Abstract
The 5′-phosphoinositol phosphatase SHIP negatively regulates signaling pathways triggered by antigen, cytokine and Fc receptors in both lymphocytes and myeloid cells. Mice with germ-line (null) deletion of SHIP develop a myeloproliferative-like syndrome that causes early lethality. Lymphocyte anomalies have been observed in SHIP-null mice, but it is unclear whether they are due to an intrinsic requirement of SHIP in these cells or a consequence of the severe myeloid pathology. To precisely address the function of SHIP in T cells, we have generated mice with T cell-specific deletion of SHIP. In the absence of SHIP, we found no differences in thymic selection or in the activation state and numbers of regulatory T cells in the periphery. In contrast, SHIP-deficient T cells do not skew efficiently to Th2 in vitro. Mice with T cell-specific deletion of SHIP show poor antibody responses on Alum/NP-CGG immunization and diminished Th2 cytokine production when challenged with Schistosoma mansoni eggs. The failure to skew to Th2 responses may be the consequence of increased basal levels of the Th1-associated transcriptional factor T-bet, resulting from enhanced sensitivity to cytokine-mediated T-bet induction. SHIP-deficient CD8+ cells show enhanced cytotoxic responses, consistent with elevated T-bet levels in these cells. Overall our experiments indicate that in T cells SHIP negatively regulates cytokine-mediated activation in a way that allows effective Th2 responses and limits T cell cytotoxicity.
Keywords: cytotoxicity, T-bet
The 5′-inositol phosphatase SHIP is a well characterized inhibitory molecule that regulates cell responses in lymphocytes and myeloid cells by its ability to hydrolyze the second messenger PI(3,4,5) trisphosphate (1–4). SHIP is recruited by engagement of the inhibitory Fc receptor in B cells and mast cells (5–8) or by the engagement of FcRs (FcεRI and FcγRIII), cytokine [interleukin 3 (IL3), granulocyte–macrophage colony-stimulating factor (GM-CSF), and TGFβ], and growth factor (steel factor and M-CSF) receptors in myeloid cells (2, 9). Once SHIP is recruited to the membrane by the signaling complexes, its enzymatic activity depletes PI(3,4,5)P3 and prevents membrane localization of PH-domain-containing factors such as Tec kinases, Akt, and PLCγ (10–13). This inhibitory effect ultimately leads to reduced calcium influx and prevents cellular activation (14).
Germ-line deletion of SHIP in mice leads to early mortality from a myeloproliferative-like syndrome characterized by profound splenomegaly and massive myeloid infiltration of the lung (15). B cells develop abnormally in SHIP-null mice, with an activated phenotype and specifically lacking the marginal zone B cell population (16, 17). The absence of marginal-zone B cells in SHIP-null mice has been explained not as a B cell primary defect, but as a consequence of macrophage dysregulation in the spleen of these animals (17). Macrophages from SHIP-null mice are skewed toward an M2-activated phenotype showing high levels of arginase I (ArgI) and Ym1. Rauh et al. (18) suggested that SHIP might repress M2 skewing by regulating macrophage responses to TGFβ.
SHIP's role in regulating T cells has been less clearly established because these cells lack expression of FcRs and cytokine receptors like IL3R and GM-CSF-R, more commonly linked to SHIP function. Several lines of evidence from experiments in vitro have pointed toward a regulatory role of SHIP in T cells. First, Edmunds et al. (19) reported that SHIP is tyrosine phosphorylated in response to TCR and CD28 ligation. Second, Freeburn et al. (20) showed that SHIP regulates PI3K effectors in Jurkat cells. Third, Tomlinson et al. (21) showed that SHIP interacts with the Tec kinase and inhibits its function in T cells. Finally, SHIP was found to be part of a negative signaling complex associated with LAT (22). T cell populations in SHIP-null mice are slightly reduced, but maintain the proper CD4/CD8 ratios both in the thymus and periphery (15). Thus, SHIP does not appear to have a significant impact on T cell development. Peripheral T cells from SHIP-null mice have been shown to be constitutively activated and to give rise to increased numbers of CD4+CD25+ regulatory T cells (23). However, given that SHIP-null mice develop severe myeloproliferative disease, it is unclear whether the differences observed in T cell function are due to a T cell-intrinsic role of SHIP or are simply the consequences of the inflammatory environment brought about by SHIP-deficient myeloid cells. To avoid the pleotropic effects of the SHIP-null deletion, we have examined T cell function when disruption of the SHIP gene occurs exclusively in the T cell lineage. We have found that T cell-specific deletion of SHIP does not alter T cell development, activation state, or the number of regulatory T cells. Instead, we have uncovered a regulatory role of SHIP in controlling Th1/Th2 bias and cytotoxic responses as a result of its inhibitory effect on cytokine-induced T-bet expression.
Results
Normal T Cell Development and TCR Signaling in Mice with a Conditional Deletion of SHIP in the T Cell Lineage.
