Abstract
Nedd4-binding partner-1 (N4BP1) has been identified as a protein interactor and a substrate of the homologous to E6AP C terminus (HECT) domain-containing E3 ubiquitin–protein ligase (E3), Nedd4. Here, we describe a previously unrecognized functional interaction between N4BP1 and Itch, a Nedd4 structurally related E3, which contains four WW domains, conferring substrate-binding activity. We show that N4BP1 association with the second WW domain (WW2) of Itch interferes with E3 binding to its substrates. In particular, we found that N4BP1 and p73α, a target of Itch-mediated ubiquitin/proteasome proteolysis, share the same binding site. By competing with p73α for binding to the WW2 domain, N4BP1 reduces the ability of Itch to recruit and ubiquitylate p73α and inhibits Itch autoubiquitylation activity both in in vitro and in vivo ubiquitylation assays. Similarly, both c-Jun and p63 polyubiquitylation by Itch are inhibited by N4BP1. As a consequence, genetic and RNAi knockdown of N4BP1 diminish the steady-state protein levels and significantly impair the transcriptional activity of Itch substrates. Notably, stress-induced induction of c-Jun was impaired in N4BP1−/− cells. These results demonstrate that N4BP1 functions as a negative regulator of Itch. In addition, because inhibition of Itch by N4BP1 results in the stabilization of crucial cell death regulators such as p73α and c-Jun, it is conceivable that N4BP1 may have a role in regulating tumor progression and the response of cancer cells to chemotherapy.
Keywords: p53, protein–protein interaction, transcription, ubiquitylation, WW domain
The conjugation of ubiquitin to protein substrates has emerged as a fundamental mechanism for regulation of many cellular activities. The specificity of the ubiquitylation reaction is conferred by the E3 ubiquitin-protein ligases (E3s), which mediate the transfer of the ubiquitin molecule from E2 ubiquitin-conjugating enzymes (E2) to substrates. Ubiquitylation controls turnover and abundance of proteins by targeting them for proteasomal or lysosomal degradation (1–4).
HECT (homologous to E6AP C terminus) domain-containing proteins are a major class of E3s, sharing a common general modular structure, with a Ca+2/lipid-binding (C2) domain involved in membrane targeting, multiple WW protein-interacting modules conferring substrate binding activity, and a HECT domain, coordinating with the E2 and providing the catalytic E3 activity (5). The reaction cycle of the HECT domain-containing E3s consists of three steps: binding to an E2 enzyme, loading the ubiquitin on themselves, and transferring ubiquitin to the target protein (1). The prototype member of the HECT family of E3 is Nedd4, mainly implicated in the regulation of fluid and electrolyte homeostasis by controlling the surface abundance of the epithelial cell sodium channel (ENaC) subunits (6, 7).
By carrying out a yeast two-hybrid screen of a midgestation mouse embryo cDNA library, we have recently identified Nedd4-binding partner-1 (N4BP1) as a developmentally expressed protein interactor and monoubiquitylation substrate of Nedd4 (8). We now know that N4BP1 can also undergo Nedd4-mediated polyubiquitylation and proteasomal degradation (P. Sharma and M.R.K., unpublished manuscript).
The HECT E3 Itch was originally identified as a gene disrupted in the non-agouti-lethal 18H mice, or Itchy mice, which suffer from severe immune and inflammatory defects (9). A number of Itch targets are central players or regulators of the immune response, including c-Jun and JunB (10, 11).
Itch E3 activity is also required for ubiquitylation and proteasomal degradation of p73 and p63 (12, 13), two structural homologues of the tumor-suppressor transcription factor p53. The p73 and p63 gene loci encode for several distinct isoforms generated by C-terminal alternative splicing (14) or through the usage of an alternative promoter (ΔN variants) (15). Itch-mediated regulation of p73 and p63 protein stability is selective for those isoforms containing the C-terminal proline-rich motif, such as the α- and β-variants (12, 13).
