Abstract
The African mosquito Anopheles gambiae is the major vector of human malaria. We report a genome-wide survey of mosquito gene expression profiles clustered temporally into developmental programs and spatially into adult tissue-specific patterns. Global expression analysis shows that genes that belong to related functional categories or that encode the same or functionally linked protein domains are associated with characteristic developmental programs or tissue patterns. Comparative analysis of our data together with data published from Drosophila melanogaster reveal an overall strong and positive correlation of developmental expression between orthologous genes. The degree of correlation varies, depending on association of orthologs with certain developmental programs or functional groups. Interestingly, the similarity of gene expression is not correlated with the coding sequence similarity of orthologs, indicating that expression profiles and coding sequences evolve independently. In addition to providing a comprehensive view of temporal and spatial gene expression during the A. gambiae life cycle, this large-scale comparative transcriptomic analysis has detected important evolutionary features of insect transcriptomes.
Keywords: comparative transcriptomics, insect development, insect evolution, microarrays
Anopheles gambiae is the major vector of human malaria in subSaharan Africa, a secondary vector of filariasis, and the key vector of O'nyong-nyong viral fever outbreaks. Effective transmission of pathogens results from extreme anthropophilic behavior and repeated bloodfeeding of A. gambiae adult females. Indeed, female mosquitoes are found typically around human habitations and bloodfeed largely on humans rather than animals. Therefore, successful malaria control campaigns to date have coincided largely with local control of Anopheles populations.
A substantial bloodmeal is required for A. gambiae egg development, the start of the next life cycle. Eggs are fertilized while traversing the genital chamber and begin embryonic development, which normally lasts 2–3 days and is similar to that of Drosophila melanogaster despite notable morphological (1) and molecular (2) differences. Four larval stages (instars) ensue, as compared with three in Drosophila, accompanied by continuous growth. In that period, larval organs are functional and adult organs incipient or slowly developing. During metamorphosis (from larva to pupa and adult) most larval organs are histolyzed, whereas others persist or grow.
To date, molecular and cell biological research on A. gambiae has used D. melanogaster as the model system. These dipterans have diverged from a common ancestor ≈250 mya. The recent introduction of high-throughput approaches including genome sequencing (3) and transcription profiling (4–6) has greatly facilitated investigation of A. gambiae biology. Here, we used an EST microarray platform, MMC1, encompassing 19,680 ESTs (7) that correspond to ≈8,872 TCLAG (transcribed cluster of A. gambiae) contigs (henceforth T-contigs) to determine genome-wide expression profiles during the A. gambiae life cycle. This study reveals transcriptional programs associated with critical developmental transition stages. It also identifies transcripts expressed in adult tissue-specific patterns in the female head, midgut, ovaries, and carcass. We further explore similarities and differences in gene expression that may underpin the variant lifestyles of Anopheles and Drosophila. This comparative transcriptomic analysis demonstrates a strong similarity in specific temporal and spatial expression patterns of orthologous gene pairs. However, this correlated expression is independent of the degree of coding-sequence conservation during evolution.
Results
Experimental Design.
The A. gambiae life cycle was sampled empirically at eight successive time periods: embryos, five larval stages, pupae, and adult females and males [supporting information (SI) Fig. 5A]. Adult tissues from head, gut, ovaries, and carcass were also collected. We investigated the mosquito developmental and adult tissue-expression profiles by competitive two-dye hybridizations on MMC1 microarrays of experimental and standard reference (SR) RNA samples. The latter were produced in vitro from ESTs that were used as substrates to produce amplicons for the spotted microarrays. They provided consistent, nonzero reference values for all array probes, allowing effective data normalization between experiments (SI Fig. 5B).
We performed three biological and one technical (dye-swap) replicates. Embryonic, pupal, female and male adult replicates, and each of the adult tissues, displayed high reproducibility of T-contig expression (SI Fig. 5C). Expression profiles at different larval periods tended to cluster by mosquito rearings rather than time period, possibly reflecting physiological differences between generations and a predominant, inherent similarity of development in larval instars. This is consistent with the continuous growth of larval and imaginal disk tissues and with the recurrence of developmental phases within each larval instar (e.g., a shift from growth to hormonally induced epithelial retraction from the old cuticle and formation of a new cuticle). We did not attempt to define separately within instar phases.
Gene Expression Differences During Development.
