Abstract
Fas-activated serine/threonine phosphoprotein (FAST) is a survival protein that is tethered to the outer mitochondrial membrane. In cells subjected to environmental stress, FAST moves to stress granules, where it interacts with TIA1 to modulate the process of stress-induced translational silencing. Both FAST and TIA1 are also found in the nucleus, where TIA1 promotes the inclusion of exons flanked by weak splice recognition sites such as exon IIIb of the fibroblast growth factor receptor 2 (FGFR2) mRNA. Two-hybrid interaction screens and biochemical analysis reveal that FAST binds to several alternative and constitutive splicing regulators, suggesting that FAST might participate in this process. The finding that FAST is concentrated at nuclear speckles also supports this contention. We show that FAST, like TIA1, promotes the inclusion of exon IIIb of the FGFR2 mRNA. Both FAST and TIA1 target a U-rich intronic sequence (IAS1) adjacent the 5′ splice site of exon IIIb. However, unlike TIA1, FAST does not bind to the IAS1 sequence. Surprisingly, knockdown experiments reveal that FAST and TIA1 act independently of one another to promote the inclusion of exon IIIb. Mutational analysis reveals that FAST-mediated alternative splicing is separable from the survival effects of FAST. Our data reveal that nuclear FAST can regulate the splicing of FGFR2 transcripts.
Keywords: TIA-1, FGFR2, exon definition
Alternative splicing of heteronuclear mRNA leads to the production of functionally distinct protein products derived from a common transcript (1). The boundaries of individual exons are recognized by consensus sequence elements found at exon/intron junctions. The recruitment of the splicing machinery to these minimal elements is influenced by exonic and intronic cis elements that promote or inhibit exon recognition (1). Fibroblast growth factor receptor-2 (FGFR2) is a well studied example of a gene that is regulated by alternative splicing. FGFR2 is produced from transcripts that include one of two adjacent exons (IIIb or IIIc) and encode receptors with distinct ligand-binding properties (2). FGFR2 splice variant choice is critical for organogenesis and prostate cancer progression (3–7). Previous studies have identified several exonic and intronic cis elements that determine whether exon IIIb or exon IIIc is included in FGFR2 mRNA transcripts (8–12). Exon IIIb has weak splice sites, and its inclusion is hampered further by an exon splicing silencer (ESS) and flanking intronic splicing silencers (ISS) that recruit hnRNPA1 (13) and pyrimidine tract-binding protein (PTB) (14), respectively. The inclusion of exon IIIb depends on a U-rich intronic splicing enhancer known as IAS1 located downstream of the 5′ splice site (15). IAS1 recruits TIA1, a regulatory factor that facilitates 5′ splice site recognition by U1 snRNP, to promote inclusion of exon IIIb (16–21).
TIA1 also functions as a translational silencer that promotes the assembly of nonfunctional 48S preinitiation complexes into cytoplasmic granules (stress granules) in cells subjected to environmental stress (22). The TIA1-interacting protein Fas-activated serine/threonine phosphoprotein (FAST) is a component of stress granules that promotes the expression of endogenous inhibitors of apoptosis to prevent stress-induced apoptosis (23). Here, we show that FAST, like TIA1, is a regulator of alternative splicing that is recruited by IAS1 to promote the inclusion of FGFR2 exon IIIb. Our knockdown experiments show that FAST and TIA1 act independently of each other in regulating FGFR2 splicing. Two-hybrid interaction screens and biochemical analysis also show that FAST binds to several other splicing regulators: HNRPK, KHDRBS1 (also known as Sam68), TIAL1 (also known as TIAR), and SF3B4 (also known as SAP49). Finally, we report that the last 188 amino acids of FAST, which are dispensable for survival activity, are absolutely required for the activation of exon IIIb.
Results
Identification of FAST-Interacting Proteins.
It has been reported that FAST binds to TIA1, an RNA-binding protein that participates in stress-induced translational arrest (22) and alternative pre-mRNA splicing (16–18). We used two different Gal4-based yeast two-hybrid systems to identify additional FAST partners, as described in Materials and Methods. We obtained 24 potential interactors that we grouped into broad functional categories [supporting information (SI) Table 1]: (i) splicing and RNA metabolism, (ii) transcription, (iii) proteasome pathway, and (iv) intracellular signaling. There are six proteins with unclear function. Because so many of FAST's interactors have a characterized role in RNA-related functions, we hypothesized that FAST is a protein that regulates events related to mRNA metabolism.
FAST Binds to Splicing Factors and Localizes to Nuclear Speckles.
