Abstract
Individual variation in fetal hemoglobin (HbF, α2γ2) response underlies the remarkable diversity in phenotypic severity of sickle cell disease and β thalassemia. HbF levels and HbF-associated quantitative traits (e.g., F cell levels) are highly heritable. We have previously mapped a major quantitative trait locus (QTL) controlling F cell levels in an extended Asian-Indian kindred with β thalassemia to a 1.5-Mb interval on chromosome 6q23, but the causative gene(s) are not known. The QTL encompasses several genes including HBS1L, a member of the GTP-binding protein family that is expressed in erythroid progenitor cells. In this high-resolution association study, we have identified multiple genetic variants within and 5′ to HBS1L at 6q23 that are strongly associated with F cell levels in families of Northern European ancestry (P = 10−75). The region accounts for 17.6% of the F cell variance in northern Europeans. Although mRNA levels of HBS1L and MYB in erythroid precursors grown in vitro are positively correlated, only HBS1L expression correlates with high F cell alleles. The results support a key role for the HBS1L-related genetic variants in HbF control and illustrate the biological complexity of the mechanism of 6q QTL as a modifier of fetal hemoglobin levels in the β hemoglobinopathies.
Keywords: β hemoglobinopathies, complex trait, F cells
Sickle cell disease and β thalassemia are among the most common genetic diseases worldwide and have a major impact on global health and mortality (1). Both these hemoglobinopathies display a remarkable diversity in their disease severity. A major ameliorating factor is an innate ability to produce fetal hemoglobin (HbF, α2γ2). HbF levels vary considerably, not only in patients with these β hemoglobin disorders, but also in healthy normal adults. The distribution of HbF and F cells (FCs, erythrocytes that contain measurable HbF) in healthy adults is continuous and positively skewed. Although the majority of adults have HbF of less the 0.6% of total hemoglobin, 10–15% of individuals have increases ranging from 0.8% to 5% (2). The latter individuals are considered to have heterocellular hereditary persistence of fetal hemoglobin (hHPFH), which refers to the uneven distribution of HbF among the erythrocytes. When coinherited with β thalassemia or sickle cell disease, hHPFH can increase HbF output to levels that are clinically beneficial (3, 4).
FC levels are strongly correlated with HbF in adults within the normal range (including hHPFH) (2), and F cells are generally used as an indirect measure of HbF within normal individuals because of the poor sensitivity for HbF assay in the lower range (see Materials and Methods). A logarithmic transformation of FC removes skewness and leads to a distribution that is approximately normal for a representative population sample (5). The heritability of HbF and FC is estimated to be 89% (5). Cis-acting variants and rare mutations at the β globin gene locus explain some of the variability (2), but >50% of the variance is unlinked to this locus (6). Our previous study of an Asian-Indian kindred in which β thalassemia and hHPFH were segregating identified a QTL that mapped to a 1.5-Mb interval on chromosome 6q23 with a logarithm of odds (lod) score of 6.3 (7, 8) (Fig. 1a). This interval contains five known protein-coding genes (ALDH8A1, HBS1L, MYB, AHI1, and PDE7B), none of which harbored mutations (nonsynonymous variants), and three (HBS1L, MYB, and AHI1) are expressed in erythroid progenitor cells (9, 10).
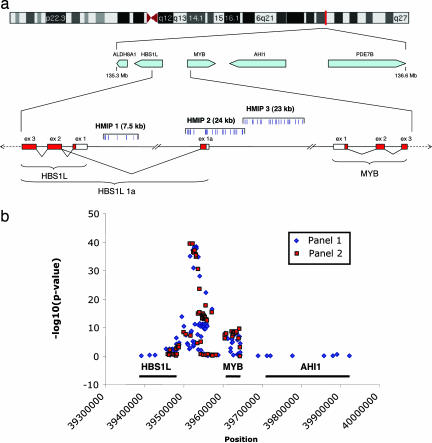
Fig. 1.
Overview of the 6q23 region and the HMIP locus. (a) Genomic organization of the 1.5-Mb candidate interval and the 126-kb segment spanning portions of HBS1L and MYB and the intergenic region on chromosome 6q23 (not to scale). The regions covered by the three trait-associated blocks (HMIP 1, 2, and 3) are indicated by square brackets with the locations of the high-scoring SNP alleles. Boxes represent both confirmed and putative exons with arrows indicating transcriptional orientation: red, coding sequence; white, 5′ UTR. (b) Positions of markers and significance (−log10 P value) of test statistics from the mixed-model ANOVA at sites within the HBS1L-MYB interval of association and flanking regions. SNPs over MYB are significantly associated with the trait but this situation reflects the linkage disequilibrium across the region.
