple, off-target proteins precluding definitive interpretation of a specific protein's function (4).
Recently, there has been significant interest in developing pharmacological methods for probing protein function without the need to generate a unique compound for each protein family (5). One common strategy uses small molecules that act as chemical inducers of dimerization (CID) by mediating the interaction of two proteins (6). One of the best-characterized CID molecules, rapamycin, binds with high affinity to the small cellular protein, FK506-binding protein (FKBP) 12, resulting in a binary complex that will heterotrimerize with a second protein, FKBP12-rapamycin-binding protein (FRB), which is a domain of mTOR kinase (7). This three-hybrid technology allows two designated cellular proteins to be brought into proximity and has been used to control many processes, including transcription and translocation (8), receptor signaling (9), and protein splicing (10).
An alternative strategy takes advantage of endogenous cellular processes, by placing the stability of a protein of interest under pharmacological control. Pioneering work in this area involved using small molecules to reduce the cellular levels of a protein of interest by inducing its association with the cellular protein degradation machinery (11, 12). More recently, Crabtree and coworkers (13) have developed complementary technologies that, instead of inducing the degradation of a target protein, rescue it from this fate. The original method used a mutant FRB, termed FRB*, whose fusion to target proteins destines them for proteasomal destruction. However, FRB* is stabilized through a ternary complex with endogenous FKBP and either rapamycin or a nontoxic derivative MaRap. Thus, proteins previously marked for degradation are rescued by the addition of the small molecule. This technology has been used to control glycogen synthase kinase-3 levels and activity in genetically modified mice. The utility of this approach has recently been expanded by replacing FRB* with destabilized mutants of FKBP that could be stabilized by a small molecule ligand termed Shld1 (14). Similar to FRB*, these FKBP mutants mediate proteasomal degradation of fused proteins of interest; however, rescue of these constructs does not require formation of a ternary complex, which presumably minimizes the effects on normal protein function.
We reasoned that an ideal degradation-mediated technology would result in the release of a native protein from the degradation signal (or “degron”) after small-molecule rescue of a chimeric protein-degron fusion. To achieve release from the degron, we somewhat counter intuitively turned to a known mediator of protein degradation, ubiquitin. Ubiquitin plays several roles in both protein trafficking and stability, primarily through its posttranslational conjugation to lysine side chains of target proteins (15). Unlike this branched ubiquitin structure, linear ubiquitin fusions are known to be translational products where the N-terminal ubiquitin moiety is rapidly and specifically cleaved from its fusion partner by dedicated ubiquitin proteases (16). These proteases have broad specificity for the residues following ubiquitin, with proline being the only amino acid resistant to cleavage. Johnsson and Varshavsky (17) have used this feature in a two-hybrid technology based on complementation and subsequent cleavage of a genetically split ubiquitin. Briefly, ubiquitin was split into N- and C-terminal fragments, corresponding to residues 1–37 and 35–76, respectively. When a point mutation (Ile-13 to Ala or Gly) was also introduced, the ubiquitin fragments would only complement and fold, allowing for cleavage of fusion constructs, when a dimerization signal brought them into close proximity.
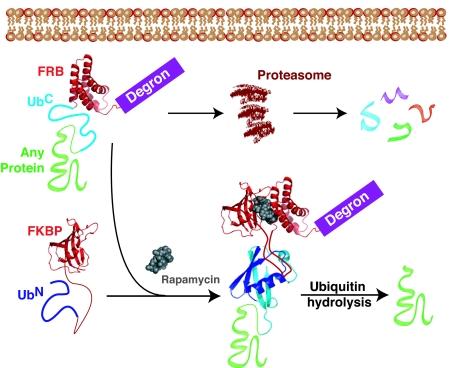
We describe here a posttranslational, small molecule-mediated technology for the manipulation of protein function. This system, split ubiquitin for the rescue of function (SURF), places the complementation of ubiquitin under the control of the FKBP/rapamycin/FRB heterotrimerization system (Fig. 1). Before complementation a protein of interest is targeted for destruction by the proteasome through the introduction of an N-terminal degron. Small-molecule-induced dimerization will result in ubiquitin complementation and folding followed by cleavage of the protein of interest, thereby releasing it from the degron and rescuing its function. This cleavage is particularly attractive as it restores the native state of the protein under investigation.
Fig. 1.
