Abstract
A stable complex between duplex DNA and an oligonucleotide is assembled with the aid of a DNA synthetic mimic, peptide nucleic acid (PNA). Homopyrimidine PNAs are known to invade into short homopurine tracts in duplex DNA forming P-loops. We have found that P-loops, formed at two closely located purine tracts in the same DNA strand separated by a mixed purine–pyrimidine sequence, merge and open the double helix between them. The opposite DNA strand, which is not bound with PNA, exposes and becomes accessible for complexing with an oligonucleotide via Watson–Crick pairing. As a result, the PD-loop emerges, which consists of locally open duplex DNA, PNA “openers,” and an oligonucleotide. The PD-loop stability and sequence specificity are demonstrated by affinity capture of duplex DNAs by using biotinylated oligonucleotides and streptavidin-covered magnetic beads. The type of complex formed by PNAs, an oligonucleotide and duplex DNA we describe, opens ways for development of various in vitro and in situ hybridization techniques with duplex DNA and may find applications in DNA nanotechnology and genomics.
Keywords: peptide nucleic acid, affinity capture, hybridization, biomagnetic separation, yeast DNA
Linear, nonsupercoiled double-stranded DNA (dsDNA) is known to be able to accommodate an additional oligonucleotide strand with much less efficiency than single-stranded nucleic acids and supercoiled DNAs. Formation of intermolecular triplexes is mostly limited to long homopurine–homopyrimidine regions (1, 2). D-loops are formed in linear dsDNA only at the ends of the DNA duplex in case of binding of long single-stranded DNA molecules (3). R-loops may be formed inside linear dsDNA, but long RNAs and transient DNA denaturation are necessary (4). A complex between an oligodeoxyribonucleotide (ODN) and linear dsDNA can be formed with assistance of the RecA protein. However, the fidelity of recognition is lower than for protein-free DNA–DNA interactions and the complex is unstable upon deproteinization (5, 6). The possibility of binding to dsDNA of a pair of complementary modified ODNs due to their self-mediated invasion into DNA duplex has recently been demonstrated. However, these complexes were formed only at the ends of dsDNA (7). In addition, a few techniques exist for formation of specific complexes between ODNs and dsDNA based on either prior DNA denaturation or degradation of one DNA strand before ODN binding. These techniques, however, require subsequent reconstruction or reparation of targeted molecules into DNA duplex (8, 9).
Here we describe a complex between linear dsDNA and an ODN with mixed sequence of purines and pyrimidines, which is formed via Watson–Crick pairing facilitated by the peptide nucleic acid (PNA) invasion. As a result, an unusual multi-component structure emerges, which we call the PD-loop. It is characterized by exceptionally high sequence specificity. The PD-loop structure opens totally new ways for hybridizing oligonucleotides with dsDNA and for selective manipulation with DNA duplexes. We demonstrate that the PD-loop makes it possible to selectively isolate a dsDNA fragment from a very complex mixture of DNA fragments. This approach has significant advantage over PCR amplification when DNA must be preserved in the intact, biologically active form, like in case of analysis of DNA epigenetic modifications in imprinted genes.
MATERIALS AND METHODS
PNA Openers.
PNA oligomerization and purification was performed as described (10, 11). The following bis-PNA openers were used in this study [their identity was confirmed by matrix-assisted laser desorption ionization time of flight (MALDI TOF) mass spectrometry]: PNA 1, H(Lys)2-TTTJTTJJ-(eg1)3-CCTTCTTT-LysNH2 (+4); PNA 2, H(Lys)2-JTTJJJJT-(eg1)3-TCCCCTTC-LysNH2 (+4); and PNA 3, H(Lys)3-TTJJTTT-(eg1)3-TTTCCTT-LysNH2 (+5).
As with peptides, PNA sequences are written from amino terminus to carboxy terminus, and T, C, and J denote here the N-1 alkylated pyrimidine nucleobases connected with N-(2-aminoethyl)glycine backbone via methylenecarbonyl linkers, respectively. H means a free amino group. NH2 means a terminal carboxamide. Lys denotes a lysine residue. Eg1 denotes the 8-amino-3,6-dioxaoctanoic acid groups, which serve as linkers connecting two PNA oligomers in bis-PNA. The J base denotes pseudoisocytosine. All bis-PNAs carry multiple positively charged Lys residues at their J-containing halves because such polycationic PNA constructions are characterized by high complex stability and high binding specificity (12). Numbers in parentheses indicate the total charge of PNA oligomers.
