Abstract
Flowering time is a fundamental trait of maize adaptation to different agricultural environments. Although a large body of information is available on the map position of quantitative trait loci for flowering time, little is known about the molecular basis of quantitative trait loci. Through positional cloning and association mapping, we resolved the major flowering-time quantitative trait locus, Vegetative to generative transition 1 (Vgt1), to an ≈2-kb noncoding region positioned 70 kb upstream of an Ap2-like transcription factor that we have shown to be involved in flowering-time control. Vgt1 functions as a cis-acting regulatory element as indicated by the correlation of the Vgt1 alleles with the transcript expression levels of the downstream gene. Additionally, within Vgt1, we identified evolutionarily conserved noncoding sequences across the maize–sorghum–rice lineages. Our results support the notion that changes in distant cis-acting regulatory regions are a key component of plant genetic adaptation throughout breeding and evolution.
Keywords: cloning, gene regulation, transformation, linkage disequilibrium
After the domestication of maize (Zea mays L.) took place in Central America (1), natural genetic variations in flowering time enabled early Native Americans to select maize adapted to a range of latitudes and lengths of growing seasons, including the very short summer season typical of the eastern Canadian region of Quebec. Under such conditions, early flowering allows seed to mature before the onset of frost. Flowering time is also a key trait of improved drought tolerance. Indeed, it has been shown that a single day of drought during flowering can decrease yield by as much as 8% (2). One way to address such losses is to develop and grow cultivars characterized by a short cycle and able to flower before predictable drought episodes.
The genetic variability available for maize breeding is essentially quantitative; i.e., it involves allelic variation at different quantitative trait loci (QTLs), which are influenced by environmental effects. Although a large body of mapping information on QTLs is available for flowering time (3), relatively little is known about the molecular basis of QTLs, with only one gene, Dwarf8, correlated thus far with quantitative effects (4, 5). Furthermore, a few mutants for flowering time have been described (6, 7), two of which, id1 (8) and dlf1 (9), have been cloned. Our results (i) show that the allelic variation responsible for the major flowering-time QTL, Vegetative to generative transition 1 (Vgt1) (10, 11) on chromosome 8, is confined to an ≈2-kb intergenic region upstream of an Ap2-like flowering-time gene, (ii) identify maize–sorghum–rice evolutionarily conserved noncoding sequences (CNSs) within Vgt1, and (iii) support a cis-acting transcription-regulatory role for Vgt1.
Results
Positional Cloning of Vgt1.
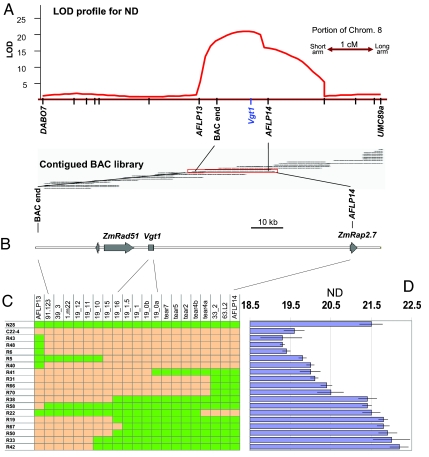
Previous work (12) mapped Vgt1 to a 1.3-cM region (Fig. 1A) on bin 8.05, based on a mapping population derived from the cross N28 × C22–4. The strain C22–4 is nearly isogenic to N28 and carries the early Vgt1 allele in an ≈7-cM introgression originating from the early maize variety Gaspé Flint. By using standard positional cloning, Vgt1 was confined to an ≈2-kb region (Fig. 1 B–D). Sequence annotation of the original BAC clone and the corresponding sequences derived from N28 and Gaspé Flint genetic backgrounds showed that Vgt1 is apparently noncoding and is located ≈70 kb (61–76 kb, depending on the genetic background) upstream of an Ap2-like gene identified here as ZmRap2.7. This gene is orthologous to Rap2.7 (also known as TOE1), a transcription factor that regulates flowering time in Arabidopsis (13, 14). No other genes were annotated between Vgt1 and ZmRap2.7. Pseudogenes due to transduplication events mediated by nonautonomous helitron elements (15) were observed in N28 and other genetic backgrounds but not in Gaspé Flint (data not shown). Within the Vgt1 region, the contrasting QTL alleles showed 29 SNPs and insertion/deletion-type polymorphisms (Indels) and one 143-bp insertion into the Gaspé Flint allele of a Mite transposon belonging to the Tourist (16) family [Fig. 4 Lower and supporting information (SI) Fig. 5].