Overview of the SURF technology. A protein of interest is genetically fused to a degron that destines the protein for destruction by the proteasome. In between the protein under investigation and the degron are FRB and the C-terminal fragment of ubiquitin (UbC). Upon addition of rapamycin a complex is formed between the small molecule and another engineered protein containing FKBP and the N-terminal ubiquitin fragment (UbN). This complex creates a shunt away from degradation by allowing dimerization and complementation of the ubiquitin moiety through interactions between FKBP and FRB. After ubiquitin folding, a protease releases the protein of interest, effectively rescuing it from destruction.
Results
Destabilized Proteins Can Be Rescued by Ubiquitin Complementation and Hydrolysis.
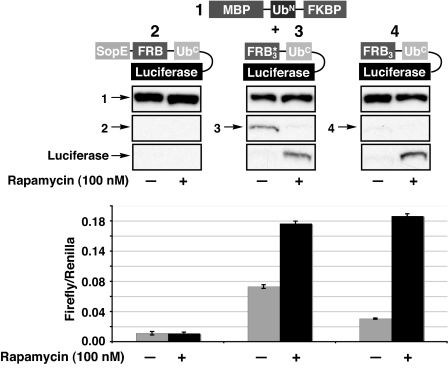
Our strategy relies on a successful balance between the rates of degradation and complementation. To examine this interplay, two general protein constructs were generated (Fig. 2). The first construct (1) contains maltose-binding protein (MBP) fused to the conditional complementation mutant of the N-terminal fragment of ubiquitin (Ile-13 to Ala, residues 1–37; see Materials and Methods) followed by FKBP. The second construct, bearing the protein of interest, has the following architecture: a degron, a single point mutant (W2101F) of FRB, here referred to simply as FRB, the C-terminal fragment of ubiquitin (residues 35–76), and the protein under investigation. Initially, we chose firefly luciferase as our model system, because of the ease and sensitivity of detection. We then chose three degrons, which we theorized would have different degradation kinetics, to examine the gross requirements for protein rescue [supporting information (SI) Table 1 for details]. The first degron (2) is a fragment (residues 1–100) of the protein SopE from Salmonella typhimurium, which has been shown to reduce the cellular half-life of proteins to which it is fused (18). The degron (3) is simply FRB* that contains the three point mutations K2095P, T2098L, and W2101F and has already seen utility as a degron (13), while construct 4 uses the single mutant of FRB mentioned above as the degron. In addition to their function as degrons, these FRB mutants allow the use of nontoxic derivatives of rapamycin as the chemical inducer of dimerization. Furthermore, we chose to use three copies of these FRB derivatives to increase the avidity for the FKBP/rapamycin complex, thus driving complementation. Previous studies in our laboratory have suggested that multiple copies of FRB act as a strong degron (10).
Fig. 2.
Evaluation of degrons. Constructs 1 and 2, 3, or 4 were transfected into HeLa cells, and the ability of rapamycin to rescue luciferase activity was measured by Western blotting (construct 1, anti-MBP; constructs 2–4, anti-HA) (Upper) and luciferase activity (Lower). All luciferase activity was normalized to an internal renilla luciferase transfection control and performed in triplicate.
HeLa cells were transfected with the complementary pairs of the above constructs and treated with rapamycin (100 nM) or DMSO vehicle for 36 h. Cleavage from the degron and rescue of luciferase function were then measured by Western blotting and luminescence activity, respectively (Fig. 2). When attached to the strong degron SopE (2), luciferase activity could not by rescued by the addition of rapamycin and no cleavage product was observed. To ensure that 2 was being expressed, cells were treated with the proteasome inhibitor ZL3VS (19), and luciferase activity was indeed observed (SI Fig. 8). From this result we concluded that the kinetics of degradation of SopE fusions must be faster than the kinetics of rapamycin-induced complementation and/or ubiquitin cleavage. Unlike the SopE fusion, detectable levels of constructs 3 and 4 were observed even in the absence of rapamycin (Fig. 2). Surprisingly, three copies of FRB resulted in a more robust degron than the corresponding FRB* construct, even though a previous study would suggest the inverse effect, at least for a single copy (13). Upon addition of rapamycin, cleavage of luciferase from both constructs 3 and 4 was very efficient, with 4 giving a better induction of activity. Therefore, the general architecture of construct 4 was used as a blueprint for all subsequent SURF-controlled proteins.
SURF Is Rapid and Dose-Dependent.