DNA Oligomers.
All nonphosphorylated ODNs (adapters, primers, tags, etc.) were from Operon Technologies (Alameda, CA). The biotinylated ODN tags used in this study were as follow: ODN 1, 5′-GAAGGTTCGAAGG-3′-biotin; ODN 2, 5′-AAGGTTCGAAG-3′-biotin; and ODN 3, biotin-5′-GAAGGCTGGAAGGA-3′.
Biotin was conjugated with ODNs through a flexible linker. Both 3′ and 5′ ends of ODNs were chosen for conjugation just to check the possible steric interference of PD-loop structure with streptavidin binding. The results (vide infra) demonstrate that either end of the ODN is accessible for protein binding. ODNs 1 and 2 were used as tags for capturing the plasmids, whereas ODN 3 was used for isolation of a yeast dsDNA fragment.
Plasmids.
Plasmids carrying the appropriate inserts were obtained by cloning of the corresponding ODNs into the BamHI site of the pUC19 vector. In all cases the inserts were verified by direct sequencing. Plasmid inserts cloned were as follows: Δ3 (pPL3 plasmid), 5′-TCCCCTTCGAACCTTCTTT-3′ and 3′-AGGGGAAGCTTGGAAGAAA-5′; and Δ11 (pPL11 plasmid), 5′-TCCCCTTCCTTCGAACCTTCCTTCTTT-3′ and 3′-AGGGGAAGGAAGCTTGGAAGGAAGAAA-5′, where Δn indicates the number of nucleotides separating adjacent bis-PNA binding sites (for PNAs 1 and 2). These PNA binding sites and their complementary sequences are in bold type.
Yeast Genomic DNA.
The yeast genomic DNA from Saccharomyces cerevisiae strain AB1380 (Genome Systems, St. Louis) was isolated in agarose inserts as described (13, 14) and stored at low temperatures. Yeast DNA was retrieved from the inserts with β-agarase I (New England Biolabs) into appropriate buffer solution, then digested with the MseI restriction enzyme and ligated with 50 pmol of MseI adapter (15): 5′-TCTCCAGCCTCTCACCGCAT-3′ and 3′-AGTGGCGTAAT-5′.
The fill-in reaction was performed at room temperature with the Klenow polymerase to generate blunt ends. The yeast target site we chose was of the Δ4 type: 5′-TTTCCTTCCAGCCTTCTTT-3′ and 3′-AAAGGAAGGTCGGAAGAAA-5′. The GenBank accession number is Z38060; coordinates are 15546–15564; and the bis-PNA binding sites (for PNAs 1 and 3) are in bold type.
Plasmid Capture Protocol.
We used full-length control (pBR322) and target (pPL3 or pPL11) plasmids linearized with the AatII restriction enzyme. Binding of a pair of PNA openers (PNAs 1 and 2) to plasmids was carried out in 25 mM Mes buffer (pH 6.1) at 37°C for 2–20 h with the PNA concentration about 0.5 μM. To avoid binding of PNA openers with the partially complementary biotinylated ODN, free openers were removed from the samples by gel filtration. After subsequent binding of biotinylated ODNs (mostly, ODN 1) at 37°C, the free ODN was removed from samples by gel filtration. Magnetic separation was performed in accordance with Dynal protocol (16) by using BioMag Streptavidin magnetic beads (PerSeptive Diagnostics, Cambridge, MA). The beads collected with a magnet were washed extensively several times, and captured DNA was released from the magnetic beads by incubation at 65°C for 20 min in TE buffer (pH 7.5) with 1 M NaCl under gentle shaking. DNA eluted from the beads was ethanol precipitated, resuspended, and typically analyzed by electrophoresis in 1% agarose gel with subsequent ethidium bromide staining and the charge-coupled device (CCD) camera detection. The quantitative analysis was done by processing the images with the IC-1000 Digital Imaging System (Innotech Scientific, San Leandro, CA).
Microbiological Analysis.