To obtain mice with T cell-specific deletion of SHIP, we crossed mice containing a loxP-flanked SHIP gene (17) with mice transgenic for Cre recombinase driven by the CD4 promoter (24). The resulting mice were designated CD4cre SHIPfl/fl mice. Nontransgenic SHIPfl/fl littermates served as controls in all experiments. We confirmed that T cells from CD4cre SHIPfl/fl mice have no detectable SHIP expression by Western blot, whereas non-T splenocytes expressed normal levels of SHIP (Fig. 1A). SHIP mRNA was reduced at least 10-fold in DP and SP thymocytes [supporting information (SI) Fig. 7]. Thus, this system efficiently and specifically deletes SHIP from T cells. CD4cre SHIPfl/fl mice were healthy and had a lifespan comparable to SHIPfl/fl littermates. Histological examination of kidneys and lungs showed no evidence of the myeloproliferative pathology that had been observed in SHIP-null mice (15).
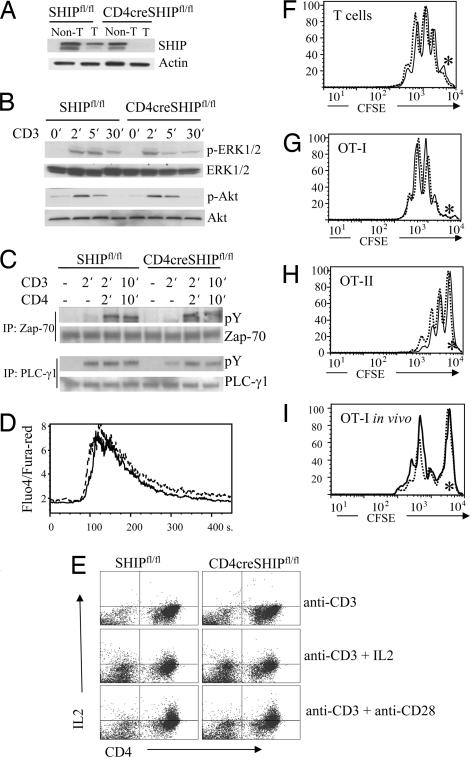
Fig. 1.
T cell-specific deletion of SHIP does not affect TCR signaling, in vitro T cell activation, or in vivo antigen-driven expansion. (A) Western blot with SHIP antibody shows efficient reduction in expression of SHIP protein in splenic T cells of CD4cre SHIPfl/fl mice. (B and C) No difference in Erk, Akt, Zap-70, or PLCγ1 activation on anti-CD3 or anti-CD3+ anti-CD4 stimulation of CD4cre SHIPfl/fl vs. SHIPfl/fl control T cells. (D) Equal calcium influx levels triggered by anti-CD3 stimulation of CD4cre SHIPfl/fl (solid line) vs. SHIPfl/fl T cells (dashed line). (E) CD4cre SHIPfl/fl T cells show the same level of IL2 production as SHIPfl/fl control cells. Purified naive CD4+ cells were incubated for 3 days with the indicated stimulus and tested for IL2 production by intracellular staining with FITC− IL2 antibodies. (F–H) In vitro proliferation of CD4cre SHIPfl/fl (solid line) or SHIPfl/fl control (dashed line) T cells alone (F) or expressing the OT-I (G) and OT-II (H) TCR transgenes. CFSE-labeled cells were incubated on anti-CD3/CD28 plates for 3 days. Asterisks mark CFSE levels of undivided cells. (I) In vivo expansion of OT-I CD4cre SHIPfl/fl (solid line) or OT-I SHIPfl/fl control (dashed line) T cells. C57BL/6 host mice were injected with ovalbumin 1 day after receiving CFSE-labeled T cells from the indicated mice. Two days later, cells were purified from the draining lymph nodes, stained with CD8-Cychrome and Vα2-PE, and analyzed by FACS. Experiments were repeated three times, with three mice per group.
The total number of lymphoid cells in the thymus, spleen, and lymph nodes were the same in CD4cre SHIPfl/fl and SHIPfl/fl mice. This result was observed in CD4cre SHIPfl/fl mice expressing three different transgenic TCRs, namely the OVA-specific MHC class I-restricted OT-I, the OVA-specific MHC class II-restricted OT-II, and the MCC-specific MHC class II-restricted 5C.C7. Flow-cytometric analysis of the thymus and peripheral tissues revealed normal percentage of T cell populations in mice with T cell-specific deletion of SHIP (SI Table 1). Mice with germ-line deletion of SHIP were shown to have an increased number of regulatory T cells (23). The number of regulatory T cells in CD4cre SHIPfl/fl mice, judged by the percentage of FoxP3+ cells in spleen, was similar to that of wild-type mice (SI Table 1). Altogether, these experiments establish that SHIP does not have an essential role in thymic selection or in T cell development in the periphery.