To explore further the role of the 6q23 QTL on HbF control, we studied two panels (824 and 1,217 individuals, respectively) of twin pairs of North European origin. In a high-resolution association study, we identified multiple genetic variants that are strongly associated with FC levels in the Caucasian controls (P = 10−75). These genetic variants reside in three linkage disequilibrium (LD) blocks within HBS1L and 5′ to HBS1L, and MYB in the intergenic region.
To delineate the functional significance of these genetic variants, we performed an expression profile of HBS1L and MYB during erythropoiesis. We observed a striking correlation of increased HBS1L expression in erythroid progenitor cells with presence of the single nucleotide polymorphisms (SNPs) associated with high trait values in the three LD blocks. The present study illustrates the power of QTL mapping for positional identification of trans-acting genetic variants influencing regulation of HbF levels, a major ameliorating factor of sickle cell disease and β thalassemia.
Results
We genotyped two panels (824 and 1,217 individuals, respectively; see Table 1) of twin pairs of North European origin recruited through the Twins U.K. Adult Twin Registry (11). FC levels were measured as described in Materials and Methods, and log-transformed to obtain an approximately normal distribution. Age, sex, and XmnI-Gγ (−158 C/T) variant upstream of the Gγ globin gene, which is associated with FC levels (6, 12), show similar associations with FC levels in both panels (Table 1). From the known genes within the 6q23 QTL interval, we selected MYB and HBS1L as candidate genes for detailed study. Both genes are expressed in erythroid precursor cells. MYB encodes a transcription factor essential for erythroid differentiation in hematopoiesis (13–15). HBS1L is the human ortholog of Saccharomyces cerevisiae HBS1 and encodes a protein with apparent GTP-binding activity, involved in the regulation of a variety of critical cellular processes (16).
Table 1.
Number of twin pairs and singletons in panels 1 and 2, within-twin correlations of loge % F cells in the two panels, parameter estimates for the fixed effects for age, sex, and XmnI-Gγ, and P values for fixed effects
| Panel | No. included in sample |
Trait correlation | Parameter estimate and P values |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Pairs | Singletons* | Individuals | Sex | Age | XmnI-Gγ Add | XmnI-Gγ Dom | |||
| Panel 1 | DZ | 311 | 3 | 625 | 0.42 ± 0.05 | 0.34 ± 0.09 | −0.006 ± 0.002 | −0.24 ± 0.04 | 0.12 ± 0.05 |
| MZ | 96 | 7 | 199 | 0.83 ± 0.03 | P = 0.0002 | P = 0.002 | P = 10−11 | P = 0.01 | |
| Panel 2 | DZ | 574 | 11 | 1,159 | 0.47 ± 0.03 | 0.39 ± 0.16 | −0.007 ± 0.002 | −0.28 ± 0.03 | 0.05 ± 0.04 |
| MZ | 29 | 0 | 58 | 0.86 ± 0.05 | P = 0.02 | P = 0.002 | P = 10−18 | n.s. | |
The first panel was used as a primary family set for genetic mapping whereas the second panel, which was collected and phenotyped during the primary mapping phase, was used for confirmation studies. The first twin panel is composed of 311 dizygotic (DZ) twin pairs, 96 monozygotic (MZ) twin pairs, and 10 singletons (for DNA or phenotypes could be obtained from only one member of the twin pair). Panel 2 consists of 574 DZ twin pairs, 29 MZ twin pairs, and 11 singletons. The fixed-effects parameter estimates are the regression coefficients with sex scored 1 for male and 2 for female, age measured in years, and genotypes at XmnI-Gγ coded 0 for CC, 1 for CT, and 2 for TT. The dominance effect at XmnI-Gγ is the estimated deviation of the CT heterozygote mean from the midpoint between the CC and TT means.
*DNA or phenotype available for only one twin in pair.