Fig. 1.
Positional cloning of Vgt1. (A) QTL logarithm of odds (LOD) profile for node number (ND) and relative position for Vgt1 and relevant markers (redrawn and integrated from ref. 12). (B) Identification of Mo17 BAC covering the Vgt1 locus and sequence annotation. Gray arrows indicate coding sequences. Details on the nature and position of all 30 sequence polymorphisms between N28 and C22–4 (carrying the Gaspé Flint allele) at Vgt1 are provided in SI Fig. 5. (C) Graphic genotypes of the parental lines N28 and C22–4 and 17 segmental QTL nearly isogenic lines (NILs) carrying crossovers around Vgt1. Column codes indicate marker names, details of which are given in SI Table 2. Orange and green colors indicate homozygosity for Gaspé Flint and N28 alleles, respectively. (D) Phenotypic values recorded for the parental lines N28 and C22–4 and the 17 QTL NILs. Blue columns indicate the total number of plant nodes (ND). Bars indicate SD.
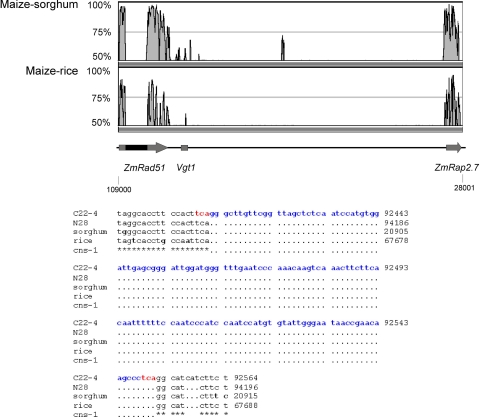
Fig. 4.
CNSs between the maize Vgt1 region and orthologous sorghum and rice sequences. (Upper) mVISTA plot of sequence identity of the maize BAC sequence spanning Vgt1 and the two most proximal genes (length of maize sequence considered was 81 kb) with the corresponding sorghum and rice orthologous sequences (50 and 35 kb, respectively). Peaks indicate region of similarity (percent of identity) for a window length set to 100 bp. Position of coding sequences and Vgt1 are indicated by the arrows and box. The lack of synteny observed within ZmRad51 is likely due to a transposon insertion in the maize gene (data not shown). Numbers refer to base pair position on maize BAC b0288K09. (Lower) Alignment of maize (C22–4/Gaspé Flint and N28), sorghum, and rice sequences corresponding to CNS1 (shown by an asterisk) and identified by using BLAST 2 Sequences. Nonconserved nucleotides within CNS1 are shown in bold. The sequence corresponding to the Mite element disrupting the C22–4/Gaspé Flint allele and the 3-bp duplicated target site are shown in blue and red, respectively. The numbers report the base pair position on BAC clones or genomic sequences (SI Materials and Methods).
Association Mapping at Vgt1.