We next tested whether SHIP regulates signaling through the TCR, but we found no differences in the level of phosphorylation of ERK, Akt, Zap-70, or PLCγ1 on polyclonal activation with anti-CD3 in SHIP-deleted cells compared with T cells from wild-type mice (Fig. 1 B and C). There were no differences in Akt or Gsk phosphorylation on CD28 engagement alone (SI Fig. 8). We found no differences in the calcium influx triggered by CD3 engagement (Fig. 1D) or in the ex vivo proliferation of freshly isolated T cells from mice with T-specific deletion of SHIP, even with single specificity TCRs in the OVA-specific OT-I/OT-II transgenics (Fig. 1 F–H). SHIP deletion had no effect on the extent of IL2 production on CD3/CD28 ± IL2 stimulation (Fig. 1E). We did not observe differences in the antigen-driven in vivo expansion of OT-I CD4cre SHIPfl/fl T cells compared with controls (Fig. 1I). The same result was obtained with OT-II CD4cre SHIPfl/fl T cells in this assay (data not shown).
Reduced Humoral Response and Inefficient Th2 Skewing in Mice with T Cell-Specific Deletion of SHIP.
The ability of SHIP-deficient T cells to provide B cell help was measured in immunization experiments. We separately tested responses to the Th2-inducing adjuvant, Alum, and to RIBI adjuvant, which induces a more Th1-type of response. CD4cre SHIPfl/fl mice showed significantly diminished NP-specific response after immunization with Alum/NP-CGG (Fig. 2A), whereas they were at least equally effective in mounting a TNP-specific antibody response against RIBI/TNP-KLH (Fig. 2B). Lymph node cells from mice immunized with Alum/NP-CGG isolated on day 7 after restimulation showed a 3-fold reduction in germinal center B cells in CD4cre SHIPfl/fl mice compared with SHIPfl/fl controls (Fig. 2C). T cells purified from immunized CD4cre SHIPfl/fl animals and restimulated in vitro with NP-CGG produced less IL5 and IL13, but more INFγ (Fig. 2D).
Fig. 2.
Humoral response in CD4cre SHIPfl/fl (filled triangles) and SHIPfl/fl (open squares) mice. (A) Mice were immunized with 50 μg of NP-CGG with Alum and pertussis toxin, and the levels of NP-specific antibodies of different subclasses in the serum were determined 2 weeks after the primary immunization (Left) or 1 week after reimmunization given on day 28 (Right). Serum was diluted 1:5,000 for primary immunizations and 1:100,000 for secondary immunizations. Lines indicate the mean antibody response. (B) Mice were immunized with 50 μg of TNP-KLH with RIBI adjuvant, and TNP-specific antibody levels in the serum were determined as in A. (C) Impaired germinal center formation in CD4cre SHIPfl/fl (Right) vs. control mice (Left) after immunization with NP-CGG plus Alum plus pertussis toxin. Gated for B220-positive cells. Results are representative from three independent experiments. (D) Cytokine production by cells purified from NP-CGG/Alum-challenged CD4cre SHIPfl/fl (black) and SHIPfl/fl mice (gray). Lymph node cells were isolated on day 7 after reimmunization and stimulated with NP-BSA for 24 h.
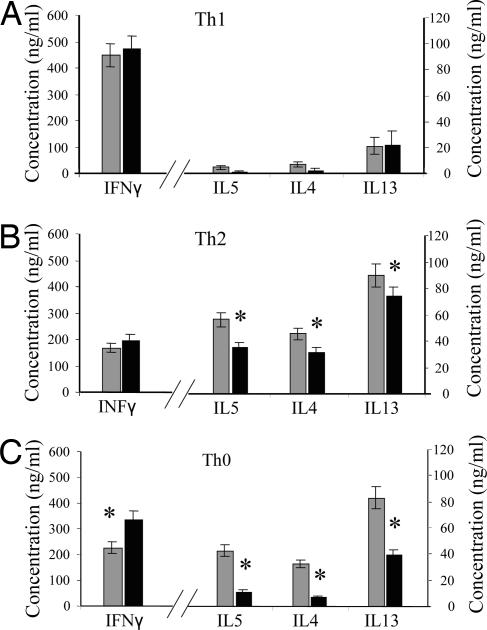
Our observation that deletion of SHIP in T cells affects the humoral response to immunization with Alum as an adjuvant and preferentially reduces the levels of Th2 cytokines suggested to us that SHIP might have a role in regulating Th1/Th2 skewing in T cells. Thus, we analyzed cytokine production of SHIP-deficient naive CD62L+CD4+ T cells in polarizing conditions in vitro. Although we found no significant differences under Th1 polarizing conditions (Fig. 3A), T cells from CD4cre SHIPfl/fl mice produced significantly lower levels of IL4, IL5, and IL13 under Th2 skewing conditions (Fig. 3B) and markedly lower levels of IL4, IL5, and IL13 and higher levels of IFNγ under nonpolarizing conditions (Fig. 3C). When tested for Th17 skewing, CD4cre SHIPfl/fl T cells cultured with IL6 and TGFβ resulted in IL17 mRNA levels comparable to that produced by SHIPfl/fl T cells (data not shown).