Polymorphisms were identified by resequencing MYB and ≈78 kb of the HBS1L-MYB intergenic region. We identified 184 markers for which the minor allele had 5% or greater frequency, 94 of which were selected for genotyping based on their positions, linkage disequilibrium patterns and intermediate association results. We added 27 markers from public databases to provide additional coverage particularly in the 3′ flanking regions of MYB and HBS1L. Altogether, 121 markers were genotyped with average spacing of 4.4 kb, and higher density (1.8 kb average) in the HBS1L–MYB intergenic region [supporting information (SI) Table 3].
Twenty-eight markers provided very strong evidence of association (P < 10−8) in the first panel, with the most significant results concentrated at sites between HBS1L and MYB (Fig. 1b). In particular, a 24-kb segment starting 33 kb upstream of HBS1L contained 12 markers showing very strong association (P values between 10−28 and 10−39, block 2 in Fig. 1a) whereas the other 13 markers from within this segment are less significantly associated with the trait (SI Table 4). Strikingly, the 12 markers with the strongest trait association have similar allele frequencies and are in complete linkage disequilibrium (except for haplotypes with frequency <2%), whereas the others exhibit different linkage disequilibrium patterns (SI Tables 5–7). We confirmed the association by characterizing a subset of 75 markers from the HBS1L-MYB interval in the second twin panel (Fig. 1b). The twelve markers with the strongest trait association in the first panel are also the most strongly associated in the second panel (Fig. 1b and SI Table 4). When the data from the two panels were combined, these markers had association P values of 10−50 to 10−75.
Markers outside of the 24-kb interval also showed consistent evidence of association in the two panels. In some instances, linkage disequilibrium between trait-associated markers appeared weak. We hypothesized that more than one variant contributes to the QTL. A stepwise statistical selection procedure led to the identification of three markers that accounted independently for a significant proportion of the trait variance even with the other markers included in the ANOVA (Table 2). The three markers were selected in the following order (with P values from the combined data for the significance calculated with previously selected markers included): rs9399137 (P = 10−75), rs52090901 (P = 10−10) and rs6929404 (P = 0.0002). The first of these SNPs (rs9399137) is 1 of 12 markers in HBS1L MYB intergenic polymorphism (HMIP) block 2 (so-labeled because of its physical position) with the strongest trait associations. We identified multiple markers in two other trait-associated blocks in strong linkage disequilibrium with rs52090901 (HMIP block 1) and rs6929404 (HMIP block 3) (Fig. 1a). Minor differences in the association statistics for markers in the same block could be attributed to rare haplotypes and/or a small amount of missing genotype data.
Table 2.
Significance tests for sex, age, XmnI-Gγ, and the three selected HBS1L-MYB markers
| Panel | Fixed effects P values |
||||||
|---|---|---|---|---|---|---|---|
| Sex | Age | XmnI-Gγ |
HBS1L-MYB |
||||
| Block 1 (rs52090901) |
Block 2 (rs9399137) |
Block 3 (rs6929404) |
|||||
| Additive | Additive | Additive | Dominance | Additive | |||
| Panel 1 | 0.0001 | 0.0005 | 10−14 | 0.006 | 10−24 | 0.0005 | 0.01 |
| Panel 2 | 0.004 | 0.0005 | 10−22 | 0.002 | 10−24 | 0.06 | 0.005 |
| Combined | 10−5 | 10−7 | 10−35 | 10−5 | 10−45 | 0.0004 | 0.0002 |
The significance tests are conditional on the presence of the nontested parameters in the model. For HBS1L-MYB, these are different from P values for the marginal test statistics in SI Table 4 because of partial LD between the markers. P values for dominance are shown only when significant. We employed a stepwise statistical procedure to select the markers shown here. New markers were incorporated into the ANOVA only if they accounted for a significant proportion of the trait variance when more strongly associated markers were already included in the model (thus accounting for linkage disequilibrium with these). We selected the marker with the most significant test statistic to incorporate at each step until no remaining markers gave a significant trait association (P > 0.01). We obtained equivalent results using either the markers genotyped in the first twin panel, or the combined data with markers that were characterized in both panels.
A Novel Transcript of HBS1L.
As part of our characterization of the HBS1L-MYB intergenic region, we confirmed by RT-PCR and sequence analysis the existence of a previously uncharacterized transcript of HBS1L that is expressed in thymus, Jurkat cells, peripheral leukocytes, and at minimal levels in erythroid progenitors. The transcript was deduced from the sequence of a thymus cDNA clone deposited in a public database (Japanese Database of Transcriptional Start Sites; DBTSS; http://dbtss.hgc.jp/; GenBank ID DB114698). This transcript contains an alternative 119-bp first exon (denoted exon 1a), which starts ≈45 kb upstream of the previously described first exon of the gene (Fig. 1a and SI Fig. 2). A 102-bp repeat-free segment that starts 129 bp upstream of the initiation codon has marked nucleotide homology with other mammals and contains binding site motifs for a putative TATA box and three members of the GATA family of transcription factors (GATA-1, -2, and -3) that regulate gene expression in hematopoietic tissue during both development and adult life (17).