Of note, several Itch targets are proapoptotic molecules displaying tumor-suppressive functions. The p53 family members and c-Jun promote apoptosis in response to genotoxic stress, such as alkylating agents or short-wavelength UV radiation (16–18).
Itch-negative regulators have remained elusive. In this paper, we show that N4BP1 binds to the WW domains of Itch and inhibits its ubiquitylation activity. As a consequence, N4BP1 stabilizes the Itch targets p73α and c-Jun and increases their transcriptional activity.
Results
N4BP1 Interacts with the WW Domain-Containing Central Region of Itch.
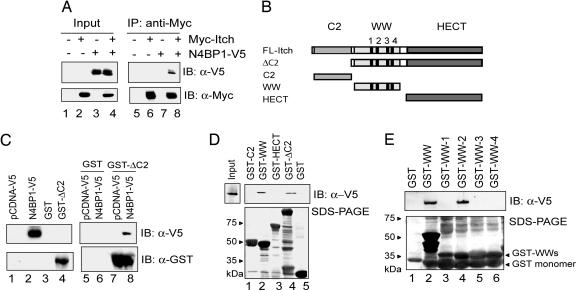
Given the common modular architecture shared by the HECT E3s, the interaction of Nedd4 and N4BP1 prompted us to investigate a possible association between N4BP1 and Itch. Coimmunoprecipitation (Co-IP) experiments revealed that N4BP1 indeed binds to Itch (Fig. 1A, lane 8). To ascertain whether the observed interaction was direct, the N4BP1/Itch association was tested in in vitro binding assays. GST pull-down assays showed that N4BP1 directly and specifically interacts with the GST-ItchΔC2 fusion protein (Fig. 1 B and C, lane 8).
Fig. 1.
N4BP1 physically binds to the WW domain-containing central region of Itch. (A) For Co-IP experiments, HCT-116 cells were transiently cotransfected with expression vectors encoding Myc-tagged Itch (Myc-Itch) and V5-tagged N4BP1 (N4BP1-V5). After 24 h, cells were lysed, and N4BP1/Itch immunocomplexes were analyzed by IP using anti-Myc followed by IB with anti-V5 antibody. (B) A schematic representation of Itch deletion fragments: the ΔC2 protein (73 kDa) harboring a deletion of the C2 domain, the C2 domain (18 kDa), the central WW domain-containing region (24 kDa), and the catalytic HECT module (40 kDa). (C) (Right) A bacterially purified GST-ItchΔC2 fusion protein was used for in vitro pull-down assays to avoid solubility problems. GST-ItchΔC2 was bound to glutathione-Sepharose beads. N4BP1-V5 was in vitro translated. Protein complexes were detected by IB with anti-V5 antibody. (Left) Inputs. (D) (Upper) Free GST control protein or the indicated GST-fused Itch fragments were loaded on glutathione-Sepharose beads, and the resin was then incubated with in vitro-translated N4BP1-V5. Resulting complexes were resolved via IB by using an anti-V5 antibody. (Lower) Filter was subsequently subjected to Coomassie staining. (E) (Upper) Individual WW domains of Itch expressed as GST fusion proteins were exposed to in vitro-translated N4BP1-V5. Protein complexes were analyzed as in C and D. (Lower) The filter was subsequently subjected to Coomassie staining.
To map the region responsible for N4BP1-Itch association, we generated GST-Itch deletion fragments, schematically depicted in Fig. 1B, and examined their ability to bind to N4BP1. The GST-fusion protein containing all four WW domains of Itch showed specific interaction with N4BP1 (Fig. 1D, lane 2). To further characterize the association between Itch and N4BP1, the four GST-WW domains of Itch were individually tested for binding to N4BP1. As shown in Fig. 1E, only WW domain 2 (WW2) displayed the ability to interact with N4BP1.