Fig. 3.
SHIP-deficient CD4 T cells polarize in culture but have decreased Th2 cytokine production. CD4cre SHIPfl/fl (dark bars) and SHIPfl/fl (light bars) CD4+ T cells were stimulated with anti-CD3 plus anti-CD28 under Th1 (A), Th2 (B), or Th0 (C) polarizing conditions for 3 days, expanded in IL2 for 4 days, and restimulated with anti-CD3 plus anti-CD28. Supernatants were collected 24 h after restimulation and cytokine expression assessed by the CBA Assay. ∗, P < 0.05.
Mice with T-Specific Deletion of SHIP Show Altered Response to S. mansoni.
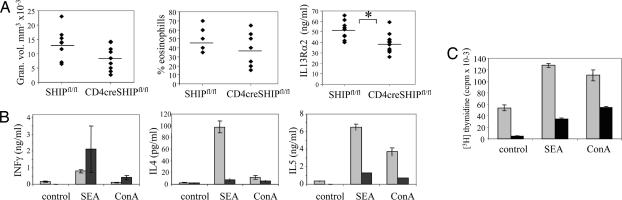
To further investigate the role of SHIP in shaping Th2 responses in vivo, we challenged our mice with eggs from S. mansoni, a type 2 response-inducing helminth. When injected i.v., S. mansoni eggs lodge in lungs and induce the development of eosinophilic granulomas as well as Th2 cytokine production by T lymphocytes. We observed that egg injection resulted in slightly smaller granulomas and somewhat reduced levels of eosinophils in the lung and significantly lower levels of IL13 Rα2 in serum of CD4cre SHIPfl/fl mice compared with SHIPfl/fl controls (Fig. 4A). In vitro challenge of T cells from the draining mediastinal lymph nodes with ConA or Schistosome egg antigen (SEA) showed a significant reduction in the production of IL4 and IL5 in the culture supernatants (Fig. 4B) and reduced proliferation (Fig. 4C) in T cells coming from egg-challenged mice with T cell-specific deletion of SHIP.
Fig. 4.
Altered responses to the Th2 polarizing agent S. mansoni eggs in mice with T cell-specific deletion of SHIP. (A) Lung granuloma volume, percentage granulome eosinophils, and serum IL13Rα2 from CD4cre SHIPfl/fl and SHIPfl/fl mice challenged with S. mansoni eggs. ∗, P < 0.05. (B) Reduced Th2 cytokine production by T cells purified from infected CD4cre SHIPfl/fl mice (dark bars) vs. SHIPfl/fl controls (light bars). Draining mediastinal lymph nodes were dissected 10 days after secondary injection with S. mansoni eggs. Cells from five mice were pooled and cultured with ConA or SEA antigen for 72 h, and supernatants were analyzed for cytokine production. (C) Reduced SEA or ConA-induced proliferation of T cells purified from lymph nodes of infected CD4cre SHIPfl/fl mice (dark bars) vs. SHIPfl/fl controls (light gray bars).
Absence of SHIP Does Not Alter the Sensitivity to TCR Signaling but Increases Basal Levels of T-bet.
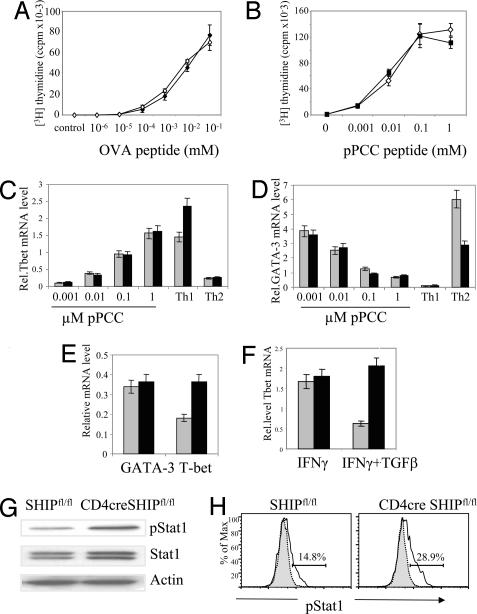
Because low-peptide concentration can skew T cell responses toward a Th2 response (25), one possible explanation for the inefficient type 2 responses in SHIP-deleted T cells could be an increase in signal strength that precludes IL4 expression at low-peptide conditions. To test this possibility, we determined the proliferation of purified naive T cells in vitro from OT-II CD4cre SHIPfl/fl (Fig. 5A) or 5CC7 CD4cre SHIPfl/fl mice (Fig. 5B) in the presence of variable concentrations of peptide. We observed that the deletion of SHIP had no effect either in the proliferative efficiency (Fig. 5 A and B) or in the early (24-h) induction of T-bet and GATA-3 transcription factors (Fig. 5 C and D) over a wide range of peptide concentrations. However, we did find consistent and significant differences, 2-fold up-regulated in CD4cre SHIPfl/fl mice, in the basal levels of T-bet in freshly purified naive CD4+CD62L+ cells (Fig. 5E). We found this increase of T-bet in CD4cre SHIPfl/fl was not due to differences in the kinetic of degradation of T-bet mRNA (SI Fig. 9).