Expression Profile of HBS1L and MYB During Erythropoiesis.
To investigate the functional significance of the trait-associated genetic variants, we used real-time quantitative RT-PCR to study the expression levels of HBS1L and MYB during erythropoiesis. As HBS1L-1a was expressed at very low levels in erythroid progenitors, it was excluded from the study. Erythroid cells obtained from 35 individuals (23 from the twin-pair panels, 2 from the Asian-Indian pedigree and 10 from other Caucasian volunteers) were cultured by using a two-phase liquid system as described (10), and RT-PCR was performed with total RNA obtained from erythroid progenitor cells on days 0 and 3 phase II erythroid culture for each individual included in the study. We hypothesized that contrasts between the extreme genotypes would be the most informative to detect effects on expression, so individuals who were homozygous at the trait-associated sites within block 2 were chosen for these studies. Alleles associated with high trait values for a block are denoted as “H” and the alleles associated with low trait values for a block as “L”. The genotype status was usually equivalent for all of the markers within a block because of the strong linkage disequilibrium. In a few instances when the genotypes were not equivalent, we classified individuals according to the predominant pattern (see legend to SI Fig. 3).
HbF and FC levels were significantly associated with genotypes in the three blocks in the samples selected for the expression study, as expected. We observed a striking relationship of increased HBS1L expression measured at day 0 associated with the presence of the H genotype in the three trait associated blocks, and a statistically less significant relationship for day 3 expression (SI Fig. 3). These results strongly suggest that the biological effects of genetic variants in one or more of these blocks include modulation of HBS1L expression.
Discussion
Our study has identified the principal genetic variants that account for the chromosome 6q QTL for F cells/HbF. These variants are distributed within three LD blocks, which we refer to as HBS1L MYB Intergenic Polymorphism (HMIP) blocks 1, 2 and 3. HMIP blocks 1, 2 and 3 span a nearly contiguous segment ≈79 kb long, starting 188 bp upstream from HBS1L exon 1 and ending 45 kb upstream of MYB (Fig. 1a). Among the 12 markers exhibiting the strongest evidence of association, one, rs52090909, is located in the 5′ UTR of exon 1a of HBS1L. The other strongly associated markers in HMIP block 2, are either in intron 1a (rs9376090, rs9399137, rs9402685 and rs11759553), or directly upstream of the 5′ UTR of HBS1L exon 1a (rs4895440, rs4895441, rs9376092, rs9389269, rs9402686, rs11154792 and rs9483788). HMIP block 1 is also located within intron 1a of HBS1L whereas HMIP block 3 is located between exon 1a of HBS1L and the first exon of MYB. Whereas markers within each of the trait-associated blocks are in strong linkage disequilibrium, there is less linkage disequilibrium and a greater diversity of frequent haplotypes between markers in different blocks (SI Table 8). The markers interspersed within a trait-associated block that are less significantly associated with the trait have lower linkage disequilibrium with the block markers (SI Tables 5–7). Each of the trait-associated blocks contains at least one marker that had also been characterized in the HapMap data set (18). As we found no significant linkage disequilibrium with HapMap markers outside of the region studied here, we concluded that the trait-associated blocks were confined to the HBS1L-MYB segment. A test of linkage in the European dizygotic (DZ) twins showed that the 6q23 QTL is completely accounted for by the markers in the three trait-associated blocks (unadjusted lod = 1.79, P = 0.002; lod adjusted for three markers that identify the trait-associated blocks = 0.0).
Based on measured haplotype analysis (SI Tables 8 and 9), we estimate that 17.6% of the trait variance is attributed to the markers in the three HBS1L-MYB blocks. An additional 11.6% of the trait variance is influenced by the Xmn I variant on chromosome 11. As the overall heritability of the FC trait in Europeans is 89% (5), it is suggested that additional genetic or other familial factors contribute substantially (residual heritability = 59.8%) to the trait variation. The genetic variants that are associated with high F cell levels are also strongly correlated to increased expression of HBS1L in cultured erythroid cells.