Statistically significant expression differences between developmental stages were established for 2,421 T-contigs with a one-factor ANOVA (SI Fig. 5D, P = 0.001). Of these, 1,571 displayed at least 2-fold differences between their respective minimum and maximum expression (Table 1 and SI Data Set 1) and were used thereafter. Embryos were the most distinct stage, displaying 624–1,009 differentially expressed contigs in pair-wise comparisons (Tukey test) with each of the other stages. Adult males were also very distinctive differing from larvae by 454–572 contigs, as compared with 214–313 differentially expressed contigs between females and larvae and 245–265 between pupae and adults. Approximately 5% of the pupae/adults differentially expressed T-contigs encode proteasome components (reflecting the extensive histolysis of larval tissues during pupal life), whereas 5% encode components of the cuticle (which is obviously different between pupae and adults). Larval periods were mostly indistinct (0–81 differences) but early larvae (La-c) tended to cluster apart from late larvae (Ld-e). It is known that precursors of adult organs (imaginal discs) develop continuously in larvae, with slow cell divisions at early instars and faster cell divisions subsequently. Most of the T-contigs that differed between larval stages encode proteins implicated in nucleic acid binding, protein metabolism, or cuticular constituents.
Table 1.
Differential expression of EST contigs during the A. gambiae life cycle
| La | Lb | Lc | Ld | Le | P | F | M | |
|---|---|---|---|---|---|---|---|---|
| E | 663 (492) | 678 (513) | 672 (513) | 816 (626) | 627 (489) | 818 (626) | 624 (505) | 1,009 (759) |
| La | 14 (12) | 19 (18) | 68 (59) | 81 (65) | 279 (232) | 272 (232) | 506 (384) | |
| Lb | 0 (0) | 49 (42) | 61 (51) | 294 (241) | 281 (238) | 572 (435) | ||
| Lc | 25 (24) | 37 (31) | 226 (192) | 214 (191) | 454 (359) | |||
| Ld | 13 (11) | 333 (296) | 313 (273) | 541 (434) | ||||
| Le | 172 (159) | 273 (238) | 466 (377) | |||||
| P | 265 (224) | 245 (206) | ||||||
| F | 179 (156) |
Numbers represent differentially expressed T-contigs in each pair-wise comparison (ANOVA Tukey test, P ≤ 0.001). The number of contigs that display at least 2-fold difference is shown in parentheses.
Developmental Programs.
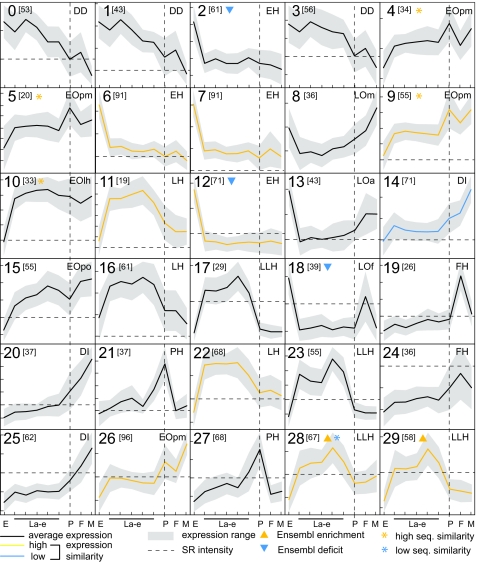
The expression profiles of the 1,571 contigs (encompassing 1,065 Ensembl genes and 783 Drosophila orthologs) were grouped by self-organizing maps (SOM) into 30 coexpression clusters with 5 × 6 node geometry (Fig. 1). Additional nodes did not reveal novel patterns, and fewer nodes yielded loose clusters. We considered each cluster individually, but, because several were closely related, we describe them in a consolidated manner according to their constituents and broad developmental dynamics (see also SI Table 3 and SI Data Set 2).
Fig. 1.
Coexpression clusters and developmental transcription programs. During the mosquito life cycle, 1571 T-contigs that display at least 2-fold difference between their maximum and minimum expression are grouped into 30 SOM coexpression clusters. Developmental program designations are at the top right of each cluster. Numbers in brackets refer to clustered contigs, solid lines refer to average expression, and gray areas refer to range of expression. y axis scale shows increments of 0.5 in log10-transformed expression values, horizontal dashed lines indicate SR signal levels, and vertical dotted lines point to the pupal stage. Arrowheads indicate clusters enriched (yellow) or deficient (blue) in Ensembl gene models. Average expression is plotted in black if the mean expression similarity of mosquito–fruitfly orthologous contigs within a cluster is comparable with that of all orthologous pairs and is plotted in yellow or blue if it is statistically above or below that standard, respectively. Yellow or blue asterisks show clusters of genes displaying CDS similarity to their fruitfly orthologs that is statistically above or below that of all orthologous pairs, respectively.