Fig. 5.
Increased basal level of T-bet in SHIP-deleted T cells due to altered cytokine signaling. (A) Proliferation of naive OT-II CD4cre SHIPfl/fl T cells (filled diamonds) vs. OT-II SHIPfl/fl controls (empty diamonds) in the presence of irradiated splenocytes and the indicated concentration of OVA peptide. (B) Proliferation of naive 5CC7 CD4cre SHIPfl/fl T cells (filled squares) vs. 5CC7 SHIPfl/fl controls (empty triangles) in the presence of irradiated P13.9 fibroblasts as described in Methods. (C and D) SHIP deletion does not alter T-bet or GATA-3 induction on antigen stimulation of T cells. Naive CD4+ CD62L+ cells from 5CC7 CD4cre SHIPfl/fl mice (black bars) and 5CC7 SHIPfl/fl control mice (gray bars) were stimulated with mitomycin C-treated P13.9 fibroblast cells that had been preloaded with 0.001–1 μM pPCC. Cells were harvested after 24 h of incubation, and relative mRNA levels (compared with L32 gene) for T-bet (C) and GATA-3 (D) were measured by real-time PCR. The experiment was performed twice with the same result. (E) T-bet and GATA-3 mRNA expression in naive freshly isolated CD4+ CD62L+ T cells from CD4cre SHIPfl/fl (black bars) and SHIPfl/fl mice (gray bars). (F) SHIP mediates TGFβ1 inhibition of IFNγ-induced T-bet up-regulation. Purified CD4+ CD62L+ T cells were incubated with IFNγ with or without TGFβ for 24 h, and the relative level of mRNA T-bet was assessed by real-time PCR. (G) Increased STAT phosphorylation in freshly purified T cells from CD4cre SHIPfl/fl vs. control SHIPfl/fl mice. (H) Increased STAT phosphorylation in IFNγ-activated T cells from CD4cre SHIPfl/fl (Right) vs. control SHIPfl/fl mice (Left). Th1-skewed T cells were incubated for 24 h with IL12 to up-regulate the IFNγ receptor level, rested for 4 h, and incubated with IFNγ for 15 min. The proportion of cells expressing pSTAT1 was calculated as the difference between anti-pSTAT intracellular flow cytometry (solid line) minus unstimulated control (gray blot).
Knowing that SHIP is a well established regulator of cytokine signaling in myeloid cells (2) and having found no effect on TCR-mediated activation in the absence of SHIP, we hypothesized that SHIP-deleted T cells might have increased levels of T-bet as a result of enhanced sensitivity to cytokine signaling. T-bet levels in T cells are increased in response to IFNγ activation and inhibited in the presence of TGFβ (26, 27). SHIP has previously been shown to regulate TGFβ function in macrophages (18). Moreover, the tyrosine phosphatase SHP-1, which inhibits signaling pathways triggered by the same immune receptors as SHIP, is essential for the TGFβ inhibition of IFNγ induction of T-bet expression (28). Freshly isolated CD4cre SHIPfl/fl and control CD4+ T cells were stimulated with a combination of anti-CD3 and anti-CD28 and with IFNγ and TGFβ separately and in combination. We observed that, although T-bet was up-regulated similarly after 24 h IFNγ induction, SHIP-deleted T cells did not respond to the inhibitory effect of TGFβ in this assay (Fig. 5F). Additionally, we found increased STAT-1 phosphorylation in unstimulated cells (Fig. 5G) or after IFNγ stimulation of CD4cre SHIPfl/fl cells for 15 min (Fig. 5H), whereas we found no differences in STAT-6 phosphorylation on IL4 treatment in the same cells (data not shown).
SHIP Regulates CD8+ Cytotoxicity.