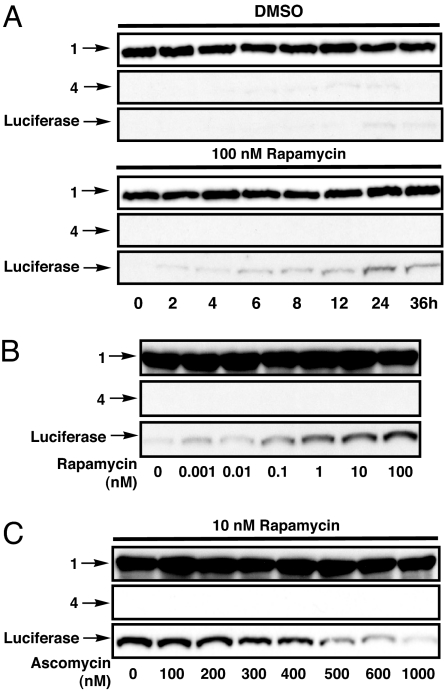
To determine the kinetics of rescue, HeLa cells were transfected with 1 and 4. After 24 h, rapamycin (100 nM) or DMSO was added to the cells and luciferase activity and cleavage were measured (SI Fig. 9A and Fig. 3A) at different time points. Appearance and activity of luciferase were observable in as little as 2 h and grew until a plateau was reached after 24 h of treatment. These kinetics seem to be governed by the rate of protein synthesis in the cell and compare well with other degradation technologies (13, 14).
Fig. 3.
Characterization of the SURF system. (A) HeLa cells were transfected with constructs 1 and 4, and rapamycin was added after 24 h expression. Luciferase cleavage was determined by Western blotting (construct 1, anti-MBP; construct 4, anti-HA) at different time points. (B) Cells were transfected and analyzed as in A to determine the effect of treatment with differing amounts of rapamycin for 30 h. (C) Cells were transfected and analyzed as in A to determine the effect of treatment with rapamycin and different amounts of ascomycin.
To explore the effects of varying rapamycin concentration, we transfected HeLa cells with our SURF constructs and treated them with different concentrations of rapamycin for 36 h. Both luciferase activity (SI Fig. 9B) and Western blotting analysis (Fig. 3B) confirmed that the amount of protein rescue could be altered by simply changing the rapamycin concentration. The rapamycin-mediated interaction of FRB and FKBP can also be reversed through the addition of another small molecule, ascomycin. Ascomycin is a competitive inhibitor of rapamycin as it binds to FKBP but prevents the formation of the ternary complex. We treated HeLa cells expressing the SURF constructs with rapamycin (10 nM) and differing amounts of ascomycin. Western blotting analysis (Fig. 3C) and luciferase assays (SI Fig. 9C) clearly show that ascomycin is able to competitively reduce the levels of rescue. Therefore, more precise control over levels of protein rescue can be achieved through the use of combinations of rapamycin and ascomycin concentrations. Additionally, by washing away rapamycin and adding ascomycin the SURF system should be effectively turned off. However, unlike other destabilized protein technologies where the protein of interest remains fused to the degron, the rate of disappearance of rescued protein depends on its intrinsic cellular stability.
SURF Is a General Method for Controlling Protein Levels.
We next explored the generality of SURF by applying it to members of three important classes of proteins: a protease, a kinase, and a transcription factor.
Caspase-3.
We chose caspase-3 as a candidate for SURF because, as a major executer of apoptosis, it is an important factor in cancer biology, with many types of cancer resistant to its effects (20, 21). Although small-molecule activators of apoptosis are available, a convenient method to titrate caspase-3 levels does not exist. Therefore, SURF control of caspase-3 may help illuminate the roles of this protease.