The WW domains mediate ligase–substrate associations through the interaction with a variety of Pro-based motifs, preferentially, PPXY (PY) (19). Although, human N4BP1 does not contain canonical PY or PPLP motifs, atypical interactions of WW domains with either noncanonical Pro-rich motifs, such as the Pro-Arg motif (20), or unrelated modular domains (21) have been reported. N4BP1 has at least two atypical Pro-rich regions that may be potential candidates for the binding site to the WW domain of Itch. Interestingly, the upstream Pro-rich motif of N4BP1 is surrounded by an Arg residue. Similarly to the WW domains, the Pro binding module SH3 displays unusual interaction modes, including the requirement of Pro and Arg residues in the target sequence or binding to Pro-independent motifs, such as the Arg- and Lys-rich motif (22). Due to the similarities in the mechanism of ligand recognition used by WW and SH3 domains, we can speculate that the binding of Itch and N4BP1 may depend on interactions between the WW2 domain and either a noncanonical Pro-rich sequence or a non-Pro-based motif.
Because we have previously shown that N4BP1 serves as a substrate for the E3 ubiquitin ligase activity of Nedd4 (8), we sought to test whether N4BP1 can also function as an ubiquitylation substrate for Itch. Unexpectedly, unlike Nedd4, Itch did not affect N4BP1 cellular ubiquitylation levels and was unable to catalyze N4BP1 polyubiquitylation in in vitro ubiquitylation reactions [supporting information (SI) Fig. 5].
N4BP1 Inhibits in Vivo Protein Ubiquitylation of Itch Substrates.
To determine the functional relevance of the interaction between N4BP1 and Itch, we sought to ascertain whether N4BP1 could regulate the ubiquitylation levels of different Itch substrates.
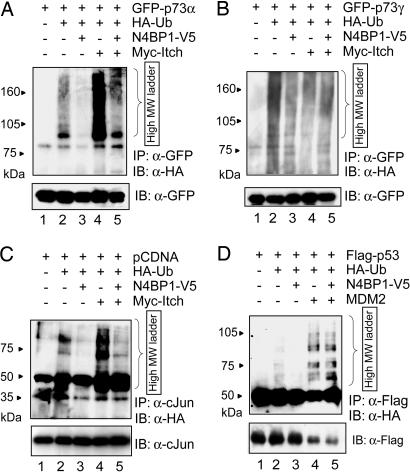
We observed that Itch-induced ubiquitylation of p73α was significantly inhibited upon coexpression of N4BP1 (Fig. 2A, lanes 4 and 5). Similarly, Itch-mediated ubiquitylation of other PY motif-proficient p73 isoforms was strongly reduced by N4BP1 (SI Fig. 6 and data not shown). In addition, the ability of Itch to polyubiquitylate the two α-variants of the p63 family member was impaired by N4BP1 (SI Fig. 6). To rule out the possibility that N4BP1 nonspecifically affects the ubiquitylation of Itch substrates, the effect of N4BP1 was tested on the p73γ isoform lacking the PY motif. The basal ubiquitylation of p73γ was not affected significantly by N4BP1 (Fig. 2B), suggesting that N4BP1 acts via an Itch-dependent mechanism. N4BP1 also strongly diminished Itch-mediated ubiquitylation of endogenous c-Jun (Fig. 2C, lanes 4 and 5), further confirming that the N4BP1 inhibitory effect arises from specific blocking of Itch-mediated ubiquitylation.
Fig. 2.
N4BP1 selectively inhibits ubiquitylation of Itch substrates. H1299 cells were transfected with GFP-p73α (A), GFP-p73γ (B), PCDNA empty vector (C), or Flag-p53 (D), along with HA-tagged ubiquitin (HA-Ub), their specific E3, Itch (A–C), or MDM2 (D) in the absence or presence of N4BP1. Twenty-four hours later, cells were treated with 40 μM proteasome inhibitor MG-132 for 1 h before harvesting. Cell lysates were subjected to IP with anti-GFP (A and B), anti-c-Jun (C), or anti-Flag (D) antibodies under denaturing conditions. Substrate-ubiquitin immunocomplexes were analyzed by anti-HA IB analysis and subsequently probed with anti-GFP (A and B), anti-c-Jun (C), or anti-Flag antibodies (D).