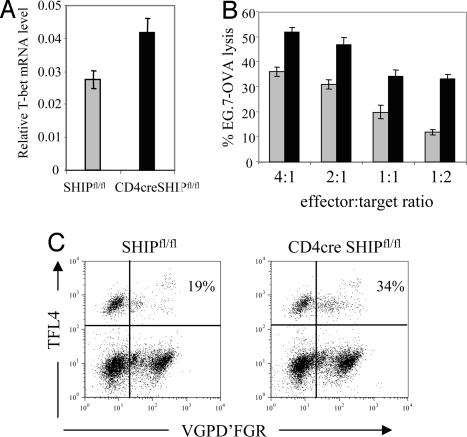
Because both CD4+ and CD8+ cells lack expression of SHIP in CD4cre SHIPfl/fl mice (Fig. 1A and SI Fig. 7), we sought to determine whether SHIP deletion results in elevated T-bet levels and altered function in CD8+ cells. As shown in Fig. 6A, we found 60% more T-bet mRNA in freshly purified SHIP-deficient CD8+ cells compared with controls. T-bet has been reported to regulate effector CD8+ T cell function (29), so we expected SHIP-deleted T cells to be more efficient in cytotoxic assays. To test cytotoxic function in CD4cre SHIPfl/fl and control OT-I cells, peripheral CD8+ cells were primed with APC/OVA peptide for 72 h, rested for 4 days, and incubated for 6 h with the mixture of CFSE-labeled EG7.OVA targets (dim) and EL-4 reference (bright) control cells. Cytolitic efficiency was significantly higher for SHIP-deleted T cells in all effector/target ratios used (Fig. 6B). This enhanced function of CD4cre SHIPfl/fl OT-I CD8+ cells was due to increased granzyme B enzymatic function (Fig. 6C).
Fig. 6.
Enhanced CTL function of SHIP-deleted CD8+ cells. (A) SHIP-deficient CD8+ T cells express 60% more T-bet mRNA. Real-time PCR was performed on mRNA from purified splenic CD8+ cells. Values are relative to the expression of L32 gene. (B) CD4cre SHIPfl/fl and SHIPfl/fl OT-I cells were stimulated with irradiated splenocytes plus peptide for 72 h, harvested, rested, incubated for 6 h with a mixture of EL-4 cells, labeled with high levels of CFSE and EG7-OVA cells, and labeled with low levels of CFSE. The ratio of the numbers of two populations of cells were determined by flow cytometry at the end of the incubation period. The percentage of killing was calculated as 1 minus the ratio of target to control over the ratio of target to control in the absence of effector T cells. (C) Granzyme-dependent killing of EG-7 OVA targets by CD4cre SHIPfl/fl (Right) and SHIPfl/fl (Left) OT-I cells measured by using the GranToxiLux kit. EG7-OVA cells are FL-4+. Upper left quadrants represent viable target cells, whereas upper right quadrants represent dying, VGPD'FGR substrate-positive target cells. Effector cells occupy the lower two quadrants.
Discussion
From the analysis of SHIP-null mice, Kashiwada et al. (23) reported that full expression of the inositol phosphatase SHIP was necessary to restrict the development of regulatory T cells. However, considering the highly pleiotropic phenotype of SHIP-null mice, it is difficult to evaluate whether there is a T cell-intrinsic role of SHIP in T cell development and function. The analysis of mice with T cell-specific deletion of the SHIP presented in this manuscript clearly establishes that deletion of SHIP in T cells does not alter T cell development, the number of regulatory T cells, or the activation status of T cells in the periphery. Our data also suggest that the inflammatory environment in SHIP-null mice is responsible for the observed increase in activated T cells and elevated number of Tregs. Our analysis of mice with conditional deletion of SHIP in the macrophage-granulocyte lineage, which also shows an activated T cell phenotype and increase numbers of Tregs, confirms this hypothesis (T.T. and S.B., unpublished data).
Although in vitro experiments earlier showed that CD28 ligation stimulates SHIP tyrosine phosphorylation (19), that SHIP is recruited to the LAT signaling complex on TCR stimulation (22) and can inhibit Tec signaling in T cells (21), in our experiments, we see no evidence of a regulatory role of SHIP in TCR- or CD28-mediated signaling. SHIP-deleted T cells proliferate as efficiently as control T cells in both polyclonal or antigen-driven activation settings, even in cells bearing the single specificities of transgenic TCRs. TCR stimulation also induced the same level of IL2 and induced T-bet and GATA-3 at the same rate at any concentration of peptide. No difference was observed when CD28 was engaged in these cells, either with addition of anti-CD28 in polyclonal activation or the use of APCs expressing CD28 ligands. Overall these experiments argue against an important role for SHIP in shaping in vivo responses through the TCR or CD28 receptors. However, experiments reported in this manuscript uncover a role for the inositol phosphatase SHIP in shaping Th2 responses in T cells; we observed significant differences in the in vitro Th2 skewing conditions, in Alum-induced humoral responses, and in type 2 responses against helminth antigens in mice with T cell-specific deletion of SHIP. Because TCR-mediated activation is normal in the absence of SHIP, we speculate that SHIP-deleted T cells are biased against a Th2 response as a result of their increased sensitivity to activation through type 1 cytokines such as INFγ. We have found, for example, that SHIP mediates the inhibitory effect of TGFβ over the INFγ-mediated activation. In agreement with this notion, SHIP-deficient T cells grown in nonskewing conditions produce higher levels of IFNγ and lower levels of IL4, IL5, and IL13. We find increased phosphorylation of STAT1 in unstimulated and short-term INFγ-stimulated SHIP-deficient T cells. This effect may be explained molecularly by the reported association of SHIP with PIAS1, a protein inhibitor of activated STAT (4, 30). Consistent with this hypothesis of increased cytokine-mediated activation, we observed elevated basal levels of the type 1 transcription factor T-bet in SHIP-deficient purified naive T cells and reduced inhibition of T-bet levels by TGFβ. As reported by Szabo et al. (31), these elevated levels of T-bet can repress the induction of Th2 responses and would bias subsequent responses toward a Th1 phenotype. The reduced Th2 responses are likely a result of an intrinsic bias toward Th1 skewing in the absence of SHIP and would account for the fact that the effect of SHIP deletion is particularly obvious in unstimulated conditions and less pronounced when cells are strongly polarized in vitro.