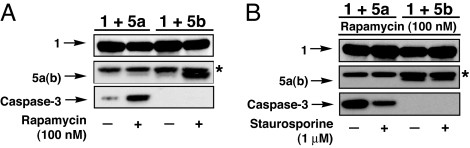
Toward this end, we generated a SURF construct containing human procaspase-3 (5a), as well as a mutant thereof (5b) bearing a proline as the residue immediately following ubiquitin, thereby precluding ubiquitin hydrolysis (SI Table 1). We hypothesized that 5b would be stabilized by rapamycin-induced dimerization with construct 1 and avoid degradation by using a mechanism similar to previously described systems (13). This “stabilization” construct should yield information concerning the importance of ubiquitin cleavage and subsequent generation of a native protein for proper functional rescue. Constructs 1 and 5a or 5b were then expressed in HeLa cells in the presence or absence of rapamycin (100 nM), and the rescue was analyzed by Western blotting (Fig. 4A). Procaspase-3 was rescued with similar efficiencies in both the cleavage (5a) and stabilization (5b) constructs. As a qualitative measure of the ability of these inducible caspase-3 systems to actively partake in an apoptotic cascade, HeLa cells expressing these SURF constructs were treated with rapamycin (100 nM) for 24 h to allow adequate rescue. At this time the cells were either treated with the apoptosis activator Staurosporine (1 μM) or the vehicle DMSO for 4 h, and the extent of proteolysis and procaspase processing was interrogated (Fig. 4B). Cleaved procapsase-3 was efficiently proteolyzed as judged by a significant decrease in the intensity of the procaspase-3 band. Stabilization construct 5b was also processed upon staurosporine treatment; however, the level of proteolysis appears to be somewhat reduced when compared with cleavage construct 5a.
Fig. 4.
SURF can rescue the protease caspase-3. HeLa cells were transfected with 1 and either a cleavage competent SURF construct 5a or stabilization mutant 5b (i.e., cleavage incompetent). (A) The capacity for rapamycin rescue (24 h treatment) was then examined by Western blotting (construct 1, anti-MBP; constructs 5a and 5b, anti-HA). (B) Processing of cleaved and stabilized caspase-3 during stauroporine (1 μM, 4 h treatment) induced apoptosis was analyzed by Western blotting (anti-HA). ∗ indicates background band.
v-Src.
Rous sarcoma virus Src (v-Src) is a constitutively active tyrosine kinase endowed with cellular transformative activity (22). v-Src is a quintessential oncogene and the first tyrosine kinase discovered, and its cellular homologue (c-Src) is involved in many cellular signaling pathways, many of which are still not fully understood (23). Thus, we chose v-Src as a model protein to explore SURF's utility for controlling kinase activities.
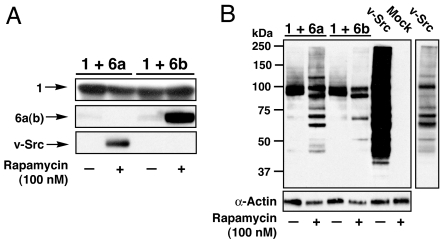
We generated a v-Src SURF construct and the corresponding stabilization mutant, 6a and 6b, respectively (SI Table 1). Constructs 1 and either 6a or 6b were coexpressed in SYF cells, which lack endogenous cellular Src kinase (24), and treated with rapamycin (100 nM) or DMSO vehicle (Fig. 5A). In the presence of rapamycin both the cleaved and stabilized forms of v-Src were efficiently rescued from degradation. The activity of these v-Src proteins against a wide array of substrates was then analyzed by a global antiphosphotyrosine Western blot (Fig. 5B). Despite the near-complete destruction of full-length 6a and 6b, two bands reacted strongly with the phosphotyrosine antibody in the absence of rapamycin. Furthermore, these bands were not present in mock-transfected cells. The full-length v-Src constructs (6a and 6b) could become self-phosphorylated and contribute to this signal. However, because of the intensity of these bands, it seems likely that additional phosphorylated proteins are present and account for most of the signal. Although the identities of these phospho-proteins are currently unknown, we speculate that they may be members of the ubiquitin–proteasome pathway involved in the recognition and trafficking of our degron. Additional studies will be needed to test this idea. More important for the current study is that when treated with rapamycin the cleavage construct 6a showed rescue of v-Src function against a variety of cellular targets. Furthermore, the pattern of phosphorylated proteins compares well to cells transfected with v-Src alone, albeit with much less intensity because of much larger amounts of v-Src in v-Src-transfected cells (data not shown). The stabilization construct 6b also showed rescue of v-Src activity. However, interestingly this construct appears to phosphorylate a much smaller pool of substrates (Fig. 5B, compare lanes 2 and 4). Thus the activity of v-Src appended to the degron is distinct from the cleaved v-Src. To further determine the functional consequences of our v-Src constructs we performed a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell proliferation assay (SI Fig. 10) (25). SYF cells were transfected with 1 and 6a or 6b, empty vector, or a plasmid containing v-Src. After treatment with rapamycin (100 nM) for 24 h, the cell proliferation rates were determined. Rescue of v-Src from the cleavage construct 6a resulted in a similar proliferation rate as cells expressing v-Src alone. In contrast, exposure of the stabilization mutant 6b did not result in an increase in cell proliferation over mock-transfected cells.