The embryo high program (EH) encompasses four clusters with strong expression in embryos. Six major functional gene classes predominate: replication, transcription, mRNA processing and regulation, cell cycle, signal transduction, cell growth, and metabolism. Drosophila orthologs of many EH contigs are well known transcriptional embryonic regulators, or are implicated in mRNA splicing and downstream processing. The presence of ubiquitin domain sequences suggests a role for ubiquitination in mosquito embryonic development (8). Two EH clusters, 2 and 12, are deficient in Ensembl gene models.
Six clusters share low embryonic expression and are further distinguished by subtler features. The single-cluster embryo low/pupa low (EOpo) program shows reduced expression in pupae, whereas EOpm (four clusters) is highly expressed in pupae and adult males, the latter suggesting expression in the testis (9). EOlh (one cluster) is different in showing high expression in larvae continuing into the pupa. In all EO programs, genes involved in metabolic reactions are prominent (overall total 25%), as are numerous proton- or electron-transport components; other contigs encode components of immune responses or cytoskeletal elements and regulators.
Two programs, LH and LLH (three clusters each), are named for their high larval or late larval expression. They differ from EOlh because of more detailed features. LH accelerates in early instars but declines gradually between Ld and pupa, reflecting growth in the larval body but not in imaginal discs, where growth remains modest. However, LLH peaks at Ld and then sharply declines between Le and pupa. LH is associated with 30% prevalence of proteins implicated in metabolic reactions (carbohydrate and lipid metabolism and proteolysis), whereas LLH is enriched in metabolic (especially proteolytic) enzymes but also cuticle components and putative defense proteins.
The female high program (FH, two clusters) is defined by prominent expression in adult females but not in males. It is enriched in putative immune components, suggesting adaptation to increase survival of the almost completely monogamous females (polygamous males are more dispensable).
The developmentally increasing program (DI, three clusters) displays low expression in embryos and larvae, followed by a strong progressive increase in pupae and adults (especially males). After emergence from the pupal cuticle, adult organs such as salivary glands, midgut epithelium, and flight and orientation organs continue to develop. Adult maturation begins earlier in males than in females, explaining the observed sex-specific expression difference (10, 11). DI encompasses proteins involved in sensory perception including odorant-binding proteins and members of the rhodopsin signaling pathway, as well as antimicrobial peptides, other immune-related proteins, and digestive enzymes.
The pupa high expression program (PH, two clusters) peaks in pupa, consistent with the restructuring of the mosquito body at metamorphosis, when many larval tissues histolyze, whereas adult structures develop, some of them acquiring an adult cuticle. Indeed, many PH genes encode structural and enzymatic components of the cuticle, including yellow family members that control pigmentation; others are implicated in ubiquitination and proteolysis, suggesting a role in histolysis.
Genes engaged in the developmentally declining program (DD, three clusters) display an overall progressive, steady decline in expression (including a detectable trough at the pupal stage); this is sharply different from the abrupt decline seen in EH. Almost 20% of DD sequences encode components of protein biosynthesis, modification, and folding; others control DNA, RNA, and nucleotide synthesis.
Finally, the larva low program, LO, is characterized by high expression in embryos and adults and encompasses three distinct expression clusters: LOa (equal expression in both adult sexes), LOm (higher in males), and LOf (higher in females). Many components of LOf have Drosophila orthologs often associated with maternal effects on embryos or involvement in asymmetric mRNA or protein localization.
Developmental Expression of Gene Functional Categories.