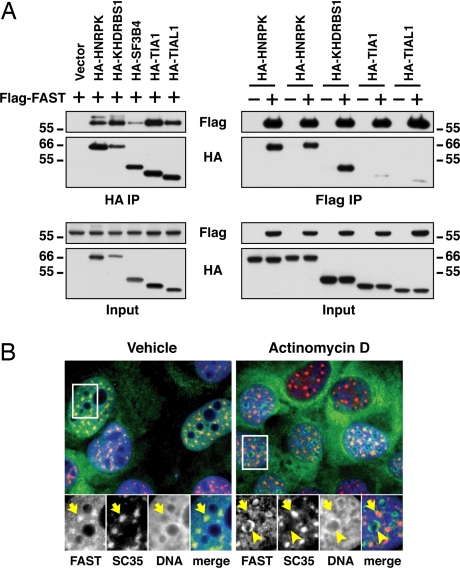
We focused our analysis on the potential for FAST to regulate splicing. To verify the interactions between FAST and the splicing factors HNRPK, KHDRBS1, SF3B4, TIA1, and TIAL1, we carried out coimmunoprecipitation assays with transfected HEK293T cells. As shown in Fig. 1A, hemagglutinin (HA)-HNRPK, HA-KHDRBS1, HA-SF3B4, HA-TIA1, and HA-TIAL1 each interact with Flag-tagged FAST. Coimmunoprecipitation in the other direction gave qualitatively similar results.
Fig. 1.
FAST binds to splicing factors and localizes to nuclear speckles. (A) After the transfection of HEK293T cells with the indicated plasmids, HA-conjugated proteins were immunoprecipitated by using rat monoclonal antibody 3F10 and Flag-FAST was purified with anti-Flag M2 agarose beads. Complexes and lysates were immunoblotted with anti-HA and anti-Flag antibodies. (B) U2OS cells stably transfected with a vector encoding FAST-YFP (green) were analyzed by immunofluorescence staining with anti-SC-35, followed by anti-mouse IgG-Cy3 antibody (red). DNA was visualized with Hoeschst 33342 (blue). Individual stainings and merge images are shown. Cells were treated for 4 h with vehicle or 5 μg/ml actinomycin D as indicated. Arrows indicate FAST-YFP colocalized with SC-35, and arrowheads indicate nucleoli.
Similar results were obtained when coimmunoprecipitations were performed by using RNase-treated cell extracts (data not shown), indicating that these interactions are not RNA-dependent.
Immunofluorescence microscopy has revealed that FAST is found in both the nucleus and at the outer mitochondrial membrane (23, 24). Here, we have further analyzed the distribution of FAST. Total lysates from wild-type mouse embryonic fibroblasts (WT-MEFs) and FAST knockout MEFs (FAST KO-MEFs) (M.S. and P.A., unpublished work) were probed with a goat anti-FAST antibody; two bands of 60 and 50 kDa were detected in WT-MEFs but not in FAST KO-MEFs. Subcellular fractionation showed that FAST is in both the cytoplasm and the nucleus (SI Fig. 9). Immunostaining of WT-MEFs and FAST KO-MEFs by using the same goat anti-FAST antibody showed similar nonspecific patterns (data not shown), leading us to generate a U2OS stable cell line expressing FAST-YFP. As shown in Fig. 1B, nuclear FAST-YFP accumulates at nuclear speckles (indicated by SC35) known to harbor components of the splicing machinery (25).
Given the close relationship between transcription and splicing events, we determined whether the localization of FAST-YFP in nuclear speckles was sensitive to transcription inhibition. Consistent with previous studies (25), the transcription inhibitor actinomycin D reduced the number of speckles, which were larger and more rounded (Fig. 1B). Treatment with actinomycin D resulted in the movement of FAST-YFP to the periphery of the nucleoli. Thus, whereas FAST-YFP is concentrated at nuclear speckles, it may not be an integral component of these foci.
FAST and FGFR2 Splicing in Vivo.
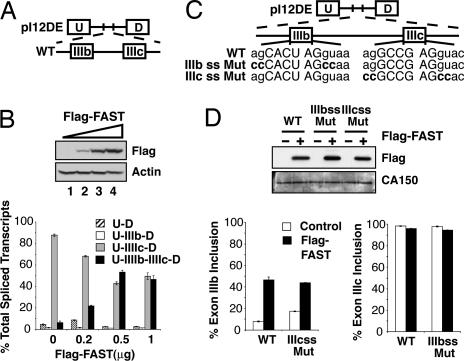
The FAST-interacting protein TIA1 has been reported to promote the inclusion of exon IIIb in the FGFR2 system, and therefore we asked whether FAST could also modulate the use of this exon. We used a splicing reporter gene that includes exons IIIb and IIIc, the cis elements that regulate their alternate splicing, and heterologous upstream (U) and downstream (D) exons (pI12DE-WT: Fig. 2A). This minigene can be alternatively spliced to produce four RNA variants: U-D, U-IIIb-D, U-IIIc-D, and U-IIIb-IIIc-D (26). Analysis of the splicing patterns of the reporter gene was performed by semiquantitative RT-PCR. pI12DE-WT was cotransfected in HEK293T cells with increasing amounts of Flag-FAST. In the control samples, the major splicing product contains only exon IIIc (Fig. 2B). Overexpression of Flag-FAST increased the production of U-IIIb-IIIc-D by 3- to 5-fold and concomitantly reduced the production of U-IIIc-D.