We also used a more sensitive method to quantify the results of affinity capture experiments. In this method, an enriched mixture of control (pBR322) and target (pPL3) linear plasmids eluted from magnetic beads were converted into circular form by T4 DNA ligase (GIBCO/BRL). Recircularized plasmids were used for transformation of competent Escherichia coli cells. Transformed bacterial cells were spread onto 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal)-containing agar plates and were grown up overnight at 37°C. Transformants carrying the pBR322 plasmid form white colonies on these selective media, whereas transformants carrying the pPL3 plasmid form blue colonies on the same plates and thus can be easily distinguished.
Yeast DNA Capture Protocol.
To isolate a specific yeast dsDNA fragment we followed mainly the basic plasmid capture protocol with minor modifications. The only significant change was that to release the captured dsDNA from magnetic beads we now incubated samples at lower temperature and under lower salt concentration: at 47°C for 20 min in TE buffer (pH 7.5) with 50 mM NaCl. This modification of the capture protocol allows retention of the PNA openers on DNA during all rounds of subsequent enrichment, thus avoiding the time-consuming step of retargeting of DNA samples with the openers in the next round of separation. Therefore, PNA openers (PNAs 1 and 3) were targeted to 250 ng of yeast DNA (1.5 × 107 copies of the yeast genome) only at the beginning of the first round. During each round of separation the captured samples were extensively washed out, released from the beads, and rebound with ODN 3; then another round was initiated. After each round, an aliquot of captured DNA was collected and amplified by using AmpliTaq DNA polymerase (Perkin–Elmer/Cetus) by 35 cycles of nonspecific PCR with an adapter-specific primer (5′-TCTCCAGCCTCTCACCGCAT-3′) and was analyzed by electrophoresis in 1.5% agarose gel.
Analysis of Captured Yeast DNA.
To analyze the captured yeast DNA fragment, we reamplified the DNA material from the bands corresponding to this fragment in the agarose gel obtained after third and fifth rounds of enrichment. The PCR amplification in both cases resulted in a homogeneous DNA fragment of the expected size. The amplified material obtained after the third round was digested by several restriction enzymes (AluI, MboI, DdeI, and RsaI). In all cases the obtained restriction maps coincided with expected ones confirming the purity and identity of the captured fragment. Sequence of the material amplified after third and fifth rounds of enrichment was confirmed by cycle sequencing (Commonwealth Biotechnologies, Alexandria, VA).
RESULTS AND DISCUSSION
Is PD-Loop Possible?
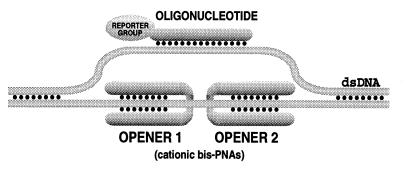
Our design (Fig. 1) is grounded on the ability of short homopyrimidine PNAs, synthetic oligomers consisting of a polyamide backbone linked to pyrimidine nucleobases, to displace one strand of dsDNA forming an exceptionally stable complex, the (PNA)2/DNA triplex, with the other strand (1, 17–29). Such structures, known as P-loops, are formed most readily when two positively charged PNA oligomers connected by a flexible linker (bis-PNAs) are used (10, 12, 14, 22, 30–33). If two bis-PNAs (dubbed here as “openers”) bind to closely located homopurine DNA tracts separated by several base pairs of a mixed purine–pyrimidine sequence, an extended open region emerges inside dsDNA. This region can serve as a target for binding of an ODN containing all four bases via Watson–Crick pairing (see Fig. 1), which cannot by itself form stable complexes within linear dsDNA or even near the end of DNA duplex (7). It is far from being obvious that such multi-component structures, the PD-loops, can be assembled because their formation is associated with mutually contradictory requirements.
Figure 1.
The PD-loop consists of duplex DNA, two PNA openers, and an oligonucleotide. In experiments described in the present study, oligonucleotides carried a reporter group (biotin).
First, the open region formed within the DNA duplex by a pair of openers is not very large, and a hybrid complex with an ODN may not fit this space. Two bulky PNA2/DNA triplexes already exist in this location, and P-loops probably adopt a compact structure (34). The subsequent binding of an ODN, which is associated with its winding around the displaced DNA single strand to form more than one turn, can be hindered sterically or kinetically. Indeed, it was shown that binding of short oligonucleotides all the way around small RNA loops is not always sterically favorable (35). Note that a complex between an ODN and the P-loop created by a PNA oligomer inside supercoiled DNA, which was described recently (36), can be facilitated by formation of a very large open region under superhelical stress.