T-bet has been reported to be important for the generation of antigen-driven CD8+ cytotoxic effector cells (29), which is consistent with our data showing that increased T-bet expression in SHIP-deficient CD8 T cells correlates with more efficient cytotoxic responses by these cells. Overall these results reveal a role of SHIP in limiting Th1 type of responses both in CD4+ and CD8+ T cells. Finally, the analysis of mice with T cell-specific deletion of SHIP provides evidence of the importance of fine regulation of cytokine signaling in T cells in preventing skewing of T cells against a type 2 response or in modifying cytotoxic responses independently of the type and affinity of antigen that is subsequently engaged.
Methods
Mice and Reagents.
The generation of SHIPfl/fl has been described (17). Anti-SHIP1, Zap-70, ERK1/ERK2, phospho-ERK1/ERK2, and actin antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); anti-PY and anti-PLCγ 1 antibodies were from UBI (Lake Placid, NY); all other antibodies were from BD Biosciences PharMingen (San Diego, CA); and pPCC peptide (residues 88–104; KAERADLIAYLKQATAK) was synthesized by AnaSpec (San Jose, CA).
In Vitro Cell Assays.
Cell activation, Western Blotting, and immunoprecipitation were carried out as previously described (32). In brief, purified peripheral T cells (2 × 107) were stimulated with 10 μg/ml of biotinylated anti-CD3ε or CD4 antibodies, followed by cross-linking with 20 μg/ml of streptavidin for various time periods. For calcium influx assays, CD4+T cells were loaded with Fluo4 (5 μg/ml) and FuraRed (5 μg/ml) (Molecular Probes, Eugene, OR), and Ca2+ mobilization was expressed as the ratio of Fluo4/Fura.
In Vitro Proliferation and in Vivo Expansion.
T cells were isolated from the spleen and lymph nodes of 5- to 7-week-old mice by using a Pan T cell isolation kit or a CD4+ isolation kit (Miltenyi Biotec, Auburn, CA). Purity of T cells was >95% in all cases. T cells were labeled with CFSE and stimulated for 72 h with plate-bound anti-CD3 (5 μg/ml) and anti-CD28 (0.5 μg/ml). After 72 h, cells were stained with propidium iodide and analyzed by FACS. To assay in vivo antigen-driven expansion, purified OT-I or OT-II T cells from CD4cre SHIPfl/fl and SHIPfl/fl mice were CFSE-labeled and transferred to a C57BL/6 host. The next day 200 μg of ovalbumin was s.c. injected, and 2 days after the draining lymph nodes were harvested, stained with CD8 or CD4-Cychrome and Vα2-PE, and acquired by a FACSCalibur.
For the Th1/Th2 in vitro assay, CD4+ T cells were stimulated with plate-bound anti-CD3 and anti-CD28 for 3 days under polarizing conditions. Th1 conditions included IL12 (10 ng/ml) and anti-IL4 (10 μg/ml). Th2 conditions included IL4 (10 ng/ml) and anti-IL12 (10 μg/ml). Cells were washed, expanded in IL2 (10 units per milliliter) for 4 days, and (1 × 106 per milliliter) restimulated with plate-bound anti-CD3 for 24 h. Supernatants were collected at 24 h, and IL4, IL5, and IFNγ concentrations were assessed.
Immunization and Serum Analysis.
Mice were immunized with 50 μg of NP-CGG in Imject Alum (Pierce Chemical, Rockford, IL) and pertussis toxin or 50 μg of TNP-KLH with the MPL+TDM System (Sigma–Aldrich, St. Louis, MO), and sera were tested by ELISA 1 and 2 weeks after immunization. Briefly, plates were coated with NP-BSA or TNP-BSA (10 μg/ml; Biosearch Technologies, Novato, CA), and bound immunoglobulins were detected by alkaline phosphotase-conjugated detection antibodies to specific mouse isotypes (Southern Biotechnology Associates, Birmingham, AL).
GATA-3 and T-bet Expression by Real-Time PCR.