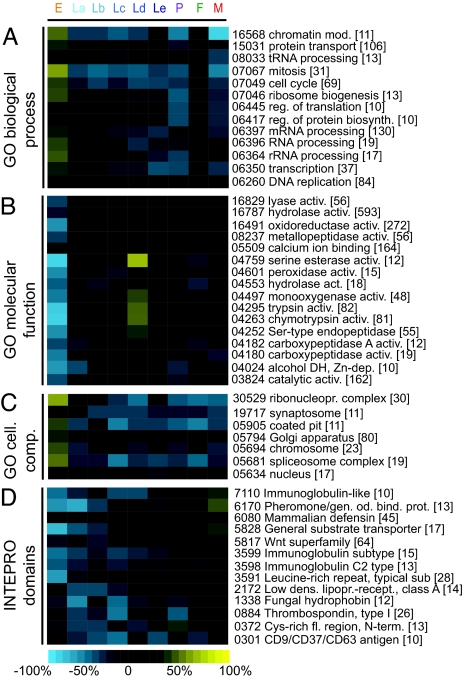
We grouped T-contigs by functional categories (GO or INTERPRO domain annotation) and, for each category, determined the percentage of contigs displaying top or bottom expression at each time period. Bottom percentages were subtracted from top percentages, and resulting data sets (representing the expression tendency of each functional category) were subjected to k-means clustering (Fig. 2 and SI Fig. 6).
Fig. 2.
Coordinated expression of related functional gene groups. Selected k-means clusters from the analysis of GO biological processes (A), INTERPRO domains (B), GO molecular functions (C), and GO cellular components (D) are presented. For each functional group, the percentage of contigs showing the lower 25% of the expression range at each time period was subtracted from the respective percentage of contigs showing the upper 25%, and resulting values (ranging from blue to yellow) were used for clustering. Numbers on the right (omitting the GO or IPR prefixes) denote functional group identifier; numbers in brackets indicate the size of each functional group in contigs.
Many contigs associated with nuclear processes, i.e., DNA replication, transcription and RNA processing, or cell-cycle processes display top expression in embryos but bottom expression in much of the remaining life cycle. These contigs mostly map to the EH program and highlight fundamental postfertilization events such as rapid succession of cell cycles associated with chromatin replication and initiation of transcription and translation for embryo patterning.
Numerous catabolic reactions show bottom expression in embryo, pupa, and adults and top expression in the Ld period, as do contigs mapping to specific subcellular organelles, e.g., microsomes, peroxisomes, and lysosomes. Catabolic clusters probably serve histolysis of larval tissues at the onset of metamorphosis. Several proteins implicated in immune reactions are associated with bottom expression in embryos and larvae but top expression in pupae and adults, indicating enhanced immune system activity during and after metamorphosis. They map mainly to the DI and EO programs, presumably reflecting adaptation to the increased infection risk of pupae and adults. Some hydrolytic functions are associated with immunity factors such as GNBPs (hydrolase activity) and CLIPs (trypsin and chymotrypsin-like activities), suggesting that immunity may have evolved in catabolic, gut-associated components that were in persistent contact with gut biota. Catabolic and numerous immunity genes are notable in the LH and, especially LLH, programs.
Coexpression Patterns in Adult Female Tissues.
We identified 898 T-contigs exhibiting differential expression between at least two tissues (SI Fig. 5D, Table 2, and SI Data Set 3). Of these, 829 contigs exceeding 2-fold difference were subjected to k-means clustering and grouped into 10 coexpression patterns; six patterns are surprisingly specific, containing sequences overexpressed in practically single tissue (Fig. 3, SI Table 3 and SI Data Set 4).
Table 2.
Differential expression of EST contigs in adult female tissues
| Carcass | Gut | Ovaries | |
|---|---|---|---|
| Head | 295 (271) | 172 (156) | 370 (322) |
| Carcass | 226 (212) | 332 (302) | |
| Gut | 177 (161) |
Numbers represent differentially expressed T-contigs in each pair-wise comparison (ANOVA Tukey test, P ≤ 0.001). The number of contigs that display at least 2-fold difference is shown in parentheses.
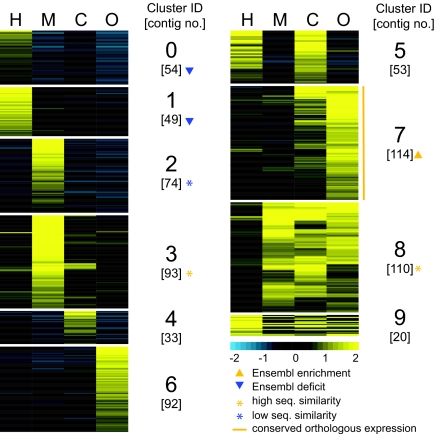
Fig. 3.