Then the requirements for stability of the ODN/DNA duplex on the one hand and the (PNA)2/DNA triplex on the other are very difficult to reconcile. Indeed, low salt is known to favor the PNA invasion into dsDNA (12, 18, 19, 30, 33, 37, 38) and stability of the PNA/DNA complexes decreases with increasing salt (39), whereas short ODN/DNA duplexes are unstable at low salt. But, as we demonstrate here, the PD-loop can be assembled and may be quite stable.
One can expect the PD-loop formation to be a very sequence-specific process, in contrast to the ODN hybridization with single-stranded DNA or RNA. Indeed, in case of PD-loop, only sites on duplex DNA flanked by two specific PNA openers are accessible for complexing with specific ODN. Most of DNA retains its duplex structure and is inaccessible for binding with the ODN having mixed purine–pyrimidine composition. P-loops formed as a result of binding of separate openers are too small to form stable complexes with an ODN. Because ODNs are short (less than 15 nt long), even one mismatch must be sufficient to make the ODN/DNA duplex unstable in case an incorrect site is open by the same pair of PNA openers.
Detection of PD-Loop.
We found the affinity capture method quite suitable for detection of stable complexes shown in Fig. 1 and also to check their specificity. To this end, biotinylated ODNs were used and the PD-loop formation was detected via selective capture by magnetic beads covered by streptavidin of full-length linear plasmid DNAs with corresponding inserts. The inserts we used consisted of mixed purine–pyrimidine sequences of various lengths flanked by two short binding sites for PNA openers (see Materials and Methods). Fig. 2 demonstrates the results of affinity capture procedure validating the PD-loop formation. Control experiments showed that in the absence of any component from the complex in Fig. 1—PNA openers, biotinylated ODN or the target site—the capturing effect disappeared (data not shown). The effect was also absent when instead of ODN 1 we used ODN 2, which differs from ODN 1 by the absence of two terminal nucleotides.
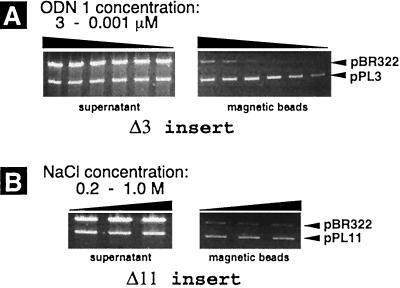
Figure 2.
Detection of the PD-loop formation by the affinity capture of linearized plasmids pPL3 and pPL11 on the streptavidin-coated magnetic beads. The gels represent DNA retained in the supernatant (Left) and DNA washed out from the magnetic beads after the affinity capture procedure (Right). The top bands in all gels correspond to the control pBR322 plasmid (without PNA binding sites), which was in 4-fold excess over pPL3 and pPL11 plasmids. (A) Effect of oligonucleotide concentration on the efficiency of capturing of the pPL3 plasmid. Complexing of ODN 1 was performed in buffer solution containing 1 M NaCl, and samples corresponding to adjacent lanes represent a 5-fold difference in the oligonucleotide concentration. (B) Effect of salt concentration on the capturing efficiency of the pPL11 plasmid. ODN 1 was at 3 μM in buffer solution containing 0.2, 0.5, and 1 M NaCl.
The following are the conclusions from our capture experiments: (i) the observed capture is caused by the PD-loop formation; (ii) PD-loops remain stable at high salt concentration (at least 1 M NaCl); (iii) PD-loops are still formed with openers’ binding sites separated by as many as 11 bp (the pPL11 plasmid); and (iv) stability of the PD-loop structure and efficiency of its formation decrease with increasing distance between openers’ binding sites.
The data in Fig. 2 also show that at high concentration of ODN significant nonspecific capture is observed. This nonspecific capture is caused by our detection method rather than by nonspecific formation of PD-loops. In fact, no capturing at all was observed in case of the control plasmid alone even in the presence of all components needed for the formation of the PD-loop complex. Our data (to be published elsewhere) indicate that this nonspecific capture is actually a random cocapture of control DNA because of its aggregation with target DNA molecules carrying the PD-loop. This aggregation is stimulated by the excess of ODN. Fig. 2 shows that the nonspecific capture at small ODN concentration is very low.