To test the role of Vgt1 allelic variation in flowering time, we performed an association analysis, based on linkage disequilibrium (LD), of a set of 95 inbred lines known to adequately represent maize-cultivated germplasm (4) and exploiting 192 SNPs and insertion/deletion-type polymorphisms (Indels). LD analysis (SI Fig. 6) showed square allele-frequency correlation (r2 values) of ≤0.2 over distances of >2 kb. Regression analysis highlighted three polymorphisms within Vgt1, G/A/indel324, Mite, and ATindel434, as those most strongly associated with flowering time (P < 0.001; Fig. 2). G/A/indel324 is a three-allele SNP (inbred lines showed an A, G, or deletion), and the most markedly associated contrast was the A vs. G or deletion. G/A/indel324(A), Mite, and ATindel434 polymorphisms had an additive genetic effect (a) of ≈4.5 days to pollen shed (DPS) and 1.8–1.9 internodes (or leaves) and explained ≈32% and 29% of the phenotypic variability of DPS and leaf number, respectively, after fitting a regression model accounting for population structure. Two other SNPs outside Vgt1, i.e., PreAdTC438 and PreAdTC443, were associated with DPS at a much lower statistical significance (0.01 < P < 0.05) and were not associated with leaf number. No association was found between flowering time and SNP within the sequenced portion of ZmRap2.7. Because of the relatively high r2 value at distances of <1 kb, it is also unlikely that important functional sequence variation could be found elsewhere within the ZmRap2.7 coding sequence, at least for the set of inbred lines considered here. In accordance with the relatively high r2 values observed over ranges of <1 kb, we identified a limited number of haplotypes in the genomic regions that we resequenced in the panel of inbred lines. At Vgt1, the inbred lines could be classified into five haplotypes (SI Fig. 7). Based on this result, we should be able to predict the presence of a flowering-time QTL at chromosome bin 8.05 in any experimental cross between two lines represented within the used set of inbred lines. Two crosses, B73 × Mo17 and H99 × Mo17, for which flowering-time data are available, were identified by searching the Maize Genetics and Genomics Database (www.maizegdb.org). B73 × Mo17 segregates at Vgt1 (haplotypes 3 and 5; SI Fig. 7) and, indeed, a QTL for flowering time was previously identified at bin 8.05 with the predicted genic effect (B73 with haplotype 3 contributes the late allele) (17). H99 × Mo17 does not segregate at Vgt1 (haplotype 5 for both inbreds; SI Fig. 7), in keeping with previous studies that assigned no QTL for flowering time to bin 8.05 when a mapping population derived from this cross was analyzed (18). Collectively, association analysis and haplotype characterization confirmed the intergenic location of Vgt1 and correctly predicted the phenotypic effect of Vgt1 on different crosses.
Fig. 2.
Association of DNA polymorphisms with flowering time across the Vgt1 chromosome region. Level of statistical association for each SNP is expressed as −Log(P). Blue diamonds and white squares indicate association with male flowering date [expressed as days to pollen shed (DPS)] and total number of nodes of the plant (ND). Red points indicate r2 LD scores for all marker pairs involving Mite (value at Mite and at Mite totally linked markers is therefore r2 = 1). The nine numbers (1, 3, 6, …, 18) under the horizontal axes indicate codes for amplicons that were sequenced for SNP identification (SI Table 3). To simplify the view, the positions of other markers used in association analysis are not shown (for full details see SI Table 3).
ZmRap2.7 Is a Flowering-Time Gene.
We used genetic engineering to test the hypothesis that ZmRap2.7 controls flowering time. Both ZmRap2.7 overexpression and down-regulation were tested by analysis of transgenic plants and their progeny in two maize genetic backgrounds, one characterized by early and one by intermediate flowering time. The effects of increased gene expression were tested by using the plasmid construct carrying the ZmRap2.7 cDNA fused to a moderately strong constitutive rice actin promoter (SI Fig. 8). In 19 of 25 regenerated (T0) plants and following generations (Fig. 3), flowering was delayed from 1 to >4 weeks, and the number of leaves increased from two to five. Gene down-regulation was tested by using an RNAi construct based on ZmRap2.7 cDNA driven by a rice actin promoter (SI Fig. 9). Transformation in an intermediate flowering-time genetic background produced an early flowering effect in three of four independent events (SI Fig. 10). Overall, the transgenic approach clearly indicated that ZmRap2.7, like its Arabidopsis ortholog (14), is a negative regulator of flowering time.
Fig. 3.