Tissue expression patterns in adult females. k-means clusters of 829 contigs showing at least 2-fold regulation between two or more tissues are presented; 477 correspond to Ensembl genes, and 318 have D. melanogaster orthologs. Numbers in brackets indicate cluster size (in contigs) and the scale bar represents log2-transformed gene expression values. Yellow and blue arrowheads indicate clusters enriched or deficient in Ensembl gene models, respectively. The ovary-enriched cluster 7 contains contigs with mean expression similarity to their fruitfly orthologs that is statistically above the mean of all orthologous pairs. Yellow or blue asterisks show clusters of genes with CDS similarity to their fruitfly orthologs that is statistically above or below that of all orthologous pairs, respectively.
The insect head carries the major sensory organs, the vision center and endocrine glands. Expression in the head includes two distinctive patterns (0 and 1) encompassing contigs of three major functional categories: rhodopsins and visual perception, odorant-binding proteins and pheromone-related proteins. A Drosophila allatostatin homolog, an adult brain peptide that blocks the synthesis of the developmental juvenile hormone, is included.
The midgut is the primary organ for nutrient absorption, synthesis and secretion of digestive enzymes, and formation of the gut-lining peritrophic membrane. It also has an endocrine role and contributes to diuresis, e.g., after a bloodmeal when blood cells are concentrated before being digested. Indeed, a quarter of T-contigs in the two midgut-specific patterns (2–3) are associated with metabolic reactions; some with previously identified midgut specific expression, and others with domains involved in vasoconstriction and diuresis.
Aside from genes putatively involved in defense reactions, annotation information is sparse for the carcass-enriched patterns 4 and 5. The surprising duality of head and carcass association in cluster 5 may be related to the presence of fat body, stationary hemocytes, and hypodermis in both body parts.
Patterns 6 and 7 encompass contigs that are strongly expressed in ovaries but differ in that cluster 7 also shows substantial expression in the carcass. Many of these contigs are associated with transcriptional programs FH and LOf; 25% are implicated in transcription regulation, translation and mRNA processing. Some contigs encode odorant-binding proteins, suggesting unorthodox functions that merit further analysis.
The triple-tissue (midgut, carcass, and ovary) expression pattern 8 is enriched in genes with metabolic functions. Annotation suggests housekeeping processes, because 20% of the genes are implicated in general polysaccharide and fatty acid metabolic reactions and 16% in protein synthesis and degradation.
Finally, the four-body-part pattern 9 differs from pattern 8 in showing the most pronounced expression in the head. It contains housekeeping genes from diverse functional classes, e.g., electron and proton transport, polysaccharide metabolism, signal transduction, etc.
Comparative Transcriptomics of the Anopheles and Drosophila Life Cycles.
Half of the genes in the Anopheles and Drosophila genomes are identified as 1:1 orthologs (12). This, in conjunction with an earlier transcriptional study of Drosophila development (13), allowed us to compare the developmental expression profiles of orthologs in the two insects, after normalization of the respective experimental designs to create comparable notional developmental phases. Pearson and smooth correlation analysis of the expression similarity of 1,039 orthologous genes showed a drastic shift toward positive correlation (Fig. 4A). A similar shift was detected with the nonparametric Spearman coefficient (data not shown). When the same data set was randomly rearranged 100 times to generate nonorthologous gene pairs, no shift was detected, and both average distributions were largely symmetric.
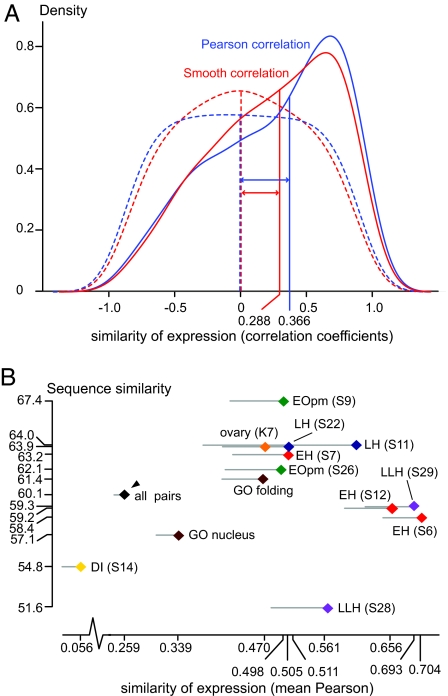
Fig. 4.