Such high specificity of the PD-loop formation is at the limit of the photometric detection of the CCD camera when the magnetic separation is followed by the gel electrophoresis assay. For accurate estimations of specificity of the PD-loop formation we used a different approach. First, the PD-loop affinity capture procedure was applied to the mixture of control and target plasmids. Then the transformation of high-competent E. coli cells by recircularized plasmid DNA washed out from the magnetic beads was performed. After that, we counted the bacterial transformants grown up on the selective media. Transformants carrying pBR322 (control) and pPL3 (target) plasmids form colonies of different color on the X-Gal-containing agar plates. Hence, the enrichment of the initial plasmid mixture by pPL3 plasmids (and, therefore, the specificity of the PD-loop formation) may be directly quantified by counting the number of colonies of each type as it was done before for the triplex affinity capture (40). This microbiological procedure allowed us to estimate the specificity of PD-loop formation close to 103.
We believe that we were able to assemble the PD-loops and to use them for DNA capture because of remarkable stability of extended P-loops. Such structures, which include two (PNA)2/DNA triplexes, are expected to be significantly more stable than regular P-loops studied before and are able to tolerate very high salt. As a result, once the extended P-loop is readily formed at low salt, it remains stable at high salt—i.e., at conditions favoring the stability of the ODN/DNA duplex. Binding of an ODN to a single-stranded DNA region of the extended P-loop additionally stabilizes the structure, making the resulting PD-loop even more stable than the extended P-loop.
Specificity of PD-Loop Formation.
To support our anticipation that the PD-loop formation must be an exceptionally sequence-specific process, we isolated a specific fragment of duplex DNA from a digest of the entire yeast genome. We chose, among many suitable sites for the PD-loop formation in the yeast genome, a unique 19-bp-long site on S. cerevisiae chromosome IX consisting of two binding sites for PNA openers 1 and 3 separated by a 4-bp-long mixed purine–pyrimidine sequence (see Materials and Methods). We used biotinylated ODN 3 as a probe. Digestion of the yeast genome with the MseI restriction enzyme yielded a 863-bp-long desired DNA fragment, which is about 1/16,000 part of the S. cerevisiae genome.
We performed five rounds of the magnetic separation procedure. Aliquots of captured DNA collected after each round of enrichment were analyzed by nonspecific PCR amplification by using a primer complementary to a special adapter ligated to both ends of all fragments of the original digest (see Materials and Methods). This assay amplified all DNA fragments captured on magnetic beads after the PD-loop formation. If the desired fragment of yeast DNA were captured by our technique, it would be detected as a fragment with the size of 903 bp (the fragment plus two adapters). Note that we used PCR only as a detection method at the end of one, two, three, four, and five rounds of enrichment. No intervening amplification of captured material was performed between the rounds.
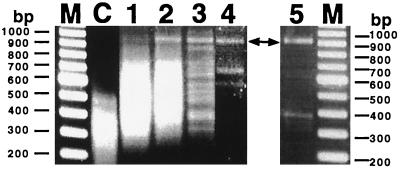
Fig. 3 shows our results, which demonstrate sequence specificity of the PD-loop structure. After three rounds of the PD-loop formation/separation procedure the variety of captured fragments dramatically reduced and the desired 903-bp-long DNA fragment could be selectively isolated. Additional rounds of enrichment, the fourth round and especially the fifth round, further reduced the variety of captured fragments, leaving very few nonspecific fragments. A band corresponding to our 903-bp fragment was the only major band that systematically appeared in the subsequent rounds of the capture procedure. Restriction analysis and sequencing performed after the third and the fifth rounds confirmed that the DNA fragment marked by arrows in Fig. 3 was the expected fragment.
Figure 3.
Demonstration of specificity of PD-loop formation by selective capture of a yeast DNA fragment. Lanes 1–5 correspond to one, two, three, four, and five rounds of PD-loop formation/separation procedure, respectively; lane M corresponds to the 100-bp DNA ladder; and lane C represents the MseI digest of the yeast genome before capturing. A targeted 903-bp-long fragment is shown by arrows.
Obviously, certain sequence limitations are inherent in the PD-loop, but they are not severe. Indeed, the presented data show that homopurine DNA binding sites for the PNA openers may be 7 bp long and may be interrupted by up to 11 bp of an arbitrary sequence. Such sites are met, statistically, after each (1/2) 27+7/11 = 750 bp. Thus, normally each gene must carry such a site, especially in case of eukaryotes. But the situation is even less restrictive than that. Our preliminary data indicate that, to form the PD-loop, the PNA openers can be as short as pentamers. If they again can be separated by up 10 bp, we expect to have one such site per each 50 bp, on average.