Fig. 2.
FAST regulates the inclusion of FGFR2 exon IIIb in vivo. (A) Schematic diagram of the minigene pI12DE-WT. (B) HEK293T cells were transfected with the minigene pI12DE-WT and increasing amounts of pcDNA3-Flag-FAST. Total lysates were probed for actin as a loading control and Flag. The quantification of the transcripts is shown below. (C) Schematic diagram of minigenes containing mutations in 5′ splice sites flanking exons IIIb (pI12DE-IIIb ss Mut) and IIIc (pI12DE-IIIc ss Mut). Mutated nucleotides are indicated in bold type. (D) HEK293T cells were transfected with the minigenes diagramed in C and pcDNA3-Flag or pcDNA3-Flag-FAST. Total lysates were probed for CA150 as a loading control and Flag. Quantification of the transcripts is shown below.
To determine whether FAST promotes the inclusion of IIIb, represses the excision of IIIc, or both, we examined the effects of FAST on the splicing of mutant reporters that are incapable of including exon IIIb (IIIb ss Mut) or exon IIIc (IIIc ss Mut) (Fig. 2C). In reporters that cannot splice out exon IIIc, FAST promoted the inclusion of exon IIIb to an extent similar to that of the wild-type reporter (Fig. 2D Left). In contrast, in reporters incapable of splicing exon IIIb, FAST did not repress the inclusion of exon IIIc (Fig. 2D Right). Therefore, FAST overexpression does not affect the inclusion of exon IIIc but activates that of exon IIIb.
Identification of FAST Domains Required for Its Splicing Activity.
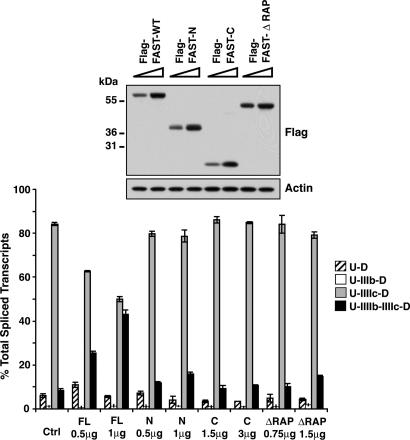
To identify the FAST domains required for its splicing activity, we constructed Flag-FAST-N (amino acids 1–369), Flag-FAST-C (amino acids 370–550) and Flag-FAST-ΔRAP (deletion of amino acids 477–535). FAST-N maintains the ability to bind to TIA1 and inhibits FAS-induced apoptosis (23). FAST-C, which we used as a putative negative control, contains the last 181 amino acids and is tethered to the mitochondria (24). FAST-C is unable to bind to TIA1 or inhibit FAS-induced apoptosis (23). RAP is a ≈60-residue domain found in FAST (amino acids 477–535) and various other eukaryotic proteins (27). It is abundant in apicomplexans and it seems to be required for the transsplicing activity of Raa3 (27).
We cotransfected increasing amounts of Flag-FAST-WT and Flag-FAST deletion mutants with a constant amount of pI12DE-WT in HEK293T cells. None of the FAST mutants altered the alternative splicing of exons IIIb and IIIc (Fig. 3). For the proper interpretation of these data, it was important to determine the distribution of the mutants in the cell by immunofluorescence. As shown in SI Fig. 10, Flag-FAST-WT and Flag-FAST-N were predominantly nuclear, Flag-FAST-ΔRAP was both nuclear and cytoplasmic, and Flag-FAST-C was predominantly cytoplasmic. We also assessed the ability of Flag-FAST-N and Flag-FAST-ΔRAP (both nuclear), to bind to HA-tagged HNRPK, HA-KHDRBS1, HA-SF3B4, HA-TIA1, and HA-TIAL1. As shown in SI Fig. 10, the deletion of amino acids 370–550 in Flag-FAST-N only abolished the interaction between FAST and SF3B4, and the deletion of amino acids 477–535 (ΔRAP) did not affect any of the interactions between FAST and the splicing factors.
Fig. 3.
FAST domains required to activate FGFR2 exon IIIb. HEK293T cells were transfected with pI12DE-WT and increasing amounts of pcDNA3-Flag-FAST-WT or pcDNA3-Flag-FAST mutants. Total lysates were probed for actin and Flag. Quantification of the transcripts is shown below.
Taken together, these data indicate that, despite their nuclear localization and their interaction with TIA1, FAST-N and FAST-ΔRAP do not have splicing activity on FGFR2. That implies that the carboxyl-terminal 181 amino acids and particularly the RAP domain of FAST are required. The results concerning the splicing activity of Flag-FAST-C are inconclusive, given their location in the cytoplasm.
FAST Requires the IAS1 Sequence for Splicing.