Comparative life cycle transcriptomic analysis. (A) Correlation of gene expression of Anopheles–Drosophila orthologous pairs. The distribution of pairs reveals a significant positive shift of expression correlation with both the Pearson (median/skewness = 0.366−0.489), and the smooth (median/skewness = 0.288/−0.416) correlation coefficients. Dashed lines indicate the referenced distribution of randomized pairs with the Pearson (median/skewness = −0.003/0.005) and smooth (median/skewness = −0.002/0.008) correlation coefficients, respectively. (B) Scatter plot of the mean expression similarity (x axis) vs. mean CDS similarity (y axis) according to Pearson correlation coefficient for developmental (S, SOMs) and tissue (K, k-means) coexpression clusters and GO functional groups. Only clusters and functional groups with significant deviation from the average expression similarity are presented. A black arrowhead indicates the average sequence and expression similarity of all 1,039 orthologous gene pairs, and gray lines show the standard error of expression similarity, plotted on one side for clarity.
We queried whether the expression similarity of orthologs was due to specific gene sets, such as particular developmental programs, tissue patterns, or functional groups. As shown in Fig. 4B and SI Tables 4–10, coexpression clusters belonging to six developmental programs or tissue patterns (EH, EOpm, DI, LH, LLH, and ovary-enriched) and two functional groups (nuclear localization and protein folding) showed significant positive or negative deviations from the median expression similarity (Wilcoxon U test ≤0.005). Next, we examined whether the degree of expression similarity between orthologs varies in parallel with their coding sequence (CDS) similarity. Such a global connection was not detected (SI Fig. 7). However, when the analysis matrix was disarticulated into coexpression clusters and functional categories (SI Tables 4–10) insights were revealed. The EOpm developmental cluster 9 displayed coherent positive deviations of both CDS and expression similarities, whereas the LLH developmental cluster 28 and the nuclear localization group showed opposite deviations, negative for CDS and positive for expression similarity.
Discussion
A major outcome of the present study is the attribution of 1,571 EST contigs to gene expression programs that apparently underpin A. gambiae development. These programs are consistent with biological processes that take place during the corresponding periods. Thus, the characteristic temporal features of the EH program reflect a rapid and specific postfertilization activation of genes implicated in embryonic development, notably associated with transcription and RNA processing and translation as well as cell cycle control and body patterning. A significant part of this program also operates in ovaries that contain mature eggs, 2 days after a bloodmeal. This is a rich source of genes worthy of attention in studies of mosquito early development. In contrast, the LH program spans several instars with limited variability, possibly reflecting their known common feature, continuous growth combined with an increasing cell division rate.
The LLH program is characterized by top expression of metabolic enzymes that usher metamorphosis, whereas the later PH program features genes implicated in adult cuticle synthesis at the metamorphic transition. In holometabola insects (which include Diptera) invention of metamorphosis was an enormous evolutionary innovation. The pupa is an outwardly quiescent state, but is in fact pivotal. Many larval tissues degenerate, and complex imaginal (adult) tissues rapidly emerge from dedicated adult-forming cells. The whole body is reshaped asynchronously as the insect transforms from an efficient metabolic factory converting food into body mass to the new lifestyle of a flying machine dedicated to sex, procreation, and, as in mosquitoes, the search for very specialized food (blood). The pivotal nature of the pupa is reflected in the identified developmental programs. In addition to pupal expression peaks in the PH program, clear expression dips are noted in the EOpo and DD program. A distinctive gene expression similarity between pupae and adult males (EOpm program) reflects precocious adult male development. Future analysis of genes implicated in these features will illuminate the key pupal phase of dipteran development.
The adult A. gambiae females are immensely important as vectors of human pathogens. The FH program is enriched in immunity genes, suggesting enhanced female protection, possibly as an adaptation to the challenges of vector competence. In addition, the various female body parts express diverse gene sets that are differentially implicated in the uptake of and susceptibility to pathogens. The head contains most of the sensory organs responsible for host tracking and bloodfeeding preference and expresses genes implicated in vision and odor-sensing. The midgut is the main organ for bloodmeal digestion and expresses enzymes involved in a variety of metabolic processes, but also represents an important barrier for ingested pathogens. A previous study has used the same microarray platform to examine midgut expression during invasion by malaria parasites (14). The combination of the two studies illuminates in considerable detail a large set of midgut-expressed genes, which might affect permissiveness to malaria.