Effect of overexpressing ZmRap2.7 on maize flowering time. Plants shown belong to a T1 family derived by selfing a transgenic T0 plant. Molecular genotypes of all plants were checked by PCR (data not shown). Segregating transgenic plants (two on left, homozygous or heterozygous for the transgenic event) are taller (have a higher number of nodes) and late flowering. Segregating nontransgenic plants (three on right) are smaller and early flowering, as typical of this genetic background.
Analysis of the Transcription-Regulatory Role of Vgt1.
The transcription-regulatory role of Vgt1 was tested by using temporal RT-PCR expression analysis of ZmRap2.7 on leaf tissues. Expression was significantly higher (P < 0.05) in N28 (i.e., the line carrying the late allele at Vgt1) and in QTL nearly isogenic lines (NILs) carrying the N28 allele compared with C22–4 and lines carrying the Gaspé Flint allele (SI Fig. 11). These results link allelic variation at Vgt1 with ZmRap2.7 transcript abundance and mirror those obtained by genetic engineering in which high ZmRap2.7 expression correlated with late flowering. The expression trend also matched the one observed for the Rap2.7 Arabidopsis ortholog (14).
We then tested the hypothesis that Vgt1 is a cis-regulatory locus by carrying out ZmRap2.7 allele-specific expression assays in N28 × C22–4 (and the reciprocal) F1 hybrid plants, which were therefore heterozygous at both Vgt1 and ZmRap2.7. In such plants, in the presence of cis regulation it is expected that the two ZmRap2.7 allelic transcripts be present in different abundances according to the Vgt1 allele present on the same chromosome; however, environmental as well as trans-acting effects should be negligible. The use of reciprocal F1 hybrids enabled us to test for genomic imprinting (i.e., a different allele is more strongly expressed in the F1 hybrid, depending on which allele was contributed by the female or male gamete). We found that the two alleles were differentially expressed across leaves. The N28/Gaspé Flint ratio of allele expression was up to 0.73, corresponding to 2.7-fold higher expression of the N28 allele (Table 1). Based on previous results (19) and our analysis of variation in replicated assays, we considered a 1.5-fold difference a threshold that implied the presence of a significant difference in allele expression. No significant difference was observed between reciprocal crosses. Additionally, allelic expression ratios in the first leaf were significantly different from those in the second (P < 0.01, two-tailed test) and third (P < 0.001, two-tailed test) leaves. All SNPs that were analyzed displayed comparable allelic expression ratios for a specific leaf (Table 1). Overall, this analysis indicated the existence of cis-regulatory mutations located in the Vgt1 region that presumably affected the developmental timing of gene expression and trans-acting factor(s) in the second and third leaves, interacting differently with each of the two allelic regulatory regions and therefore revealing the occurrence of the cis variation.
Table 1.
ZmRap2.7 allele-specific expression in leaves
| Exp. 1 |
Exp. 2 |
||||
|---|---|---|---|---|---|
| N28 × C22–4 | N28 × C22–4 | C22–4 × N28 | |||
| N28/C22–4 SNP (position) | 1st leaf | 2nd leaf | 3rd leaf | 3rd leaf | 3rd leaf |
| T/C (199) | 0.58 (0.03) | 0.62 (0.03) | 0.64 (0.01) | — | — |
| A/C (2265) | 0.52 (0.02) | 0.58 (0.02) | 0.61 (0.03) | 0.73 (0.04) | 0.67 (0.11) |
| G/C (2293) | 0.52 (0.01) | 0.57 (0.01) | 0.59 (0.01) | 0.73 (0.04) | 0.71 (0.07) |
| Average over SNPs | 0.55 (0.04) | 0.60 (0.03) | 0.62 (0.03) | 0.73 (0.04) | 0.69 (0.09) |
Expression proportion of N28 allele with standard error (in parentheses) is always shown (expression proportion of C22 allele = 1 − expression proportion of N28 allele). Experiment 1 (Exp. 1) was a temporal analysis (three developmental stages) from plants derived from N28 × C22–4, whereas experiment 2 (Exp. 2) involved one single developmental stage with plants derived from the two reciprocal crosses (N28 × C22–4 and C22–4 × N28). The average expression proportion for each leaf is an average of proportions obtained for all SNPs for a specific leaf.