To further validate the specificity of the functional interaction of Itch and N4BP1, we assessed the effect of N4BP1 against the structurally distinct RING finger E3 MDM2. As a substrate for the E3 ubiquitin ligase activity of MDM2, we used p53 (23). Fig. 2D shows that no alteration of p53 ubiquitylation by MDM2 was observed in the presence of N4BP1.
Ablation of N4BP1 Increases Protein Levels and Transcriptional Activity of Itch Protein Substrates.
We then investigated whether N4BP1 would affect p73α and c-Jun protein levels. Overexpression of N4BP1 promoted the accumulation of p73α as well as of endogenous c-Jun protein levels (SI Fig. 7). In addition, N4BP1 increased their half-life from ≈6 to >9 h and from 2 to 5 h for p73 and c-Jun, respectively (SI Fig. 7). Unlike p73α and consistent with an Itch-dependent mechanism, the decay rate of p53 (SI Fig. 7) and p73γ (SI Fig. 8) was unaffected by N4BP1.
We next sought to determine the contribution of endogenous N4BP1 to the regulation of Itch substrate protein stability. We recently have generated N4BP1 knockout mice (R.M. and M.R.K., unpublished manuscript) and have isolated primary mouse embryonic fibroblasts (MEFs) from N4BP1−/− mutants for this study. N4BP1 loss indeed significantly diminished p73 endogenous protein levels in MEFs (Fig. 3A). In keeping with the results shown in Fig. 2D, p53 protein levels were essentially unaltered in N4BP1−/− MEFs compared with their wild-type counterpart.
Fig. 3.
Genetic and RNAi-mediated knockdown of N4BP1 diminish protein levels and transcriptional activation of Itch substrates. (A) Cell extracts from N4BP1+/+ and N4BP1−/− MEFs were examined by IB using anti-p73 and anti-p53 antibodies. The same blots were reprobed with anti-N4BP1 and anti-β-tubulin antibodies. (B) N4BP1+/+ and N4BP1−/− MEFs were UV-treated (60 J/m2) and lysed at 0, 4, 8, and 24 h after irradiation. Total extracts were probed with anti-c-Jun and anti-β-actin antibodies. Levels of c-Jun are represented as fold of induction over untreated controls. (A and B) Transcriptional activation of p73α (A) and c-Jun (B) in N4BP1+/+ and N4BP1−/− MEFs was measured as described in SI Methods. (C and D) HCT-116 (3) cells were transfected with control Lamin A/C or N4BP1 (N4BP1_3 HP) siRNAs. Cells were harvested 72 h after transfection. Cellular lysates were analyzed by IB using anti-p73 and anti-p53 (C) or anti-cJun and anti-JunB antibodies (D). Lamin A/C and β-actin are shown as transfection and loading control, respectively.
JNK-mediated phosphorylation of c-Jun prevents its ubiquitin-dependent degradation, thus contributing to its transcriptional activation after cellular stress, such as UV irradiation (24). Interestingly, we found that UV-induced stabilization of c-Jun was significantly hampered in N4BP1−/− MEFs (Fig. 3B).
Similarly, RNAi-mediated knockdown of N4BP1 in human cells, by using two different siRNA oligos, resulted in decreased expression of p73 (Fig. 3C and data not shown) and Jun family members, at both steady state and after UV irradiation (Fig. 3D and data not shown). Altogether, these observations demonstrate that suppression of endogenous N4BP1 promotes increased ubiquitylation and protein degradation of Itch substrates.
In keeping with these findings, we observed that transcriptional activation of p73α and c-Jun was impaired in both N4BP1−/− MEFs (Fig. 3 A and B) and human cells upon RNAi-mediated silencing of N4BP1 (data not shown). Furthermore, overexpression of N4BP1 specifically enhanced the transactivation ability of p73α and c-Jun, but not of p73γ- and p53 (SI Fig. 9 and data not shown). These data suggest that N4BP1 is a previously unrecognized regulator able to finely tune p73 and c-Jun transcriptional function.