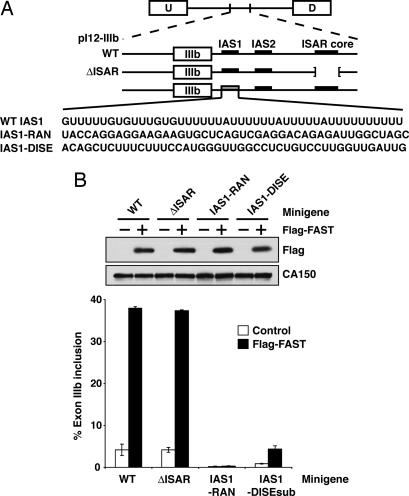
We next determined the effect of FAST on alternative splicing of exon IIIb reporters containing mutations in the cis-acting elements known to regulate the inclusion of exon IIIb. IAS1, a sequence immediately downstream of exon IIIb, is known to promote exon inclusion (16). IAS2 and ISAR, downstream intronic sequences that form a stem structure, activate IIIb inclusion and repress IIIc (26, 28). The downstream intronic splicing enhancer (DISE) is an element downstream of exon IIIc that can replace IAS1 in its ability to activate IIIb inclusion (P. Seth and M.A.G.-B., unpublished work). Analysis of mutant reporters allowed us to determine whether the ability of FAST to promote the inclusion of exon IIIb required these cis elements.
We cotransfected Flag-FAST with pI12-IIIb-WT and the indicated pI12-IIIb mutants (Fig. 4A) in HEK293T cells, and exon IIIb inclusion was measured by RT-PCR. As shown in Fig. 4B, Flag-FAST activates the splicing of the WT minigene ≈9-fold. Flag-FAST activates the splicing of the minigene ΔISAR to a similar extent. In contrast, the replacement of IAS1 sequence with a random sequence (minigene IAS1-RAN) abolishes the ability of Flag-FAST to activate splicing, which indicates that FAST requires the IAS1 element to promote exon IIIb inclusion. When we replaced IAS1 sequence with a similar intronic splicing enhancer (DISE), DISE was capable of supporting a 5-fold activation of exon IIIb by overexpressed FAST but was incapable of fully replacing IAS1.
Fig. 4.
FAST requires IAS1, but not ISAR, to activate FGFR2 exon IIIb. (A) Schematic diagram of pI12-IIIb minigenes. The parent minigene is shown, with exon IIIb and its regulatory cis-elements. The expanded diagram shows the wild-type sequence of the 5′ splice site and IAS1 along with the mutant sequences are shown. (B) HEK293T cells were transfected with the minigenes diagramed above and 1 μg of pcDNA3-Flag or pcDNA3-Flag-FAST. Total lysates were probed for CA150 and Flag. Quantification of exon IIIb inclusion transcripts is shown below.
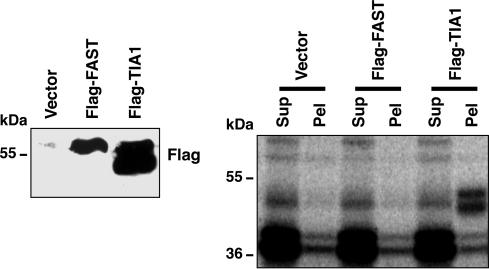
Given the requirement of the RAP domain and the IAS1 sequence for the splicing activity of FAST, we tested the ability of FAST to bind to IAS1. HEK293T cells were transfected with Flag-FAST and Flag-TIA1. TIA1 acted as a positive control because it is known to be recruited by IAS1 (16). Nuclear extracts were prepared and UV cross-linked with a32P-labeled IAS1 RNA probe. As shown in Fig. 5, Flag-FAST was unable to coimmunoprecipitate IAS1 RNA probe. Therefore, FAST indirectly interacts with IAS1 for its splicing activity.
Fig. 5.
FAST does not coimmunoprecipitate with IAS1 RNA probe. HEK293T cells were transfected with the indicated plasmids, and protein levels were monitored by Western blot. Nuclear extracts were UV cross-linked with 32P-labeled IAS1 RNA probe and immunoprecipitated. Supernatants (Sup) and pellets (Pel) from the coimmunoprecipitation were analyzed by autoradiography.
FAST Splicing Activity Is Independent of TIA1 and TIAL1.
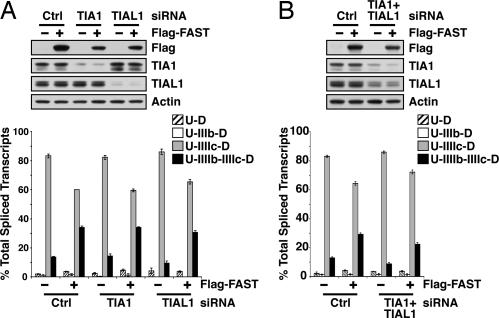
Because FAST and TIA1 both require IAS1 to activate exon IIIb, we sought to determine whether FAST activity depended on TIA1. HEK293T cells were treated with siRNA targeting TIA1 or its homologue TIAL1 and then transfected with Flag-FAST and pI12DE-WT. As shown in Fig. 6A, there was no significant change in Flag-FAST-induced inclusion of exon IIIb in cells with significantly reduced levels of TIA1 or TIAL1. This was confirmed by using TIA1 KO-MEFs, where the cotransfection of Flag-FAST and the minigene pI12DE-WT resulted in a similar activation of exon IIIb as compared with WT-MEFs (data not shown). Moreover, FAST activation was not deterred in cells with reduced levels of both TIA1 and TIAL1 (Fig. 6B). This result suggests that FAST can function in the absence of TIA1 and TIAL1.