Vgt1 Contains Maize–Sorghum–Rice CNS.
We observed that two genes flanking Vgt1, Rap2.7 and Rad51, are microcolinear (i.e., they are found in pairs and maintain polarity of coding direction) among maize, sorghum, rice, and Arabidopsis. This observation supports their orthologous nature and provides groundwork for the identification and analysis of orthologous CNSs in the intergenic space. Comparative analysis of maize, sorghum, and rice sequences (≈81-, 50-, and 31-kb long, respectively) spanning the two genes identified only two interspersed (i.e., >2 kb from any coding sequence) CNSs. These two CNSs, CNS1 and CNS2, are both within Vgt1. Two additional CNSs within Vgt1, CNS3 and CNS4, were identified in the maize–sorghum comparison only, a finding in accord with the closer, phylogenetic relationship. Fig. 4 shows the maize–sorghum and maize–rice sequence identity and the detailed structure of CNS1. Details on CNS2, CNS3, and CNS4 are given in SI Fig. 12. Interestingly, CNS1 is disrupted in the Gaspé Flint Vgt1 allele by the Mite insertion used as a flowering-linked marker during positional cloning and found to be highly associated with flowering time throughout the maize germplasm. A comparison of maize with Arabidopsis did not reveal the presence of intergenic CNS.
Discussion
Early and recent reviews on the genetics of maize flowering time recognized the chromosome region bin 8.05, bearing Vgt1, as a “hot spot” for flowering-time QTLs and genes (3, 20). Although the genetic resolution reported in those studies was never at the gene level, it is likely that at least some of the reviewed QTLs were due to allele segregation at Vgt1. The haplotype information at Vgt1 produced in our study enabled us to test whether haplotype segregation at Vgt1 predicts previously mapped flowering-time QTLs as shown for two mapping populations identified from a literature review.
A recurring issue within the plant genetics community is whether and how QTL cloning can be carried out without the costly exercise of producing and testing nearly isogenic experimental populations (21). Indeed, in our work, we observed a strong coincidence of results obtained by positional cloning and association mapping. The r2 level (r2 ≤ 0.2 for distances of >2 kb) observed at this genomic region was within the range already observed at other loci for the same collection of lines (22) and would guarantee a gene-like resolution in a genome-wide association approach. Because of the observed LD and the genome dimension of maize (2.4 × 109 bp) (23), however, such an approach would have located Vgt1 only if a technology providing a scan of several millions of SNPs (i.e., one or two informative SNPs per kilobase) spread over coding and noncoding regions were currently available and cost-effective. On the other hand, the lack of statistically significant effects on flowering time observed at ZmRap2.7 does not support an association mapping approach based on candidate genes, which worked in other cases (24, 25).
All our results are consistent with Vgt1 being or containing long-range, cis-regulatory element(s) of the downstream ZmRap2.7 gene. The Arabidopsis ortholog of ZmRap2.7 has been shown to be down-regulated by the microRNA miR172 (14). miR172 is also present in maize (26), and the target site for miR172 is present in ZmRap2.7 (data not shown), which is therefore likely to be also regulated by an miR172-mediated trans-acting mechanism. Instead, based on our findings, it appears that an important part of ZmRap2.7 natural variation of expression also exploited by artificial selection is represented by DNA sequence polymorphisms at the Vgt1 cis-regulatory region.
The functional role of Vgt1 is further substantiated by the observation that Vgt1 contains the most highly CNSs between maize and rice (evolutionarily separated ≈50 million years ago) (27) across the entire large intergenic region upstream of ZmRap2.7. This result supports the notion that comparing large orthologous genomic sequences could quickly extend our knowledge of the molecular basis of transcriptional regulation. Our findings are also in keeping with previous observations that CNSs are often found upstream of transcription factors (28) and are shorter and less conserved in plants than in animals (29–31).