CONCLUSION
A complex between duplex DNA, an ODN, and a pair of PNAs described here opens the way for development of various in vitro and in situ hybridization techniques for dsDNA and may find applications in genomics and DNA nanotechnology. Although in the present study we used the capture procedure exclusively to demonstrate that the PD-loop can be assembled and that the PD-loop formation is a highly sequence-specific process, the procedure itself may prove to be useful for selective dsDNA isolation. Its advantages over PCR amplification consist in the lack of introduced mutations and in the potential to capture large chunks of the genome. But probably the most important advantage consists in the possibility of isolating imprinted genes carrying postsynthetic modifications (e.g., methylation) of nucleotides. Such modifications are normally lost during PCR amplification, although recent data emphasize their important role in normal development as well as disease (41–43).
Acknowledgments
We thank Alexei Veselkov for his participation in the project at its initial stage. Support by the National Institutes of Health (Grants GM52201 and GM54434 to M.F.-K.) and by PerSeptive Biosystems is appreciated.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PNA, peptide nucleic acid; dsDNA, double-stranded DNA; ODN, oligodeoxynucleotide; X-Gal, 5-bromo-4-chloro-3-indolyl β-d-galactoside; CCD, charge-coupled device.
References
- 1.Frank-Kamenetskii M D, Mirkin S M. Annu Rev Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 2.Soyfer V N, Potaman V N. Triple-Helical Nucleic Acids. New York: Springer; 1996. [Google Scholar]
- 3.Wetmur J G. Crit Rev Biochem Mol Biol. 1991;26:227–259. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]
- 4.Thomas M, White R L, Davis R W. Proc Natl Acad Sci USA. 1976;73:2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West S C. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- 6.Malkov V A, Sastry L, Camerini-Otero R D. J Mol Biol. 1997;271:168–177. doi: 10.1006/jmbi.1997.1164. [DOI] [PubMed] [Google Scholar]
- 7.Kutyavin I V, Rhinehart R L, Lukhtanov E A, Gorn V V, Meyer R B, Jr, Gamper H B., Jr Biochemistry. 1996;35:11170–11176. doi: 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- 8.Shepard A R, Rae J L. Nucleic Acids Res. 1997;25:3183–3185. doi: 10.1093/nar/25.15.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Life Technologies. GIBCO/BRL Products and Reference Guide. Gaithersburg, MD: Life Technologies; 1997/1998. pp. 1914–1915. [Google Scholar]
- 10.Egholm M, Christensen L, Dueholm K L, Buchardt O, Coull J, Nielsen P E. Nucleic Acids Res. 1995;23:217–222. doi: 10.1093/nar/23.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen L, Fitzpatrick R, Gildea B, Petersen K H, Hansen H F, et al. J Peptide Sci. 1995;3:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn H, Demidov V V, Frank-Kamenetskii M D, Nielsen P E. Nucleic Acids Res. 1998;26:582–587. doi: 10.1093/nar/26.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith C L, Klco S R, Cantor C R. In: Genome Analysis: A Practical Approach. Davies K, editor. Oxford: IRL; 1988. pp. 41–72. [Google Scholar]
- 14.Veselkov A G, Demidov V V, Nielsen P E, Frank-Kamenetskii M D. Nucleic Acids Res. 1996;24:2483–2488. doi: 10.1093/nar/24.13.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broude N E, Chandra A, Smith C L. Proc Natl Acad Sci USA. 1997;94:4548–4553. doi: 10.1073/pnas.94.9.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dynal. Dynabeads Biomagnetic Separation System: Technical Handbook. Oslo: Dynal; 1992. pp. 1–3. [Google Scholar]
- 17.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 18.Hanvey J C, Peffer N C, Bisi J E, Thomson S A, Cadilla R, et al. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 19.Cherny D Y, Belotserkovskii B P, Frank-Kamenetskii M D, Egholm M, Buchardt O, Berg R H, Nielsen P E. Proc Natl Acad Sci USA. 1993;90:1667–1670. doi: 10.1073/pnas.90.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchardt O, Egholm M, Berg R, Nielsen P E. Trends Biotechnol. 1993;11:384–386. doi: 10.1016/0167-7799(93)90097-S. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen P E, Egholm M, Buchardt O. J Mol Recognit. 1994;7:165–170. doi: 10.1002/jmr.300070303. [DOI] [PubMed] [Google Scholar]
- 22.Betts L, Josey J A, Veal J M, Jordan S R. Science. 1995;270:1838–1841. doi: 10.1126/science.270.5243.1838. [DOI] [PubMed] [Google Scholar]
- 23.De Mesmaeker A, Altmann K-H, Waldner A, Wendeborn S. Curr Opin Struct Biol. 1995;5:343–355. doi: 10.1016/0959-440x(95)80096-4. [DOI] [PubMed] [Google Scholar]
- 24.Hyrup B, Nielsen P E. Bioorg Med Chem. 1996;4:5–23. doi: 10.1016/0968-0896(95)00171-9. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson M, Nielsen P E. Q Rev Biophys. 1996;29:369–394. doi: 10.1017/s0033583500005886. [DOI] [PubMed] [Google Scholar]
- 26.Dueholm K L, Nielsen P E. New J Chem. 1997;21:19–31. [Google Scholar]
- 27.Nielsen, P. E. & Haaima, G. (1997) Chem. Soc. Rev. 73–78.