Fig. 6.
FAST does not require TIA1 or TIAL1 to activate to FGFR2 exon IIIb. (A) HEK293T cells were transfected with siRNAs targeting TIA1 or TIAL1 as well as the minigene pI12DE-WT and pcDNA3-Flag-FAST (1 μg). Proteins were extracted and monitored by Western blot analysis, and RNAs were analyzed for single and double-inclusion transcripts. (B) HEK293T cells were simultaneously transfected with TIA1 and TIAL1 siRNAs as well as pI12DE-WT and pcDNA3-Flag-FAST (1 μg) and analyzed as described for A.
TIA1/L1 Splicing Activity Is Independent of FAST.
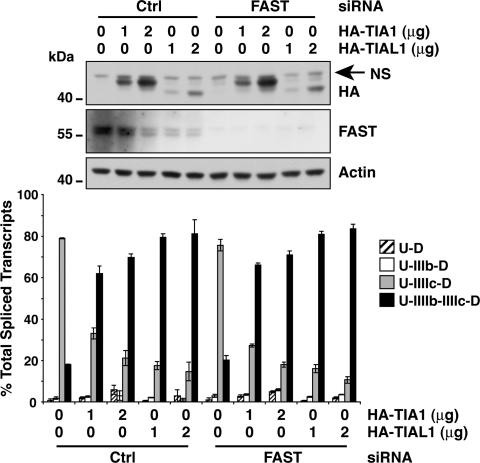
We used a similar analysis to determine whether the alternative splicing activity of TIA1 and TIAL1 depends on FAST. HEK293T cells were treated with siRNA targeting FAST and then transfected with HA-TIA1 or HA-TIAL1. As shown in Fig. 7, the ability of HA-TIA1 and HA-TIAL1 to promote the inclusion of exon IIIb was not deterred in cells with reduced levels of FAST. Therefore, the abilities of either TIA1/L1 or FAST to promote the inclusion of exon IIIb are independent of each other.
Fig. 7.
TIA1 and TIAL1 do not require FAST to activate to FGFR2 exon IIIb. HEK293T cells were transfected with pI12DE-WT, the indicated plasmids, and siRNAs. Proteins were extracted and monitored by Western blot analysis, and RNAs were analyzed for single and double-inclusion transcripts.
FAST Is Not Essential for FGFR2 Alternative Splicing.
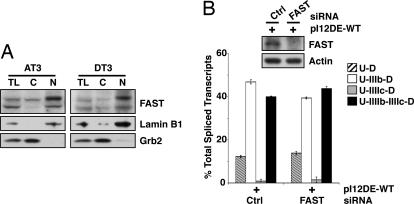
To determine whether FAST is essential for FGFR2 alternative splicing, we used the rat prostate cancer cell lines AT3 and DT3. The DT3 cell line is a well differentiated carcinoma that solely expresses FGFR2 IIIb, whereas the AT3 line is poorly differentiated and exclusively expresses FGFR2 IIIc (7). As shown in Fig. 8A, AT3 and DT3 express similar levels of FAST, negating the possibility that FGFR2 splicing is influenced by differences in FAST expression.
Fig. 8.
FAST is dispensable for exon IIIb activation. (A) Total (TL), cytoplasmic (C), and nuclear (N) protein from AT3 and DT3 cells were probed for FAST, Grb2 as a cytoplasmic marker and Lamin B1 as a nuclear marker. (B) DT3 cells were transfected with siRNAs targeting FAST and pI12DE-WT. Proteins were extracted and monitored by Western blot analysis, and RNAs were analyzed for single and double-inclusion transcripts.
We next determined whether targeted knock-down of FAST affects alternative splicing of FGFR2 in DT3 cells. Cells were cotransfected with FAST siRNA and the minigene pI12DE-WT. In the control samples, the major splicing product of the minigene reporter contains both exon IIIb and exon IIIc (double inclusion). In cells expressing reduced amounts of FAST, the double-inclusion transcript is expressed at a similar level (Fig. 8B). Thus, FAST is not absolutely required for the activation of exon IIIb in DT3 cells.