Interestingly, however, FC levels and HBS1L expression were not significantly correlated in this sample set despite the association of both traits with the same genetic variants. Examination of the samples showed that it was principally due to the inclusion of two individuals with high FC values who harbor the LL genotype and exhibit low HBS1L expression. The presence of such samples is not unexpected given the selection on genotype, and the fact that most of the FC trait variance (82%) is not accounted for by the HBS1L-MYB locus.
In a previous study of 26 individuals selected to have high or low HbF, we found a negative correlation between FC levels and HBS1L expression (10). The previous sample partially overlaps with the present data set, but it contains 13 (50%) individuals with the block 2 H/L genotype, and only 13 with H/H or L/L genotypes. In an attempt to reconcile the results in these two data sets, we reexamined HBS1L expression by repeating all of the RT-PCR experiments. Using the new data from all 47 individuals in the combined sample set, we found significant association of block 2 genotypes with FC levels (P = 0.007) and with HBS1L expression (day 0: P = 0.01; day 3: P = 0.03). After adjustment for genotype effects under an additive model, the residual FC trait and HBS1L expressions values were negatively correlated (day 0: ρ = −0.31, P = 0.04; day 3: ρ = −0.39, P = 0.01) as reported in the original subset. We conclude that multiple factors affect both the FC trait and HBS1L expression, and that these factors include, but are not limited to the genetic variants within HBS1L-MYB region. The sampling scheme used for ascertainment (e.g., selection on genotype or phenotype) may impact the magnitude and the direction of the observed relationships.
The biological complexity underlying gene regulation in this region is further illustrated through analysis of MYB expression. Although MYB expression was not significantly related to the genotype status (SI Fig. 4) or to FC levels in the block 2 H/H vs. L/L samples, MYB expression at day 3 was positively correlated to HBS1L expression (SI Table 10). Moreover, significant correlation remained after adjustment of HBS1L for the associated HBS1L-MYB genotypes. Thus, it would seem that the correlation of HBS1L and MYB expression is principally due to factors outside of the HBS1L-MYB locus.
The location of the most significantly associated variants and their correlation with HBS1L expression implicate HBS1L in the F Cell QTL. HBS1L (16), is a putative member of the “GTPases” superfamily (19), which bears a close relationship to the eEF-1A (eukaryotic elongation factor 1A, or EF 1α) and eRF3 (eukaryotic release factor 3) families (16, 20). GTPases, which bind and hydrolyze GTP, are involved in regulating a variety of critical cellular processes, including protein synthesis, cytoskeleton assembly, protein trafficking and signal transduction (19). Recently it has been shown that another GTP-binding protein, the secretion-associated and RAS-related (SAR) protein may be a key molecule in the induction of γ-globin expression by hydroxyurea (21). The role of HBS1L on FC levels is not immediately apparent and could be manifested indirectly through its effect on the expression of various cytokines and transcription factors that impact erythroid cell growth (15).
The present study illustrates how genetic approaches can contribute new knowledge to the regulation of human hemoglobin through dissection of the quantitative genetic variation. The identification of novel transacting genetic variants that are associated with modulation of HbF and FC levels is a key step toward resolving some of the outstanding biological questions in the field and has the potential for novel diagnostic and therapeutic applications.
Materials and Methods
Subjects and Phenotyping.
Study participants consisted of monozygotic and same-sex DZ twin pairs of North European descent. The study participants were phenotyped for F-cell levels and genotyped for the XmnI-Gγ site and 121 other markers. The twin pairs who were not selected for HbF or F-cell levels or any disease or trait, were recruited from the TwinsUK Adult Twin Registry (www.twinsuk.ac.uk) (11). The average age of the participants was 47.6 years of age, ranging from 18 to 79 years of age. The average FC level of the sample was 4.06% of total erythrocytes (SD 3.15%; range 0.23% to 36.7%).
Blood samples were collected in EDTA, and F cells were enumerated by flow cytometry of 20,000 cells by using a monoclonal anti-γ globin chain antibody conjugated with fluorescein isothiocyanate (FITC) (22). Current methods of quantifying HbF are not sensitive enough for measuring levels in the 0–1% range, the range usually encountered in normal subjects. Therefore, in normal subjects, the trait is represented by F cells measured by using a monoclonal antibody against γ chains of HbF (α2γ2).