Itch undergoes self-ubiquitylation in vivo (10, 11). Similarly to its substrates, Itch autoubiquitylation activity was strongly inhibited by coexpression of N4BP1 (SI Fig. 10). The catalytically inactive Itch mutant (Itch-C830A) was only slightly ubiquitylated in vivo, likely because of the endogenous Itch E3 activity, which was also inhibited by N4BP1 (SI Fig. 10). However, the steady-state levels and the decay rate of endogenous Itch were not affected by N4BP1 in different cell lines (SI Fig. 10 and data not shown) nor were they altered in N4BP1 knockout MEFs and on N4BP1 RNAi (Fig. 3D and data not shown). These findings suggest a nonproteolytic regulatory function for Itch self-ubiquitylation.
N4BP1 Competes with Itch Substrates for Binding to the WW2 Domain.
Because the inhibitory action exerted by N4BP1 is not achieved via an alteration of Itch protein stability (Fig. 3D and SI Fig. 10), we have explored alternative molecular mechanisms. The inability of N4BP1 to serve as a substrate for the E3 ubiquitin ligase activity of Itch (SI Fig. 5) also rules out the possibility that N4BP1 may compete with other substrates for Itch-catalyzed polyubiquitylation.
Although minimally diffusely distributed in both the nucleus and the cytoplasm, N4BP1 is typically localized in discrete nuclear speckles (8). This observation prompted us to test whether the N4BP1/Itch interaction would alter Itch subcellular localization, thus sequestering Itch away from its protein substrates. Nevertheless, coexpression of N4BP1 and Itch did not change the diffuse cellular distribution of Itch (data not shown).
Lastly, we explored the possibility that N4BP1 may directly interfere with the ability of Itch to ubiquitylate its protein substrates. In in vitro ubiquitylation assays, we found that Itch self-ubiquitylation was robustly and dose-dependently repressed by the addition of N4BP1 (Fig. 4A). This finding and the observation that N4BP1 directly interacts with the WW domains of Itch (Fig. 1 D and E), strongly suggest that N4BP1 may decrease ubiquitylation of Itch and its protein targets by competing with the substrates for Itch binding. To explore the possibility of a titration mechanism, we tested the ability of N4BP1 to affect the Itch/p73 interaction in in vitro competition assays and found that N4BP1 was indeed able to interfere with the formation the p73–Itch complex (Fig. 4B, lane 4 vs. lane 6). To ascertain and validate the competition mechanism, we investigated the affinity of p73α for binding to the single WW domains of Itch. Intriguingly, we found that, similarly to N4BP1, p73α interacted with WW2 of the E3 (Fig. 4C), indicating that the overlapping of binding sites is responsible for N4BP1 preventing ubiquitylation of Itch protein substrates.
Fig. 4.
N4BP1 directly inhibits protein ubiquitylation through competition with substrates for Itch binding. (A) In vitro self-ubiquitylation activity of ΔC2Itch was tested in the absence (lane 2) or in the presence (lanes 5–7) of increasing doses of bacterially purified GST-N4BP1. The catalytic-defective ΔC2Itch-C830A mutant (lane 1) and free GST (lane 4) were used as negative controls. Reaction mixtures were analyzed by IB with anti-Ub and anti-GST antibodies. (B) (Upper) Competition experiments were conducted using GST-ItchΔC2 fusion protein bound to glutathione-Sepharose resin and incubated with a constant amount of in vitro-translated HA-p73α and N4BP1-V5 at 6-fold excess. The beads were washed and subjected to SDS/PAGE and IB with anti-p73 and anti-N4BP1 antibodies. (Lower) Binding of p73 to Itch was normalized to the amount of glutathione-Sepharose resin as assessed by anti-GST IB. (C) GST fusion proteins of the individual WW domains of Itch were bound to glutathione-Sepharose beads and incubated with in vitro-translated HA-p73α. Protein complexes were analyzed by IB analysis using anti-p73 antibody.