- 28.Corey D R. Trends Biotechnol. 1997;15:224–229. doi: 10.1016/S0167-7799(97)01037-8. [DOI] [PubMed] [Google Scholar]
- 29.Frank-Kamenetskii M D. Phys Rep. 1997;288:13–60. [Google Scholar]
- 30.Demidov V V, Yavnilovich M V, Belotserkovskii B P, Frank-Kamenetskii M D, Nielsen P E. Proc Natl Acad Sci USA. 1995;92:2637–2641. doi: 10.1073/pnas.92.7.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffith M C, Risen L M, Greig M J, Lesnik E A, Sprankle K G, Griffey R H, Kiely J S, Freier S M. J Am Chem Soc. 1995;117:831–832. [Google Scholar]
- 32.Veselkov A G, Demidov V V, Frank-Kamenetskii M D, Nielsen P E. Nature (London) 1996;379:214. doi: 10.1038/379214a0. [DOI] [PubMed] [Google Scholar]
- 33.Demidov V V, Frank-Kamenetskii M D, Nielsen P E. In: Biological Structure and Dynamics. Sarma R H, Sarma M H, editors. Vol. 2. Schenectady, NY: Adenine; 1996. pp. 129–134. [Google Scholar]
- 34.Almarsson Ö, Bruice T C, Kerr J, Zuckermann R N. Proc Natl Acad Sci USA. 1993;90:7518–7522. doi: 10.1073/pnas.90.16.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ecker D J, Vickers T A, Bruice T W, Freier S M, Jenison R D, Manoharan M, Zounes M. Science. 1992;257:958–961. doi: 10.1126/science.1502560. [DOI] [PubMed] [Google Scholar]
- 36.Smulevitch S V, Simmons C G, Norton J C, Wise T W, Corey D R. Nat Biotechnol. 1996;14:1700–1704. doi: 10.1038/nbt1296-1700. [DOI] [PubMed] [Google Scholar]
- 37.Peffer N J, Hanvey J C, Bisi J E, Thomson S A, Hassman F C, Noble S A, Babiss L E. Proc Natl Acad Sci USA. 1993;90:10648–10652. doi: 10.1073/pnas.90.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittung P, Nielsen P E, Nordén B. J Am Chem Soc. 1996;118:7049–7054. [Google Scholar]
- 39.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen P E, Nordén B, Graslund A. J Am Chem Soc. 1996;118:5544–5552. [Google Scholar]
- 40.Ito T, Smith C L, Cantor C R. Proc Natl Acad Sci USA. 1992;89:495–498. doi: 10.1073/pnas.89.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reik W, Maher E R. Trends Genet. 1997;13:330–335. doi: 10.1016/s0168-9525(97)01200-6. [DOI] [PubMed] [Google Scholar]
- 42.Hastie N. Nature (London) 1997;389:785–787. doi: 10.1038/39732. [DOI] [PubMed] [Google Scholar]
- 43.Sun F-L, Dean W L, Kelsey G, Allen N D, Reik W. Nature (London) 1997;389:809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]