The molecular mechanism of Vgt1 action on ZmRap2.7 expression cannot currently be predicted and it deserves further investigation. However, CNSs have so far been putatively associated, both in animals and plants, with chromosome-level structural or regulatory regions, chromatin matrix attachment regions, long-range enhancers/silencers, transcription factor-binding sites, and possibly other features and functions (32), all of which could play a role in the cis regulation of gene expression.
Ultimately, the discovery of QTLs due to polymorphisms at intergenic noncoding regions in Vgt1 and other naturally occurring phenotypes (33–35) confirms that such sequences represent an important component of quantitative genetic variation.
Materials and Methods
Plant Materials.
N28 and C22–4 (parental lines) and a large mapping population obtained by their cross have been previously described (10, 12). C22–4 is an early derivative of N28 obtained by the cross N28 × Gaspé Flint followed by 20 generations of backcrossing that used N28 as recurrent parent and selection for early flowering (10, 12).
Positional cloning, genomic sequence production and annotation, identification of the SNPs used for association mapping, genetic engineering, RT-PCR for semiquantitative PCR, and cDNA preparation for the allele-specific PCR are described in SI Materials and Methods.
Association Mapping.
Ninety-five inbred lines belonging to the set assembled by Buckler and coworkers (4) were used for this part of the work. The 95 lines were grown in 2002 and 2003 in replicated field trials (plots of 15 plants, three repetitions) at the Experimental Station of the University of Bologna, Italy. Days to pollen shed (DPS) and number of plant nodes (ND) were recorded as described (12). Six lines did not reach flowering, likely because of photoperiod sensitivity. ND was successfully collected for the whole set of lines. N28 and C22–4 were added to the field evaluation and to the haplotype analysis but were excluded from the computation of LD and marker-trait associations. The complete list of markers and their origin and nature are given in SI Table 2. Statistical analyses for LD and association were carried out by using the program TASSEL, version 1.0.4. Of the entire matrix of 95 lines and 192 markers, only markers with the rare allele at a frequency of >0.1 were included. Analyses were carried out by using both logistic regression and generalized linear model (GLM) regression modes. Only results obtained by using the GLM are shown because logistic regression produced very similar outputs. Analyses were run by including population structure information as provided in ref. 4. P values were obtained based on an experimental-wise permutation test (for details see the TASSEL tutorial at www.maizegenetics.net/tassel).
Comparative Sequence Analysis.
The maize, sorghum, rice, and Arabidopsis orthologous genomic sequences encompassing Rap2.7 and Rad51 were identified by standard BLAST searches at www.ncbi.nlm.nih.gov/GenBank/index.html (or www.phytozome.org/sorghum for sorghum) using ZmRap2.7 and ZmRad51 as queries. The definition of plant CNSs adopted here followed criteria defined in previous studies (27, 28): 100% identity over a portion of 15-bp intervals or >70% identity over 20 bp or longer intervals. The search for CNSs was carried out by pairwise analysis of maize with sorghum, rice, and Arabidopsis, using both mVISTA (at http://genome.lbl.gov/vista/index.shtml) and BLAST 2 Sequences (at www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi). For maize, the sequence considered was the portion from 28,000 to 109,000 of BAC b0288K09 (B73 inbred background; available at www.genome.arizona.edu/fpc/maize/gbrowse/) as submitted (similar results were obtained when the genomic sequence derived from Mo17 BAC bacm.pk066.l14 was used). For sorghum, the portion was from 630,000 to 680,000 of contig Super_27, as of February 14, 2007, at www.phytozome.org/sorghum. For rice, the portion was from 51,000 to 86,000 of GenBank accession no. AC093088 (chromosome 5). For Arabidopsis, the portion was from 12,233,000 to 12,247,000 of chromosome 2, as displayed at www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=3702&chr=2. For VISTA, parameters were set as follows: shuffle-LAGAN alignment modality with a “calculated window” of 100 bp and “consensus identity” of 50%. For BLAST 2 Sequences, parameters were set as “cost to open a gap” of 2, “cost to extend a gap” of 1, and “word size” of 7 as suggested (289) and by relaxing the “expectation” value to 1,000,000. Comparisons of maize and Arabidopsis did not reveal the presence of intergenic CNSs.