Discussion
Previous studies have shown that FAST interacts with TIA1 (29) and that FAST/TIA1 interactions play a role in the regulation of both protein translation and cell survival (23). The ability of TIA1 to also regulate alternative splicing suggests that FAST might participate in this process. TIA1 promotes the inclusion of an exon encoding the transmembrane domain of the death receptor Fas (17–19). TIA1/L1 also promotes the inclusion of alternative exons encoded by Drosophila male-specific-lethal 2 (17), myosine phosphatase targeting subunit 1 (21), calcitonin/CGRP (30), and FGFR2 (16). In each case, TIA1/L1 is recruited to cis elements downstream of a weak splice site. The carboxyl terminus of TIA1 then binds to U1 snRNP protein U1-C, allowing the recruitment of this snRNP to the 5′ splice site (18).
Like U1-C (18), FAST binds to the glutamine-rich carboxyl terminus of TIA1 (23), suggesting that nuclear FAST might play a role in the regulation of alternative splicing. Consistent with this possibility was the finding that FAST interacts with multiple splicing factors in both yeast two-hybrid and coimmunoprecipitation analyses (Fig. 2). Furthermore, FAST-YFP appears at SC35+ nuclear speckles, reservoirs of factors required for cotranscriptional splicing occurring at chromosomes (25).
Our results show that FAST can regulate alternative splicing of an FGFR2 reporter construct. FAST, like TIA1, promotes the inclusion of exon IIIb in an IAS1-dependent manner. A truncation mutant encoding the N terminus (including the TIA1-binding domain) of FAST is inactive in this assay, suggesting that recombinant FAST does not function by binding to TIA1 to affect its interactions with other proteins such as U1-C. This conclusion is strengthened by the observation that TIA1/L1 knockdown has no significant effect on FAST-mediated inclusion of exon IIIb. We conclude that FAST functions in a TIA1-independent manner.
The IAS1-dependence of FAST-mediated inclusion of exon IIIb is surprising, given its TIA1/L1-independence and our finding that FAST does not directly bind to IAS1. It is possible that FAST allows U1-snRNP to bind the 5′ splice site in the absence of TIA1 by interacting with other IAS1-binding factors.
AT3 cells exclude exon IIIb, whereas DT3 cells include exon IIIb. Our results indicate that the levels of nuclear and cytoplasmic FAST are similar in both cells, indicating that FAST is unlikely to be the determining factor in exon IIIb expression in these cells. This is consistent with the fact that FAST can promote exon IIIb inclusion in the absence of the ISAR element, which is a cell type-specific ISE. Moreover, targeted knockdown of endogenous FAST in DT3 cells did not affect the inclusion of exon IIIb. Finally, we analyzed the lung, a tissue that highly expresses FGFR2 IIIb (31), of FAST KO mice and observed normal levels of FGFR2 IIIb inclusion (data not shown). This suggests that FAST is one of many functionally redundant splicing regulators for FGFR2.
FAST is a member of a family of proteins that share a leucine-rich domain and the putative RNA-binding domain RAP (27). Indeed, preliminary results have revealed that the FAST homologue TBRG4 can bind to TIA1 and regulate its function (M.S. and P.A., unpublished work). Therefore, a FAST homologue could allow for the inclusion of exon IIIb in the absence of FAST.
Our results introduce FAST as a regulator of alternative splicing. We have demonstrated its ability to regulate the splicing of the FGFR2 gene, and we predict that it will also regulate the splicing of additional genes encoding exons with weak 5′ splice sites. Interestingly, while this manuscript was under revision, JM Izquierdo et al. (32) described a TIA1-dependent system whereby FAST influences Fas alternative splicing. Perhaps FAST influences splicing activity through various mechanisms depending on the reporter system used. It will also be important to determine whether other proteins possessing the FAST leucine-rich domain can regulate alternative splicing. Finally, the physiologic importance of FAST will have to await analysis of mutant organisms lacking the expression of this multifunctional protein.
Materials and Methods
Yeast Two-Hybrid Screening.
Two complementary protocols were used. The full-length cDNA of FASTK was cloned into the pMA424 vector to produce a fusion protein Gal4 DNA-binding domain (Gal4 DBD)-FASTK. The yeast strain GGY1::171 was then cotransformed with (Gal4 DBD)-FASTK and a human B cell cDNA library that express fusion proteins with the Gal4 activation domain (Gal4 AD; pSE1107-based). About 1 million transformants were screened for each bait for their ability to grow on plates lacking histidine, leucine, and uracil and for β-galactosidase activity. In the second protocol, a Gateway system-based protocol was used (33). The yeast strain MaV103 expressing Gal4 DBD-FASTK as bait (pPC97-based) was transformed with either a human spleen or a brain Gal4 AD-cDNA library (pPC86-based). About 5 million transformants were screened by uracil and histidine (in the presence of 20 mM 3-amino-1,2,4-triazole)-positive nutritional selection. Positive yeast colonies were then tested for β-galactosidase activity and for negative selection in the presence of 5-fluorotic acid and uracil. All interactions were carefully retested in fresh yeast cells by gap repair as described (33). Next, positive colonies for three phenotypic assays were retrieved by PCR amplification.