The study was approved by the local Research Ethics Committee (LREC No: 01-332 and LREC No: 01-083) of King's College Hospital, London. XmnI-Gγ genotyping was performed on genomic DNA as described (23).
SNP Discovery.
A systematic investigation of genetic variants between HBS1L-MYB was made by resequencing this 125-kb region by using DNA from 32 European control subjects. The genomic sequence encompassing the region (NT_025741.13, 39,480,452–39,606,881, 126,430 bp) was excised with 1-kb each of adjacent sequences at both ends. PCR primers were designed by PRIMER3 to generate a total of 139 PCR amplicons (ranging from 759 bp to 1,725 bp, with an average length of 1,208 bp) with an overlap of >160 bp between adjacent amplicons. In addition, 428 internal primers were also used for sequencing. Resequencing of the human MYB gene was performed with 50 PCR amplicons generated by PRIMER3 to cover the 15 exons and parts of the introns. PCR was undertaken in 15-μl reaction volume by using 1 unit of ExTaq DNA polymerase (TaKaRa Biomedicals, Paris, France) and 25 ng of genomic DNA. The PCR profile consisted of an initial melting step of 5 min at 94°C, followed by 35 cycles of 5 s at 98°C, 30 s at 60°C, and 2 min at 72°C; and a final elongation step of 10 min at 72°C. PCR products were purified by using Bio-gel P100 Gel (Bio-Rad Inc, Hercules, CA). PCR products were sequenced by using the Bigdye Terminator cycle sequencing chemistry method. Reactions were purified by using Sephadex G-50 Superfine (Amersham Biosciences, Uppsala, Sweden) before applying to the ABI 3730 DNA Analyzers. Detection of genetic variants was performed with in-house software (the Genalys program available at http://www.cng.fr).
Erythroid Cell Cultures and Expression Analysis of HBS1L and MYB by Quantitative Real-Time PCR.
Erythroid cells were cultured by using a two-phase liquid system [modified from Fibach et al. (24)]. Mononuclear cells were isolated from peripheral blood by centrifugation on a gradient of Ficoll-Hypaque and cultured for 7 days in phase I medium which consisted of serum-free StemSpan (Stem Cell Technologies, U.K.) supplemented with 1 μg/ml cyclosporin A, 25 ng/ml interleukin-3 (IL-3), 50 ng/ml human stem cell factor (Sigma, U.K.), and 0.01% BSA. Cells were incubated at 37°C, 5% CO2. After 7 days, nonadherent cells were collected and reseeded at a concentration of 2.5 × 105 cells/ml in phase II medium [StemSpan supplemented with 10−7 M dexamethasone (Sigma, U.K.), 50 ng/ml stem cell factor, and 2 units/ml human recombinant erythropoietin (EPO; Sigma, U.K.). The cultures were diluted once or twice to maintain the cell concentration lower than 1 × 106 cells per ml in phase II. Cell samples were collected from phase II cultures on days 0 and 3.
Total RNA was isolated from erythroid cells by using Tri-reagent (Sigma, U.K.) and quantified by absorbance at 260 nm. cDNA was synthesized by using SuperScript III reverse transcriptase (Invitrogen, U.K.) from 1 μg of total RNA. Primers and probes were designed by using Primer Express 2.0 program and synthesized by Applied Biosystems. Quantitative RT-PCR was carried out in an ABI 7900 HT Sequence Detection System by using TaqMan master mix and the protocol of the manufacturer (Applied Biosystems). Sequences of the primers and probes were as follows: MYB probe, 6-FAM-TGCTACCAACACAGAACCACACATGCA-TAMRA; MYB forward primer, 5′-ATGATGAAGACCCTGAGAAGGAAA-3′; MYB reverse primer, 5′-AACAGGTGCACTGTCTCCATGA-3′; HBS1L probe, 6-FAM-CTATAACTACGATGAAGATTTT-TAMRA; HBS1L forward primer, 5′-TCTACAGACTGGCCGTAGAGATCA-3′ (in exon 2); HBS1L reverse primer, 5′-CCCGGCATCGGAATGTT-3′ (in exon 1).
All data were normalized by using the endogenous hypoxanthine phosphoribosyltransferase (HPRT) control. Assays for HPRT are available from the Applied Biosystem database. To quantify gene expression, a relative standard method was used. The quantities of targets and of the endogenous HPRT were determined from the appropriate standard curves. The target amount was then divided by the HPRT amount to obtain a normalized value. One of the experimental samples on day 0 (HPRT normalized) was designated as the calibrator, and given a relative value of 1.0. All quantities (HPRT normalized) were expressed as n-fold relative to the calibrator.