Allele-Specific Expression Assay.
PCR primers that flanked the marker polymorphisms were designed by using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and were as follows: exon I left primer, 5′-CACCAGTTCGCCAGGTAGTT, and exon I right primer, 5′-CGTTGAGCTAGATCCCTCCTC; exon X left primer 5′-CGACGATGCTCCCTCTGA, and exon X right primer 5′-AAGACGAAGAAGGGGTGAGG. Single base extension (SBE) primers, with a minimum length of 18 nt, were designed for three SNPs. One was in exon 1, SNP T/C, at position 199, SBE_1, 5′-CAGGAGCAGGAGATGCAG-3′. Two were in exon 10, SNP A/C, at position 2265, SBE_2, 5′-CGATGCTCCCTCTGAGCT-3′, and SNP G/C, at position 2293, SBE_3, 5′-GGCGGATGCTCAGCCACGA-3′. Positions are given starting from the ATG start codon of ZmRap2.7. For PCR amplification of the region containing SNPs, we combined templates (20 ng for DNA and 2 μl for cDNA or a no-reverse transcriptase control) with Taq Gold (1.25 units; Applied Biosystems, Foster City, CA), manufacturer's buffer (2 mM MgCl2/200 μM dNTPs/1% DMSO) (Sigma, St. Louis, MO), and 1 μM each locus-specific primers (Sigma–Genosys, St. Louis, MO). Thermocycling conditions consisted of an initial denaturation step at 95°C for 10 min, followed by 38 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 7 min. Amplicon sizes were verified by agarose gel electrophoresis. Amplified samples were incubated with ExoSAP-IT (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions before the primer extension reaction. Primer extension was carried out with the SNaPshot Multiplex Ready Reaction mix (Applied Biosystems). Reactions were performed in a total volume of 10 μl containing 3 μl of treated PCR products diluted 1:10, 2.5 μl of SNaPshot premix, 1 μl of 0.2 μM SNP-specific primer, and 3.5 μl of nuclease-free H2O. Primer extension thermocycling conditions consisted of 25 cycles at 96°C for 10 s, 50°C for 5 s, and 60°C for 30 s. After primer extension, reactions products were purified by shrimp alkaline phosphatase (SAP) (Amersham Biosciences) according to the manufacturer's instructions. Cleaned products were combined with 0.25 μl of GeneScan-120 LIZ size standard mix and 9.75 μl of formamide and run on an Applied Biosystems 3730 DNA Analyzer. Peaks of dye intensities corresponding to extensions of SBE primers were determined by inspecting output from the 3730 DNA Analyzer. The ratios between peak heights were expressed as N28/(N28 + C22–4). Mixes of the genomic DNA of N28/C22–4 were prepared in 1:1, 3:1, and 1:3 proportions, and SBE reactions on these templates were run with the cDNA and no-reverse transcriptase control samples. The genomic mixes allowed for the construction of a titration curve by linear regression from which the ratio for the cDNA samples was extrapolated. The obtained ratios were normalized on the basis of the peak height ratio measurements obtained from SBE on hybrid genomic DNA, representing a perfect 50:50 ratio of the two alleles. Allelic expression estimates are an average of measurements obtained in three separate RNA extractions, respective cDNA synthesis, and multiple PCRs and SBE reactions.
Supplementary Material
Acknowledgments
We thank Edward Buckler (Cornell University, Ithaca, NY) for making available the seed of the maize inbred line collection used for association mapping.
Abbreviations
- QTL
quantitative trait locus
- CNS
conserved noncoding sequences
- LD
linkage disequilibrium
- SBE
single base extension.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF659467 and EF659468).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704145104/DC1.