Plasmids.
Procedures for plasmid construction are indicated in SI Table 2. pCI-puro was a generous gift from S. D. Scott (Biotechnology and Biological Sciences Research Council, U.K.). pcDNA3-Flag is a derivative of pcDNA3 (Invitrogen, Carlsbad, CA) containing a Flag tag upstream of a modified polylinker (sequences available upon request). The plasmid encoding FAST-YFP was described (34). The FGFR2 splicing minigene reporters were described (8, 14, 26). Constructs pI12-IIIb-IAS1-RAN and pI12-IIIb-IAS1-DISE were created by chimeric PCR (P. Seth and M.A.G.-B., unpublished work).
Antibodies.
Sources of commercial antibodies are as follows: goat polyclonal anti-FAST, goat polyclonal anti-TIA1, goat polyclonal anti-TIAL1, rabbit polyclonal anti-KHDRBS1, goat polyclonal anti-HNRPK, goat polyclonal anti-SF3B4, rabbit polyclonal anti-Lamin B1, and rabbit polyclonal anti-Grb2 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); rat monoclonal anti-HA was from Roche (Indianapolis, IN); purified rabbit polyclonal anti-HA was from Covance (Berkeley, CA); mouse monoclonal anti-Flag M2, anti-Flag M2-conjugated agarose beads, rabbit polyclonal anti-Flag, and mouse monoclonal anti-splicing factor SC-35 was from Sigma–Aldrich (St. Louis, MO); mouse monoclonal anti-actin was from Chemicon International (Temecula, CA); rabbit polyclonal anti-histone H3 was from Abcam (Cambridge, MA); human anti-mitochondria autoantisera was from ImmunoVision (Springdale, AR); and all secondary antibodies for immunofluorescence and Western blotting (Cy2, Cy3, Cy5, and horseradish peroxidase-conjugates) were from Jackson ImmunoResearch (West Grove, PA). A polyclonal antibody directed at human CA150 was described (8).
Cell Culture.
HEK 293T cells, U2OS, AT3, DT3, and mouse embryonic fibroblasts (MEFs) were grown in Dulbecco's modified Eagle's medium (low glucose) supplemented with 10% FBS (HyClone, Logan, UT). For producing stable U2OS transfectants expressing FAST-YFP, U2OS cells were transfected with pEF-FAST-YFP, selected with G418, and screened for FAST-YFP expression by immunoblotting and immunofluorescence. Clonal transfectants were also established by limited dilution method. MEFs were prepared from E9.5 embryos with and without targeted disruption of the FAST gene (M.S. and P.A., unpublished work). FAST−/− MEFs have been characterized by Southern blotting, Northern blotting and Western blotting.
Protein Interactions.
Protein interactions were examined by coimmunoprecipitation as described (23).
Protein/RNA Interactions.
HEK 293T cells were transfected with Flag-tagged constructs by using Lipofectamine 2000, and nuclear extracts were prepared as described (35). Radiolabeled RNA containing the 5′splice site of exon IIIb and the IAS1 element was produced by in vitro transcription with α[32P]UTP as follows. First, a DNA fragment was generated by PCR using the following primers: 5′-ATTAATACGACTCACTATAGGGAGAAGGCTCACTGTCCTGCCC-3′ (forward) and 5′-TCTTTTTTAACTCCTATATTCAG-3′ (reverse). From this PCR fragment, the RNA was transcribed and gel-purified by using standard techniques. RNA (300,000 cpm) was incubated with 40 μg of nuclear extract in a reaction containing 2 mM MgCl2, 65 mM KCl, and 50 nM Heparin. Reactions were incubated for 15 min at 30°C. Then, reactions were placed on ice and irradiated twice with 0.5 J in a UV Stratalinker 2400 (Stratagene, La Jolla, CA). RNase A (10 μg; Sigma) was added to each reaction, and the reactions were incubated at 37°C for 1 h. Immunoprecipitations of the cross-linked flag-tagged proteins were performed. Samples were run on a 12.5% SDS/PAGE gel, and the gel was dried and exposed to a phosphorscreen overnight.
Immunofluorescence.
Cells were stained and processed for fluorescence microscopy as described (22). Conventional fluorescence microscopy was performed by using a microscope (model Eclipse E800; Nikon, East Rutherford, NJ) with epifluorescence optics and a digital camera (model CCD-SPOT RT; Diagnostic Instruments, Sterling Heights, MI). The images were compiled by using Photoshop (Adobe Systems, Mountain View, CA) software.
siRNA Transfection.
HEK-293T and DT3 cells were transfected with siRNA duplexes as described (8). The following siRNA target sequences were chosen: D0 (control),5′-GCAUUCACUUGGAUAGUAA-3′; F4 (FAST), 5′-GUCAGCUCAUCAUCCGAAA-3′; T1 (TIA1), 5′-GGAAGUCAAAGUGAAUUGG-3′; R1 (TIAL1), 5′-GGAGGUCAAAGUAAACUGG-3′.