Fig. 5.
SURF can rescue the tyrosine kinase v-Src. SYF cells were tranfected with 1 and either the cleavage construct 6a or stabilization mutant 6b. (A) Rescue of v-Src was explored by Western blotting (construct 1, anti-MBP; constructs 6a and 6b, anti-v-Src; 24 h rapamycin treatment). (B) Recovered v-Src activity was examined by a global tyrosinephosphate Western blotting. The pattern of proteins phosphorylated by rescued v-Src was compared with cells tranfected with a v-Src plasmid (vSrc lanes, far right = lower exposure to allow comparisons).
Smad3.
Smad3 is a latent transcription factor involved in the TGF-β signaling pathway (26, 27). In the basal state, Smad3 is unphosphorylated and primarily cytoplasmic. Activation of the TGF-β receptor (which is composed of two receptor kinases TβR-I and TβR-II) results in phosphorylation of Smad3 on two serine residues at its extreme C terminus. Phospho-Smad3 then accumulates in the nucleus where it forms heterotrimers with a related protein, Smad4, and regulates gene transcription. Activated TGF-β receptors are also involved in Smad-independent signaling cascades (27). Thus, stimulation of cells with TGF-β can lead to pleiotropic signaling effects, making it difficult to understand the precise mechanistic basis of a phenotype. Therefore, SURF control of Smad3 might provide a convenient tool to isolate the canonical TGF-β pathway from other receptor-mediated signaling cascades.
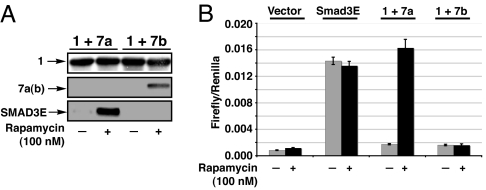
In preliminary studies, we generated a constitutively active version of Smad3 in which the two phospho-serine sites were replaced by glutamic acids. Based on structural studies (28), this double mutant was expected to mimic the acidic bis-phosphate surface in the active Smad3. Indeed, this construct was shown to drive Smad-dependent transcription in cultured cells in a receptor-independent manner (SI Fig. 11). We then generated the SURF construct 7a and its stabilization proline mutant 7b (SI Table 1). Treatment of HeLa cells expressing 1 and 7a or 7b with rapamycin (100 nM) resulted in escape from destruction for both the cleavage (7a) and stabilization (7b) constructs (Fig. 6A). Rescue of the cleavage construct resulted in a 10-fold increase in transcriptional output, as measured by the production of a luciferase reporter driven by the 3TP-binding element, when compared with cells treated with DMSO vehicle. This increase is consistent with an almost complete recovery of Smad3E activity as compared with cells transfected with Smad3E alone as a positive control (Fig. 6B). Isolation of Smad3E activity from endogenous Smads, by pretreatment with SB43152 (10 μM), an inhibitor of the TGF-β receptor kinase (29), resulted in a 15- to 20-fold induction upon rapamycin treatment (SI Fig. 12). In contrast to the cleavage construct, stabilization of the SURF proline mutant, 7b, yielded no increase in luciferase output, despite its cellular presence as judge by Western blotting.
Fig. 6.
SURF can rescue Smad3E activity. HeLa cells were transfected with 1 and either a cleavage-competent SURF construct 7a or stabilization mutant 7b. Rapamycin-stimulated rescue was then examined by Western blotting (construct 1, anti-MBP; constructs 7a and 7b, anti-HA; 24 h rapamycin treatment) (A) and luciferase production from a 3TP-luciferase-reporter plasmid (B). All luciferase activity was normalized to an internal renilla luciferase transfection control and performed in triplicate.
Kinetic Examination of Autoinduction of TGF-β by Smad3.
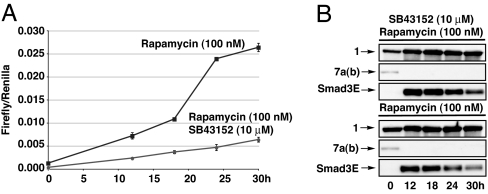
It has been known for almost two decades that TGF-β1 has the ability to activate its own translation and secretion (30), and more recently Smad3 has been identified as a key component in this feed-forward activation (31). In principle, SURF controlled Smad3 provides a window into the role of the canonical pathway in this feed-forward mechanism because it should be possible to compare the kinetics of Smad-dependent transcription with and without autocrine stimulation of the receptor. Accordingly, HeLa cells transfected with 1 and 7a were either left untreated and then induced with rapamycin (100 nM) or pretreated with SB43152 (10 μM) for 16 h and then induced with rapamycin (100 nM) and SB43152 (10 μM). Fresh media containing the appropriate small molecules were then added every 6 h. The extent of luciferase production from the Smad3 driven 3TP-luciferase reporter plasmid was then ascertained at different times (Fig. 7A).