RNA Analysis.
RNA was obtained from Clontech-Europe, U.K. or prepared from cultured cells by using Tri Reagent (Sigma, U.K.) according to manufacturer's instructions. One microgram of total RNA was reverse transcribed by using SuperScript III RT (Invitrogen, U.K.) and oligo(dT) primers. One hundred nanograms of cDNA was then used in a 25-μl PCR containing TaqGold (Applied Biosystems, U.K.) at 2.5 mM MgCl2 and 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s.
Genotyping.
Markers in the target region were selected for genotyping from the dbSNP (www.ncbi.nlm.nih.gov/SNP/) and HapMap (www.hapmap.org/) databases, or from the sequencing experiments described above. Most markers were genotyped by Taqman (Applied Biosystems, Foster City, CA). Taqman reactions were performed according to the manufacturer's instructions by using 5.0 ng of purified and quantified genomic DNA. Plate reading was conducted on ABI Prism 7900HT sequence Detection System, and analysis was undertaken with SDS 2.0 software. A small number of markers were genotyped by direct sequencing with techniques as described above, or by using the tetra primer ARMS method (25). The T homopolymer upstream of MYB (Bpil) was genotyped on a microsatellite genotyping platform from Applied Biosystems, by using an ABI Prism 3100 Genetic Analyzer.
Statistical Methods.
The relationship of the quantitative trait with age, sex, and marker genotypes was evaluated by using the mixed-model ANOVA procedure (PROC MIXED) from SAS version 8.2 (SAS Institute Inc., Cary, NC) with restricted maximum likelihood estimation. Monozygotic (MZ) and DZ twins were assumed to have distinct trait variances and covariances. The combined data from Panel 1 and Panel 2 were analyzed assuming common trait variances and equal covariances for MZ and DZ twins in the two panels. Age, sex and marker genotypes were incorporated as fixed effects for analysis. Likelihood ratio tests were used to evaluate hypotheses involving equality of the variances and covariances in different subsets of the data, and to test the fit of the additive genetic model. Haplotype estimates were obtained with the MERLIN and fugue programs (26) and haploview programs (27).
PAP (version 4.2; http://hasstedt.genetics.utah.edu/) was used to estimate effects and to obtain likelihood ratio test statistics in the measured haplotype analysis by modifying the measured genotype procedure (qmlprmv). Briefly, the phenotype trait was simultaneously adjusted for age, sex and the XmnI-Gγ marker whilst fitting a measured genotype model. The variance, correlations for DZ and MZ twins, haplotype means and dominance terms were estimated by maximum likelihood conditional on the observed genotypes at the sites included in the model, the adjusted trait phenotype and the family structure. MZ twins were constrained to be identical-by-descent at the HBS1L-MYB locus by inclusion of a completely linked and fully informative indicator marker. The mean associated with the combination of two haplotypes, Hi and Hj, was written as Mi + Mj, except when considering dominance. In the latter case, the mean was expressed as Mi + Mj + DS for haplotype combinations with presence of hypothesized dominant allele at site S. Under the between-site additive model, the haplotype mean was written as the sum of means associated with the alleles at each site, plus a site-specific dominance term when it was included in the model. Likelihood ratio tests were used to test specific hypotheses involving nested models. A variance-components linkage analysis of FC levels in the DZ twins was performed with the MERLIN program (26) allowing for linkage disequilibrium between markers (28). Tests of population stratification (admixture) were performed with the QTDT program (29).
Supplementary Material
Acknowledgments
We thank C. Steward for help in preparation of the manuscript, and M. Foglio, S. Heath, D. Zelenika, A. Boland, and C. Julier for valuable advice and technical assistance. This work was supported by Medical Research Council Grant G0000111, ID51640 (to S.L.T.) and the French Ministry of Higher Education and Research (M.L.). TwinsUK is supported by the Wellcome Trust and Framework V European Union Grant GenomEU Twin.
Abbreviations
- QTL
quantitative trait locus/loci
- HbF
fetal hemoglobin
- FC
F cells
- lod
logarithm of odds
- hHPFH
heterocellular hereditary persistence of fetal hemoglobin
- LD
linkage disequilibrium
- HMIP
HBS1L MYB intergenic polymorphism
- HPRT
hypoxanthine phosphoribosyltransferase
- MZ
monozygotic
- DZ
dizygotic.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611393104/DC1.