Numerous EST contigs included in our analysis do not overlap with existing gene models (7). In fact, one-third (505 of 1,571) of the highly regulated T-contigs in specific developmental programs show no overlap with present gene models. The developmental programs EH and LOf and the adult head-enriched pattern show a pronounced deficit of gene models, whereas the LLH program and the ovary-enriched pattern show a greater than average coverage of gene models. This suggests that numerous embryonic, maternal and head-related genes might be missed or imperfectly predicted by automatic prediction algorithms. This feature seems paradoxical because embryonic expression is well characterized in Drosophila and other insects, indicating that novel genes implicated in early development may remain to be discovered. In clear contrast, metamorphosis-associated and ovary-enriched genes have good automated prediction probability.
Interspecies comparisons of orthologous gene expression have been reported in plants (15, 16), between rodent and human cancer cells (17), and between Caenorhabditis elegans and D. melanogaster (18). In insects, expression patterns of only a limited set of orthologs have been compared between the ant Camponotus festinantus and D. melanogaster (19). Guided by methodological conclusions of previous studies addressing the issue of gene expression comparisons between microarray platforms (20–24), we performed the first large-scale comparative analysis of insect development between A. gambiae and D. melanogaster. Our analysis shows a strong positive correlation of expression for 1,039 orthologous gene pairs and reveals that orthologs frequently share similar developmental expression patterns. Importantly, the expression similarity is not globally linked to the CDS similarity of orthologs, suggesting different evolutionary pressures exerted on CDS and expression properties of orthologs during the 250 myr separating these two insects.
The degree of orthologous expression similarity varies between different developmental programs or functional groups. In general, orthologs engaged in early developmental events such as egg fertilization, embryo formation, and body patterning display highly similar expression, whereas some later events engage genes of significantly lower similarity in expression. The success of insects is based in part on their great diversification for different niches in adult life. Retention of successful genetic solutions for early development may have provided a stable background, on which innovations for the adult life could be accepted with minimal disruption. However, this trend allows for exceptions: orthologs in the late EOpm program show high expression similarity as well as high CDS similarity. Apparently, adult male differentiation (probably for development of the male gonad) is highly conserved in insects. Our study points toward a new concept of defining corresponding genes: expression-based “orthoregulation” alongside sequence-based orthology.
Materials and Methods
Biological Material.
Approximately 400 laboratory-reared A. gambiae mosquitoes of the G3 strain were fed at days 3 or 8 of adulthood on CD1 mice and produced the experimental generations P1 and P2. The P3 generation was the progeny of P1 mated females fed on mice 3 days after emergence. Adult head, gut, and carcass (freed of head, gut, wings, and legs) tissues were collected from 1-day-old females; ovaries were from 5- to 6-day-old females that were bloodfed 48 h earlier.
Analysis of Functional Groups.
Gene Ontology (GO) terms and INTEPRO domains represented in at least 20 distinct T-contigs were assembled into functional groups. For each group and time period, the percentage of T-contigs showing bottom expression (the lower 25% of each contig's overall expression range) was subtracted from the percentage of T-contigs showing top (upper 25%) expression, and resulting values were subjected to k-means clustering.
Comparative Transcriptomic Analysis.
The raw microarray data from a D. melanogaster life cycle study (13) were downloaded from http://genome.med.yale.edu/Lifecycle/Data_download and analyzed by using the same criteria as for the A. gambiae data set, except for the negative spike-in control criterion. Expression profiles of ESTs were averaged to the respective gene, and data were normalized as for Anopheles. The 3,571 genes with reliable measurements in at least 130 of 151 hybridizations and with a t test P value <0.05 in at least 1 of 75 developmental time periods were considered further.
The Anopheles and Drosophila data sets were divided into seven notional developmental periods. Anopheles male and female stages were compared with 24-h-old Drosophila adult males (Am24h) and females (Af24h), respectively. Based on correlation analysis (data not shown), the mosquito embryo period was compared with the average of Drosophila embryonic time points E056–E0112 and the mosquito pupa period with the average of Drosophila metamorphosis time points M04–M12. By using a sliding window procedure, three (early, middle, and late) comparable phases of larval development were defined from the Anopheles (La–Lc, Lb–Ld, and Lc–Le) and the Drosophila (L24–L57, L43–L84, and L67–L105) studies.
Orthologous gene pairs were constructed from best reciprocal hits and information from syntenic regions (12). The combined expression matrix of orthologs was normalized to the median of each gene and the 50th percentile of each notional time period. Correlation analysis with Pearson, smooth, and Spearman coefficients was performed in GeneSpring.