Discussion
To gain insight into the regulatory mechanisms of Itch E3 ubiquitin ligase activity, we explored its potential interaction with N4BP1, a molecular partner and ubiquitylation substrate of Nedd4. Our findings highlight the importance of the N4BP1/Itch interaction for the functional regulation of Itch target substrates.
The C-terminal WW domains (WW3, WW4) of Nedd4 are known to directly mediate the association with the substrate, providing high-affinity binding to the proline-rich motifs, whereas a regulatory function has been ascribed to the first WW domains (25, 26). However, the binding affinities of Itch substrates for its WW domains have not been elucidated yet.
Our results demonstrated that N4BP1 specifically associates with the WW domain-containing central region of Itch and point out that both N4BP1 and p73α interact with the WW2 domain of Itch. As a consequence, N4BP1 strongly inhibits Itch-catalyzed polyubiquitylation by preventing the interaction with its substrates, thereby reducing the transfer of ubiquitin molecules to Itch protein targets. Hence, N4BP1 interferes with the proteolytic pathway of both p73α and c-Jun, leading to protein stabilization and increased transcriptional activity. Interestingly, a similar but nonoverlapping competition mechanism, which is based on the ability of the adapter protein Yes-associated protein 1 (Yap1) to recruit p73 via its WW domains, has been recently described (27). Binding of Yap1 to the PY motif of p73 prevents access to Itch and results in p73 protein stabilization. Similarly, binding of the WW domain-containing oxidoreductase (WWOX) to the same PY motif antagonizes the coactivation ability of Yap to mediate p73-mediated transcription (28).
The competition mechanism and the ability of the catalytically defective mutant, Itch C830A, to be in vivo ubiquitylated by endogenous Itch strongly suggest that the autoubiquitylation occurs through an in trans reaction. Interestingly, Itch C830A can be properly in vitro polyubiquitylated by the wild-type E3 (data not shown), demonstrating that Itch catalyzes the transfer of ubiquitin from its catalytic cysteine to a nearby E3 molecule. In line with our observations, Gallagher et al. (11) have recently demonstrated that both the Itch WW and HECT domains have the ability to interact with the full-length protein. Although the WW–HECT-mediated association is engaged in negative regulatory intramolecular interactions, a fraction of Itch may also be available for intermolecular interactions as well as for substrate binding. Alternatively, intramolecular interactions could be replaced by intermolecular interactions in response to cellular stress.
Developing therapeutic approaches for cancer treatment targeting protein degradation is currently an attractive research avenue. An effective therapeutic approach would be targeting specific components of the ubiquitin system, such as the E3 enzymes. For instance, the inhibition of the E3 activity of Itch could be used to increase chemosensitivity of tumor cells by selectively up-regulating p73, p63, and c-Jun basal protein levels. The importance of Itch down-regulation becomes evident in response to DNA damage-based chemotherapeutic drugs, in which reduction of its protein levels leads to p73α stabilization and increased proapoptotic function (ref. 12 and our unpublished observations). The identification of N4BP1 as a specific inhibitor of Itch-mediated ubiquitylation of tumor suppressor molecules may provide an alternative means to selectively block Itch function and thereby regulate tumor progression and the response of cancer cells to chemotherapy.
It remains a challenge for future research to elucidate the relative contribution of N4BP1 on Itch-mediated regulation of protein proteasomal degradation in both physiological and pathophysiological conditions.
Materials and Methods
Cell Culture and Transfection Conditions.
MEFs were derived from 13.5-day-old N4PB1 wild-type and knockout embryos (R.M. and M.R.K., unpublished manuscript). Cells were maintained in DMEM supplemented with 100 μg/ml penicillin and streptomycin and 10% FBS (Sigma, St. Louis, MO) in 5% CO2 at 37°C. Transfections were performed using Effectene (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Immunoblot Analysis and IP.