References
- 1.Weatherall DJ, Clegg JB. Bull WHO. 2001;79:704–712. [PMC free article] [PubMed] [Google Scholar]
- 2.Thein SL, Craig JE. Hemoglobin. 1998;22:401–414. doi: 10.3109/03630269809071538. [DOI] [PubMed] [Google Scholar]
- 3.Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, Klug PP. N Engl J Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 4.Ho PJ, Hall GW, Luo LY, Weatherall DJ, Thein SL. Br J Haematol. 1998;100:70–78. doi: 10.1046/j.1365-2141.1998.00519.x. [DOI] [PubMed] [Google Scholar]
- 5.Garner C, Tatu T, Reittie JE, Littlewood T, Darley J, Cervino S, Farrall M, Kelly P, Spector TD, Thein SL. Blood. 2000;95:342–346. [PubMed] [Google Scholar]
- 6.Garner C, Tatu T, Game L, Cardon LR, Spector TD, Farrall M, Thein SL. GeneScreen. 2000;1:9–14. [Google Scholar]
- 7.Craig JE, Rochette J, Fisher CA, Weatherall DJ, Marc S, Lathrop GM, Demenais F, Thein SL. Nat Genet. 1996;12:58–64. doi: 10.1038/ng0196-58. [DOI] [PubMed] [Google Scholar]
- 8.Garner C, Mitchell J, Hatzis T, Reittie J, Farrell M, Thein SL. Am J Hum Genet. 1998;62:1468–1474. doi: 10.1086/301859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Close J, Game L, Clark BE, Bergounioux J, Gerovassili A, Thein SL. BMC Genomics. 2004;5:33. doi: 10.1186/1471-2164-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang J, Best S, Menzel S, Silver N, Lai MI, Surdulescu GL, Spector TD, Thein SL. Blood. 2006;108:1077–1083. doi: 10.1182/blood-2006-01-008912. [DOI] [PubMed] [Google Scholar]
- 11.Spector TD, MacGregor AJ. Twin Res. 2002;5:440–443. doi: 10.1375/136905202320906246. [DOI] [PubMed] [Google Scholar]
- 12.Sampietro M, Thein SL, Contreras M, Pazmany L. Blood. 1992;79:832–833. [PubMed] [Google Scholar]
- 13.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh IH, Reddy EP. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 15.Cantor AB, Orkin SH. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 16.Wallrapp C, Verrier S-B, Zhouravleva G, Philippe H, Philippe M, Gress TM, Jean-Jean O. FEBS Lett. 1998;440:387–392. doi: 10.1016/s0014-5793(98)01492-6. [DOI] [PubMed] [Google Scholar]
- 17.Ko LJ, Engel JD. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The International HapMap Consortium. Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. Nature. 2005;437:1299–1320. [Google Scholar]
- 19.Bourne HR, Sanders DA, McCormick F. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 20.Inge-Vechtomov S, Zhouravleva G, Philippe M. Biol Cell. 2003;95:195–209. doi: 10.1016/s0248-4900(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 21.Tang DC, Zhu J, Liu W, Chin K, Sun J, Chen L, Hanover JA, Rodgers GP. Blood. 2005;106:3256–3263. doi: 10.1182/blood-2003-10-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorpe SJ, Thein SL, Sampietro M, Craig JE, Mahon B, Huehns ER. Br J Haematol. 1994;87:125–132. doi: 10.1111/j.1365-2141.1994.tb04881.x. [DOI] [PubMed] [Google Scholar]
- 23.Craig JE, Sheerin SM, Barnetson R, Thein SL. Br J Haematol. 1993;84:106–110. doi: 10.1111/j.1365-2141.1993.tb03032.x. [DOI] [PubMed] [Google Scholar]
- 24.Fibach E, Manor D, Oppenheim A, Rachmilewitz EA. Blood. 1989;73:100–103. [PubMed] [Google Scholar]
- 25.Ye S, Dhillon S, Ke X, Collins AR, Day IN. Nucleic Acids Res. 2001;29:E88–8. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Abecasis GR, Wigginton JE. Am J Hum Genet. 2005;77:754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abecasis GR, Cardon LR, Cookson WO. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.