References
- 1.Doebley J. Annu Rev Genet. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- 2.Shaw HS. In: Corn and Corn Improvement. Sprague GF, editor. Madison, WI: Am Soc Agron; 1977. pp. 591–623. [Google Scholar]
- 3.Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A. Genetics. 2004;162:2169–2185. doi: 10.1534/genetics.104.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES., IV Nat Genet. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- 5.Camus-Kulandaivelu L, Veyrieras JB, Madur D, Combes V, Fourmann M, Barraud S, Dubreuil P, Gouesnard B, Manicacci D, Charcosset A. Genetics. 2006;172:2449–2463. doi: 10.1534/genetics.105.048603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neuffer MG, Coe EH, Wessler SR. Mutants of Maize. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. [Google Scholar]
- 7.Chardon F, Hourcade D, Combes V, Charcosset A. Theor Appl Genet. 2005;112:1–11. doi: 10.1007/s00122-005-0050-z. [DOI] [PubMed] [Google Scholar]
- 8.Colasanti J, Yuan Z, Sundaresan V. Cell. 1998;93:593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- 9.Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON. Plant Physiol. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips RL, Kim TS, Kaeppler SM, Parentoni SN, Shaver DL, Stucker RI, Openshaw SJ. Proc Ann Corn and Sorghum Res Conf. 1992;47:135–150. [Google Scholar]
- 11.Vladutu C, McLaughlin J, Phillips RL. Genetics. 1999;153:993–1007. doi: 10.1093/genetics/153.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salvi S, Tuberosa R, Chiapparino E, Maccaferri M, Veillet S, van Beuningen L, Isaac P, Edwards K, Phillips RL. Plant Mol Biol. 2002;48:601–613. doi: 10.1023/a:1014838024509. [DOI] [PubMed] [Google Scholar]
- 13.Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. Proc Natl Acad Sci USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aukerman M, Sakai H. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 16.Bureau TE, Wessler SR. Proc Natl Acad Sci USA. 1994;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beavis WD, Smith OS, Grant D, Fincher R. Crop Sci. 1994;34:882–896. [Google Scholar]
- 18.Austin DF, Lee M. Genome. 1996;39:957–968. doi: 10.1139/g96-120. [DOI] [PubMed] [Google Scholar]
- 19.Pastinen T, Hudson TJ. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y-R, Schertz KF, Paterson A. Genetics. 1995;141:391–411. doi: 10.1093/genetics/141.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES. Plant J. 2005;44:1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- 22.Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES., IV Proc Natl Acad Sci USA. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett MD, Laurie DA. Maydica. 1995;40:199–204. [Google Scholar]
- 24.Palaisa KA, Morgante M, Williams M, Rafalski A. Plant Cell. 2003;15:1795–1806. doi: 10.1105/tpc.012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson LM, Whitt SR, Ibanez AM, Rocheford TR, Goodman MM, Buckler ES., IV Plant Cell. 2004;16:2719–2733. doi: 10.1105/tpc.104.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. Proc Natl Acad Sci USA. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellogg EA. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas BC, Rapaka L, Lyons E, Pedersen B, Freeling M. Proc Natl Acad Sci USA. 2007;104:3348–3353. doi: 10.1073/pnas.0611574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplinsky NJ, Braun DM, Penterman J, Goff SA, Freeling M. Proc Natl Acad Sci USA. 2002;99:6147–6151. doi: 10.1073/pnas.052139599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, Moose SP. Plant Cell. 2003;15:1143–1158. doi: 10.1105/tpc.010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lockton S, Gaut BS. Trends Genet. 2005;21:60–65. doi: 10.1016/j.tig.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Dermitzakis ET, Reymond A, Antonarakis SE. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 33.Stam M, Belele C, Ramakrishna W, Dorweiler JE, Bennetzen JL, Chandler VL. Genetics. 2002;162:917–930. doi: 10.1093/genetics/162.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark RM, Wagler TN, Quijada P, Doebley J. Nat Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- 35.Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.