Cells were lysed in RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) (29) containing protease and phosphatase inhibitors (Roche, Indianapolis, IN). Immunoblot (IB) analysis was performed under standard procedures (29). The following antibodies were used: rabbit polyclonal anti-N4BP1 (8), monoclonal anti-p73 (Abcam, Cambridge, MA), anti-p53 (clone DO-1; Santa Cruz Biotechnology, Santa Cruz, CA), anti-HA (clone H11; Covance, Richmond, CA); rabbit polyclonal anti-GFP (Clontech, Mountain View, CA); anti-Flag (clone M2; Sigma); goat polyclonal anti-GST (Promega, Madison, WI); monoclonal anti-Itch (BD Biosciences, San Jose, CA); monoclonal anti-Myc (Cell Signaling, Beverly, MA); monoclonal anti-c-Jun (BD Biosciences), anti-JunB (clone N-17; Santa Cruz Biotechnology), monoclonal anti-V5 (Invitrogen, Carlsbad, CA). For IP, cells were lysed in Nonidet P-40 lysis buffer (29). Samples were precleared with protein A/G-Sepharose beads and then immunoprecipitated for 2 h at 4°C with 0.5–1 μg per sample of the appropriate antibodies preadsorbed on protein A/G-Sepharose beads. Immunocomplexes were washed four times in lysis buffer and eluted by boiling in SDS loading buffer.
GST-Pulldown Assays.
GST-tagged recombinant proteins were purified using glutathione beads (GE Healthcare, Piscataway, NJ). V5-tagged N4BP1 (N4BP1-V5) and HA-tagged p73 (HA-p73) were produced in vitro using the T7-Rabbit reticulocyte system (Promega). Binding reactions typically contained 1–10 μg of the Sepharose-immobilized GST fusion proteins or GST and 3–15 μl of the in vitro translated protein in binding buffer (20 mM Tris·HCl, pH 7.5/200 mM NaCl/0.1% Triton X-100). The reactions were incubated with gentle inversion for 1 h at 4°C, followed by five washes with binding buffer. Complexes were resolved by SDS/PAGE and probed with the indicated antibodies.
In Vivo Ubiquitylation Assay.
In vivo ubiquitylation assays were performed as described in ref. 29. Briefly, cells were transiently transfected with indicated expression vectors for 24 h. Cells were treated with 40 μM proteasome inhibitor MG-132 (Calbiochem, San Diego, CA) for 1 h before harvesting and then were lysed in denaturing RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). IPs were performed as described above. Polyubiquitylated species were detected using anti-HA antibody.
In Vitro Ubiquitylation Assay.
The ubiquitylation reaction mixture and conditions for the assay were carried out as described in ref. 12. Briefly, the ubiquitylation reaction mixture contained 25 mM Tris·HCl, 100 mM NaCl, 1 mM DTT, 2.5 mM ATP, 4 mM MgCl2, 2 μl of Escherichia coli BL21 bacterial extracts overexpressing wheat E1, 0.1 μg of UbcH7, 1 μg of purified recombinant ItchΔC2, and 5 μg of Flag-tagged ubiquitin. After incubation for 90 min at 30°C, the reactions were terminated by boiling in SDS loading buffer and resolved by SDS/PAGE, followed by IB with anti-Flag.
RNAi-Mediated Silencing of N4BP1.
The predesigned Lamin A/C and N4BP1 siRNAs oligos were purchased from Dharmacon (Lafayette, CO) and, Qiagen (N4BP1_3 HP) and Ambion (Austin, TX) (N4BP1-141575), respectively. Target cells were transfected with the siRNA duplexes by using Oligofectamine (Invitrogen) at a final concentration of 100 nM.
Supplementary Material
Acknowledgments
We thank Dr. Angelo Peschiaroli for helpful discussions and for critical review of the manuscript. This work was supported by European Union Grant LSHB-CT-019067; the Fondo per gli Investmenti della Ricerca di Base Grants RBNE01KJHT_004 and RBNE01NWCH_008; the Ministero dell'Istruzione dell'Università e della Ricerca/Programma di Ricerca di Rilevante Interesse Nazionale Grant 004064744_003; Associazione Italiana per la Ricerca sul Cancro Grant 1338; Istituto Superiore di Sanitá Grant 530/F-A19; Grant Telethon GGPO4110 (to G.M.); the Kimmel Scholar Award (to R.I.A.); and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research (M.R.K.).