Methods for EST construction, sequencing and clustering, microarray construction, preparation of experimental and SR RNA samples and hybridizations, imaging, and data analysis of microarrays are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank E. Furlong for the updated version of the Drosophila microarray annotation, M. Gross for suggestions about microarray analyses, and L. Ettwiller for discussions about regulatory sequence-finding algorithms. This work was supported by National Institute of Allergy and Infectious Diseases–National Institutes of Health (NIAID/NIH) Grants U01AI48846 and 2PO1AI044220-07, European Commission Research and Training Networks Grant HPRN-CT-2000-00080 and the European Molecular Biology Laboratory. It was also part of the activities of the BioMalPar European Network of Excellence supported by European Grant LSHP-CT-2004-503578 from the Priority 1 “Life Sciences, Genomics and Biotechnology for Health” in the 6th Framework Program. This work was greatly facilitated by the AnoEST Project, which was funded by NIAID/NIH VectorBase Contract NIAID-DMID-04-34.
Abbreviations
- EH
embryo high
- EO
embryo low
- EOpo
embryo low/pupa low
- EOpm
embryo low/pupa and males high
- EOlh
embryo low/larva high
- LH
larva high
- LLH
LATE larva high
- FH
female high
- DI
developmentally increasing
- PH
pupa high
- DD
developmentally declining
- LO
larva low
- LOa
larva low/equal expression in both adult sexes
- LOm
larva low/higher expression in males
- LOf
larva low/high expression in females.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in ArrayExpress (accession nos. E-TABM-186 and E-TABM153) and are graphically available through VectorBase at www.vectorbase.org/ExpressionData.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703988104/DC1.
References
- 1.Monnerat AT, Machado MP, Vale BS, Soares MJ, Lima JB, Lenzi HL, Valle D. Mem Inst Oswaldo Cruz. 2002;97:589–596. doi: 10.1590/s0074-02762002000400026. [DOI] [PubMed] [Google Scholar]
- 2.Goltsev Y, Hsiong W, Lanzaro G, Levine M. Dev Biol. 2004;275:435–446. doi: 10.1016/j.ydbio.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 4.Dimopoulos G, Christophides GK, Meister S, Schultz J, White KP, Barillas-Mury C, Kafatos FC. Proc Natl Acad Sci USA. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dana AN, Hong YS, Kern MK, Hillenmeyer ME, Harker BW, Lobo NF, Hogan JR, Romans P, Collins FH. BMC Genomics. 2005;6:5. doi: 10.1186/1471-2164-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JM. Insect Mol Biol. 2005;14:365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- 7.Kriventseva EV, Koutsos AC, Blass C, Kafatos FC, Christophides GK, Zdobnov EM. Genome Res. 2005;15:893–899. doi: 10.1101/gr.3756405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, III, Grant PA. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- 9.Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanayev RA, Makunin IV, Kafatos FC, Zhimulev IF. Proc Natl Acad Sci USA. 2005;102:8269–8274. doi: 10.1073/pnas.0502702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Meillon B, Sebastian A, Khan ZH. Bull World Health Org. 1967;36:7–14. [PMC free article] [PubMed] [Google Scholar]
- 11.Haddow AJ, Gillett JD, Corbet PS. Ann Trop Med Parasitol. 1959;53:123–131. doi: 10.1080/00034983.1959.11685909. [DOI] [PubMed] [Google Scholar]
- 12.Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, et al. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 13.Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- 14.Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC. Curr Biol. 2005;15:1185–1195. doi: 10.1016/j.cub.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 15.Ma L, Chen C, Liu X, Jiao Y, Su N, Li L, Wang X, Cao M, Sun N, Zhang X, et al. Genome Res. 2005;15:1274–1283. doi: 10.1101/gr.3657405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao Y, Ma L, Strickland E, Deng XW. Plant Cell. 2005;17:3239–3256. doi: 10.1105/tpc.105.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, et al. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 19.Goodisman MA, Isoe J, Wheeler DE, Wells MA. Evol Int J Org Evol. 2005;59:858–870. [PubMed] [Google Scholar]
- 20.Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- 21.Tan PK, Downey TJ, Spitznagel EL, Jr, Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC. Nucleic Acids Res. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, et al. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, Gabrielson E, Garcia JG, Geoghegan J, Germino G, et al. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 24.Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Nat Methods. 2005;2:337–344. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.