Fig. 7.
Kinetic analysis of Smad3E transcriptional activation and rescue. HeLa cells were transfected with 1 and 7a and treated with rapamycin or a combination of rapamycin and SB43152. (A) Smad3E activity was determined by luciferase activity at different time points. All luciferase activity was normalized to an internal renilla luciferase transfection control and performed in triplicate. (B) Smad3E rescue was determined by Western blotting (anti-HA) at different time points.
In cells treated with the kinase inhibitor an almost linear increase in luciferase activity is seen throughout the entire time course. Noninhibited cells display the same linear increase at early time points; however, the rate of this increase is approximately double that of inhibited cells. One possibility for this increase is that Smad3 activity may affect parallel signaling pathways, thereby increasing the signal resulting from TGF-β already present in the growth media. Alternatively the difference may be an effect of treatment with rapamycin itself or off-target effects of SB43152. It has been shown that FKBP12 binds the TGF-β type I receptors blocking their phosphorylation by TβR-II (32). Rapamycin disrupts this interaction by blocking the binding surface on FKBP12 (9). Both a TβR-I mutant defective in FKBP12 binding and treatment of wild-type receptors with FKBP agonists generates modest increases in basal activity (see mock-transfected cells in Fig. 6B).
In contrast to the early time points, cells that were not treated with the kinase inhibitor exhibited a dramatic increase in luciferase activity that commenced between 18 and 24 h. This increase was not observed in cells treated with SB43152. We believe this is the result of increased secretion of TGF-β leading to autocrine receptor activation (29, 30), thereby revealing a timeframe for Smad3-mediated autoinduction. These data agree well with the published kinetics of TGF-β-induced expression of TGF-β mRNA (≈6 h) and secreted TGF-β (≈16 h) (30).
To further characterize this time course, samples from each time point were analyzed by Western blotting (Fig. 7B). In contrast to the build-up of luciferase (Fig. 3A) over the same timeframe, the amount of cleaved Smad3E decreased. This phenomenon may contribute to the linear kinetics of the transcriptional luciferase reporter (Fig. 7A) and could be the result of two possible scenarios. The efficiency of Smad3E rescue could decrease at the later time points. This scenario is unlikely, however, when taken in context with the luciferase-SURF data (Fig. 3A) and that fresh media containing rapamycin were added every 6 h to avoid possible rapamycin metabolism. Assuming that Smad3E rescue is constant at all time points, this decrease could be explained if Smad3E positively regulates its own destruction, perhaps by stimulation of the ubiquitin-proteasome pathway (33).
Discussion
In summary, we have developed a posttranslational technology for controlling protein function, SURF. We were able to observe ubiquitin complementation and cleavage of protein fusions when the FRB* and FRB degrons were used, but not with the strong SopE degron. In contrast to SopE, both FRB* and FRB can be stabilized by interations with rapamycin bound to FKBP. It is likely that this stabilized intermediate is essential, as it allows time for ubiquitin complementation and processing of the protein fusion. Luciferase was used as a model protein to examine the kinetics and tunability of SURF. Like other degradation technologies (13, 14), the kinetics of SURF are limited to the rate of protein synthesis in the cell. SURF is highly responsive to the concentration of the small-molecule activator rapamycin, and complementation and cleavage can be abrogated by the addition of another small-molecule ascomycin. SURF should therefore allow for highly tunable protein rescue by simple adjustment in rapamycin and ascomycin concentrations. Furthermore, the use of the W2101F mutant of FRB is permissive for nontoxic rapamycin analogues, allowing for the possible extension of this technology to living systems. Unlike other degradation technologies, the protein of interest is released from the degron in SURF. Therefore its native structure and stability is restored, which is attractive for the interrogation of protein function and turnover.