RNA Isolation and RT-PCR Assay for Transfected Minigenes.
Splicing products of minigenes were analyzed by semiquantitative RT-PCR analysis as described (14).
Supplementary Material
Acknowledgments
We thank Puneet Seth for the construction of pI12-IIIb-IAS1-RAN and pI12-IIIb-IAS1-DISE and Florence Poy for technical help. This work was supported by National Institutes of Health Grants R01 GM63090 (to M.A.G.-B.), R01 AR51472 (to P.A.), and a Postdoctoral Supplement AI50167S1 (to M.S.).
Abbreviations
- FAST
Fas-activated serine/threonine phosphoprotein
- FGFR2
fibroblast growth factor receptor 2
- HA
hemagglutinin.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704964104/DC1.
References
- 1.Blencowe BJ. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Miki T, Bottaro DP, Fleming TP, Smith CL, Burgess WH, Chan AM, Aaronson SA. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns RC, Fairbanks TJ, Sala F, De Langhe S, Mailleux A, Thiery JP, Dickson C, Itoh N, Warburton D, Anderson KD, Bellusci S. Dev Biol. 2004;265:61–74. doi: 10.1016/j.ydbio.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 4.Carstens RP, Eaton JV, Krigman HR, Walther PJ, Garcia-Blanco MA. Oncogene. 1997;15:3059–3065. doi: 10.1038/sj.onc.1201498. [DOI] [PubMed] [Google Scholar]
- 5.De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. Development (Cambridge, UK) 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 6.Hajihosseini MK, Wilson S, De Moerlooze L, Dickson C. Proc Natl Acad Sci USA. 2001;98:3855–3860. doi: 10.1073/pnas.071586898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehan WL. Mol Cell Biol. 1993;13:4513–4522. doi: 10.1128/mcb.13.8.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baraniak AP, Chen JR, Garcia-Blanco MA. Mol Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hovhannisyan RH, Carstens RP. Mol Cell Biol. 2005;25:250–263. doi: 10.1128/MCB.25.1.250-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hovhannisyan RH, Warzecha CC, Carstens RP. Nucleic Acids Res. 2006;34:373–385. doi: 10.1093/nar/gkj407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner EJ, Baraniak AP, Sessions OM, Mauger D, Moskowitz E, Garcia-Blanco MA. J Biol Chem. 2005;280:14017–14027. doi: 10.1074/jbc.M414492200. [DOI] [PubMed] [Google Scholar]
- 12.Wagner EJ, Curtis ML, Robson ND, Baraniak AP, Eis PS, Garcia-Blanco MA. RNA. 2003;9:1552–1561. doi: 10.1261/rna.5840803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carstens RP, Wagner EJ, Garcia-Blanco MA. Mol Cell Biol. 2000;20:7388–7400. doi: 10.1128/mcb.20.19.7388-7400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Gatto F, Breathnach R. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stevenin J, Breathnach R. Mol Cell Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J. Mol Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 18.Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Le Guiner C, Lejeune F, Galiana D, Kister L, Breathnach R, Stevenin J, Del Gatto-Konczak F. J Biol Chem. 2001;276:40638–40646. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
- 21.Shukla S, Dirksen WP, Joyce KM, Le Guiner-Blanvillain C, Breathnach R, Fisher SA. J Biol Chem. 2004;279:13668–13676. doi: 10.1074/jbc.M314138200. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, Simarro M, Kedersha N, Anderson P. Mol Cell Biol. 2004;24:10718–10732. doi: 10.1128/MCB.24.24.10718-10732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Kedersha N, Chen S, Gilks N, Lee G, Anderson P. Biochem Biophys Res Commun. 2004;318:95–102. doi: 10.1016/j.bbrc.2004.03.188. [DOI] [PubMed] [Google Scholar]
- 25.Lamond AI, Spector DL. Nat Rev Mol Cell Biol. 2003;4:605–12. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 26.Carstens RP, McKeehan WL, Garcia-Blanco MA. Mol Cell Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee I, Hong W. Trends Biochem Sci. 2004;29:567–570. doi: 10.1016/j.tibs.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Baraniak AP, Lasda EL, Wagner EJ, Garcia-Blanco MA. Mol Cell Biol. 2003;23:9327–9337. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Q, Taupin JL, Elledge S, Robertson M, Anderson P. J Exp Med. 1995;182:865–874. doi: 10.1084/jem.182.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H, Hasman RA, Young KM, Kedersha NL, Lou H. Mol Cell Biol. 2003;23:5959–5971. doi: 10.1128/MCB.23.17.5959-5971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orr-Urtreger A, Bedford MT, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 32.Izquierdo JM, Valcarcel J. J Biol Chem. 2007;282:1539–1543. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- 33.Walhout AJ, Vidal M. Methods. 2001;24:297–306. doi: 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- 34.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.