Abbreviations
- C2
Ca+2/lipid-binding
- E3
E3 ubiquitin–protein ligase
- HECT
homologous to E6AP C terminus
- IB
immunoblot analysis
- IP
immunoprecipitation
- MEF
mouse embryonic fibroblast
- N4BP1
Nedd4-binding partner-1
- WWn
WW domain n.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701773104/DC1.
References
- 1.Pickart CM. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 4.Rose I. Cell Death Differ. 2005;12:1198–1201. doi: 10.1038/sj.cdd.4401710. [DOI] [PubMed] [Google Scholar]
- 5.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey KF, Dinudom A, Komwatana P, Jolliffe CN, Day ML, Parasivam G, Cook DI, Kumar S. J Biol Chem. 1999;274:12525–12530. doi: 10.1074/jbc.274.18.12525. [DOI] [PubMed] [Google Scholar]
- 8.Murillas R, Simms KS, Hatakeyama S, Weissman AM, Kuehn MR. J Biol Chem. 2002;277:2897–2907. doi: 10.1074/jbc.M110047200. [DOI] [PubMed] [Google Scholar]
- 9.Perry WL, Hustad CM, Swing DA, O'Sullivan TN, Jenkins NA, Copeland NG. Nat Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 10.Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, Karin M. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 11.Gallagher E, Gao M, Liu YC, Karin M. Proc Natl Acad Sci USA. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, Cesareni G, Melino G. EMBO J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce C, Melino G. Proc Natl Acad Sci USA. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grob TJ, Novak U, Maisse C, Barcaroli D, Luthi AU, Pirnia F, Hugli B, Graber HU, De Laurenzi V, Fey MF, et al. Cell Death Differ. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- 16.Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, et al. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 17.Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H, Lena AM, Candi E, Terrinoni A, Catani MV, et al. EMBO J. 2005;24:2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborn MT, Chambers TC. J Biol Chem. 1996;71:30950–30955. doi: 10.1074/jbc.271.48.30950. [DOI] [PubMed] [Google Scholar]
- 19.Sudol M, Recinos CC, Abraczinskas J, Humbert J, Farooq A. IUBMB Life. 2005;57:773–778. doi: 10.1080/15216540500389039. [DOI] [PubMed] [Google Scholar]
- 20.Komuro A, Saeki M, Kato S. J Biol Chem. 1999;274:36513–36519. doi: 10.1074/jbc.274.51.36513. [DOI] [PubMed] [Google Scholar]
- 21.Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, Fang D, Hunter T, Liu YC. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 22.Kang H, Freund C, Duke-Cohan JS, Musacchio A, Wagner G, Rudd CE. EMBO J. 2000;19:2889–2899. doi: 10.1093/emboj/19.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haupt Y, Maya R, Kazaz A, Oren M. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 24.Musti AM, Treier M, Bohmann D. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 25.Shcherbik N, Kumar S, Haines DS. J Cell Sci. 2002;115:1041–1048. doi: 10.1242/jcs.115.5.1041. [DOI] [PubMed] [Google Scholar]
- 26.Henry PC, Kanelis V, O'Brien MC, Kim B, Gautschi I, Forman-Kay J, Schild L, Rotin D. J Biol Chem. 2003;278:20019–20028. doi: 10.1074/jbc.M211153200. [DOI] [PubMed] [Google Scholar]
- 27.Levy D, Adamovich Y, Reuven N, Shaul Y. Cell Death Diff. 2006;14:743–751. doi: 10.1038/sj.cdd.4402063. [DOI] [PubMed] [Google Scholar]
- 28.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. Cancer Res. 2005;65:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 29.Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, Pandolfi PP. J Exp Med. 2004;199:1545–1557. doi: 10.1084/jem.20031943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.