We used SURF to control the levels and activity of members of three important classes of protein: a protease, a kinase, and a transcription factor. Rescue of caspase-3, a cysteine protease involved in apoptosis and important in many types of cancer, was observed, and the cleaved native caspase-3 was apparently processed by upstream caspases during staurosporine-induced apoptosis. The tyrosine kinase activity of v-Src was also efficiently recovered. SURF was also able to rescue Smad3E, effectively restoring transcriptional activity upon the addition of rapamycin. In the midst of the complicated TGF-β signaling pathway, we were able to isolate Smad3-driven transcription by using two pharmacological agents, rapamycin and SB43152. Kinetic evaluation of Smad3E regulated transcription allowed us to examine the timeframe of Smad3's role in the TFG-β feed-forward loop. Further analysis of this system with mutant TGF-β receptors may allow us to examine the effects of Smad3, as well as, rapamycin treatment on this important signaling pathway. In addition, analysis of Smad3E rescue over time uncovered the possibility that Smad3 may regulate its own degradation. Although we can only speculate as to the mechanism of this negative feedback, we do note that the E3 ubiquitin ligase complex ROC1-SCFFbw1a is responsible for the ubiquitin-dependent degradation of Smad3 in a TGF-β-dependent manner (33). Although outside the scope of the current work, which focuses on the SURF methodology, the possibility that active Smad3 may up-regulate its own destruction via the ubiquitin-proteasome pathway merits further study.
In the case of the v-Src, Smad3E, and, to a lesser extent, caspase-3 stabilization mutants, which rescued their proteins through stabilization and not cleavage, limited or altered activity was rescued even though protein levels did increase. This finding highlights the complementary utility of SURF to existing degradation technologies that require permanent fusion of degron domains to proteins of interest. SURF represents an attractive alternative when the protein of interest is known to take part in protein–protein interactions or multimeric complexes as rescue is inextricably linked to generation of the native protein. In the future this cleavage-dependent rescue technology could be expanded to include other methods of protein regulation such as mislocalization or autoinhibition (34).
Materials and Methods
Plasmid Construction.
Constructs 1-7 were prepared by using standard molecular cloning techniques. Full details are given in SI Text.
Cell Culture Experiments.
HeLa and SYF cell lines were cultured in DMEM (Gibco, Carlsbad, CA) supplemented with 10% FBS (Sigma, St. Louis, MO) at 37°C and 5% CO2 according to standard procedures. Cells were transfected by using FuGENE-6 (Roche Applied Science, Indianapolis, IN). Plasmid DNA [250 ng of SURFn and SURFc constructs and 25 ng of reporter construct and/or 25 ng of the renilla control pRL-SV40 (Promega, Madison, WI) per 1 ml, 12-well plate] was mixed in a 1:3 ratio with the FuGENE reagent in DMEM and was then applied to cells growing in DMEM/10% FBS. In the time-course experiments, transfection was allowed to proceed for 24 h before replacement with fresh media containing DMSO (Sigma; 0.1% final concentration) or with rapamycin (Sigma; 100 μM in DMSO, 100 nM/0.1% DMSO final concentration) for the indicated time points. For all other experiments the above transfection mixture was applied to cells growing in DMEM/10% FBS containing DMSO (0.1% final concentration), rapamycin (Sigma) and/or ascomycin (Sigma) in DMSO (0.1%–0.2% final concentration) where appropriate. After 12 h, fresh medium for continued drug treatment was added except for the Smad3E time course where fresh medium was added every 6 h.
Luciferase Measurements.
After the indicated drug treatments, cells were washed twice with PBS and lysed with 1× passive lysis buffer (Promega) for 15 min at room temperature. After centrifugation for 10 min at 10 × g, luciferase readings were performed in a Sirius luminometer (Berthold, Nashua, NH) using a Promega dual-luciferase assay kit.
Western Blotting.
Detailed Western blotting procedures are given in SI Text.
MTT Cell Proliferation Assay.
The MTT cell proliferation assay was purchased from American Tissue Culture Collection (Manassas, VA) and performed per the manufacturer's instructions. Briefly, SYF cells were tranfected with appropriate plasmids as above. After 24 h to allow for protein expression, the cells were trypsinized and counted, and ≈1 × 104 were plated in 96-well plates in DMEM containing 10% FBS and rapamycin (100 nM) or DMSO where appropriate. After an additional 24 h to allow for rescue, the MTT cell proliferation assay was performed.
Supplementary Material
Acknowledgments
We thank J. Massagué (Memorial Sloan-Kettering Cancer Center, New York) for pHA-Smad3, p3TP-luc, and pSmad3; M. E. Hahn for helpful comments on the manuscript; E. Stebbins (The Rockefeller University) for Salmonella Ty. genomic DNA; and I. Martinez and P. Cole (Johns Hopkins Unversity, Baltimore, MD) for an antiphosphotyrosine Western blotting procedure. This work was supported by National Institutes of Health Grants EB001991 and GM072015 (to T.W.M.), an American Cancer Society postdoctoral fellowship (to M.R.P.), and a Howard Hughes Medical Institute fellowship (to E.C.S.).
Abbreviations
- FKBP
FK506-binding protein
- FRB
FKBP12-rapamycin binding
- SURF
split ubiquitin for the rescue of function
- MBP
maltose-binding protein
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700816103/DC1.
References
- 1.Ryding AD, Sharp MG, Mullins JJ. J Endocrinol. 2001;171:1–14. doi: 10.1677/joe.0.1710001. [DOI] [PubMed] [Google Scholar]
- 2.Novina CD, Sharp PA. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 3.Shogren-Knaak MA, Alaimo PJ, Shokat KM. Annu Rev Cell Dev Biol. 2001;17:405–433. doi: 10.1146/annurev.cellbio.17.1.405. [DOI] [PubMed] [Google Scholar]
- 4.Bain J, McLauchlan H, Elliott M, Cohen P. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banaszynski LA, Wandless TJ. Chem Biol. 2006;13:11–21. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber SL. Bioorg Med Chem. 1998;6:1127–1152. doi: 10.1016/s0968-0896(98)00126-6. [DOI] [PubMed] [Google Scholar]
- 7.Choi J, Chen J, Schreiber SL, Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 8.Liberles SD, Diver ST, Austin DJ, Schreiber SL. Proc Natl Acad Sci USA. 1997;94:7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockwell BR, Schreiber SL. Chem Biol. 1998;5:385–395. doi: 10.1016/s1074-5521(98)90072-2. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz EC, Saez L, Young MW, Muir TW. Nat Chem Biol. 2007;3:50–54. doi: 10.1038/nchembio832. [DOI] [PubMed] [Google Scholar]
- 11.Schneekloth JS, Jr, Fonseca FN, Koldobskiy M, Mandal A, Deshaies R, Sakamoto K, Crews CM. J Am Chem Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 12.Janse DM, Crosas B, Finley D, Church GM. J Biol Chem. 2004;279:21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- 13.Stankunas K, Bayle JH, Gestwicki JE, Lin YM, Wandless TJ, Crabtree GR. Mol Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- 14.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 16.Baker RT, Tobias JW, Varshavsky A. J Biol Chem. 1992;267:23364–23375. [PubMed] [Google Scholar]
- 17.Johnsson N, Varshavsky A. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubori T, Galan JE. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 19.Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh H. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 21.Riedl SJ, Shi Y. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 22.Frame MC, Fincham VJ, Carragher NO, Wyke JA. Nat Rev Mol Cell Biol. 2002;3:233–245. doi: 10.1038/nrm779. [DOI] [PubMed] [Google Scholar]
- 23.Yeatman TJ. Nat Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 24.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 25.Ohno M, Abe T. J Immunol Methods. 1991;145:199–203. doi: 10.1016/0022-1759(91)90327-c. [DOI] [PubMed] [Google Scholar]
- 26.Siegel PM, Massague J. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Massague J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 28.Chacko BM, Qin B, Correia JJ, Lam SS, de Caestecker MP, Lin K. Nat Struct Biol. 2001;8:248–253. doi: 10.1038/84995. [DOI] [PubMed] [Google Scholar]
- 29.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 30.Van Obberghen-Schilling E, Roche NS, Flanders KC, Sporn MB, Roberts AB. J Biol Chem. 1988;263:7741–7746. [PubMed] [Google Scholar]
- 31.Zhang M, Fraser D, Phillips A. Am J Pathol. 2006;169:1282–1293. doi: 10.2353/ajpath.2006.050921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YG, Liu F, Massague J. EMBO J. 1997;16:3866–3876. doi: 10.1093/emboj/16.13.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuchi M, Imamura T, Chiba T, Ebisawa T, Kawabata M, Tanaka K, Miyazono K. Mol Biol Cell. 2001;12:1431–1443. doi: 10.1091/mbc.12.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mootz HD, Blum ES, Muir TW. Angew Chem Int Ed. 2004;43:5189–5192. doi: 10.1002/anie.200460941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.