Naive CD4+ T cells (5 × 105) were cultured for 24 h in 1 ml of RPMI in a 24-well plate with 1.25 × 105 mitomycin C-treated P13.9 fibroblast cells stably expressing I-Ek, CD80, and CD54, and preloaded with 0.001–1 μM pPCC. Stimulated cells were lysed in TRIzol (Invitrogen, Carlsbad, CA), and total RNA was isolated by using the RNeasy Mini Kit (Qiagen, Valencia, CA). One microgram of total RNA was reverse-transcribed by using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Quantitative PCR was performed by using the real-time iCycler PCR platform (Bio-Rad). The sequences of primers for GATA-3 and T-bet are described previously (33). The levels of mRNA transcription factors were normalized to that of L32 ribosomal RNA. PCR primers for L32 were 5′-AGAGGACCAAGAAGTTCATCAGGC-3′ and 5′-CTCCTTGACATTGTGGACCAGGAA-3′.
CTL Assay.
OT-I CD4cre SHIPfl/fl and OT-I SHIPfl/fl cells were stimulated with irradiated splenocytes + OVA peptide for 72 h, harvested, rested for 4 days, incubated for 6 h with a mixture of EL-4 cells, labeled with a high level of CFSE and EG7-OVA cells, and labeled with a low level of CFSE. The EL-4/EG7-OVA ratio was determined by flow cytometry at the end of the incubation period. The percentage of killing is calculated as 1 minus ratio of target to control over the ratio of target to control in the absence of effector cells. Granzyme-dependent killing was assessed by GranToxiLux kit from Oncoimmunin (Gaithersburg, MD) by flow cytometry. EG7-OVA target cells were fluorescently labeled and then coincubated for 45 min with effector cells in the presence of a fluorogenic granzyme B substrate. Cleavage of substrate results in increased green fluorescence detected by flow cytometry.
Responses to Schistosome Eggs.
The induction of synchronous egg-induced granulomas was performed as described previously (34). S. mansoni eggs were separated from the livers of infected mice (Biomedical Research Institute, Rockville, MD) and enriched for mature eggs. For the sensitization, mice were immunized with 5,000 eggs i.p. and then challenged with 5,000 eggs i.v. 14 days later. Animals were killed on day 8 after challenge, and lungs were removed for histology and RNA extraction. Granulomas (30 per mouse on average) and serum IL13 Rα2 were measured as previously described (34, 35). All histologic examinations were scored by the same individual in a blinded fashion.
Lung-associated lymph node cells (thoracic/mediastinal) were extracted from the mice, plated in 24-well plates (3 × 106/ml), and stimulated with SEA at 20 μg/ml or with Con A at 5 mg/ml, and supernatants were collected after 72 h to measure the levels of IL4, IL5, IL13, and IFNγ. T cell proliferation was assessed by [3H]TdR incorporation in the last 8 h of culture. Cytokine levels were calculated with the CBA kit (BD Biosciences).
Supplementary Material
Acknowledgments
We thank A. Cheever for help with histology, B. Scott for managing the mouse colony, D. Littman (Memorial Sloan-Kettering Cancer Center, New York, NY) for providing CD4cre mice, Dr. R. Germain (National Institutes of Health) for the I-Ek-expressing P13.9 cell line, and P. Schwartzberg and S. Pierce for critical reading of the manuscript. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviation
- SEA
Schistosome egg antigen.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704853104/DC1.
References
- 1.Ono M, Okada H, Bolland S, Yanagi S, Kurosaki T, Ravetch JV. Cell. 1997;90:293–301. doi: 10.1016/s0092-8674(00)80337-2. [DOI] [PubMed] [Google Scholar]
- 2.Kalesnikoff J, Sly LM, Hughes MR, Buchse T, Rauh MJ, Cao LP, Lam V, Mui A, Huber M, Krystal G. Rev Physiol Biochem Pharmacol. 2003;149:87–103. doi: 10.1007/s10254-003-0016-y. [DOI] [PubMed] [Google Scholar]
- 3.Rauh MJ, Kalesnikoff J, Hughes M, Sly L, Lam V, Krystal G. Biochem Soc Trans. 2003;31:286–291. doi: 10.1042/bst0310286. [DOI] [PubMed] [Google Scholar]
- 4.Rohrschneider LR, Fuller JF, Wolf I, Liu Y, Lucas DM. Genes Dev. 2000;14:505–520. [PubMed] [Google Scholar]
- 5.Ono M, Bolland S, Tempst P, Ravetch JV. Nature. 1996;383:263–266. doi: 10.1038/383263a0. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Oliveira-Dos-Santos AJ, Mariathasan S, Bouchard D, Jones J, Sarao R, Kozieradzki I, Ohashi PS, Penninger JM, Dumont DJ. J Exp Med. 1998;188:1333–1342. doi: 10.1084/jem.188.7.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tridandapani S, Kelley T, Cooney D, Pradhan M, Coggeshall KM. Immunol Today. 1997;18:424–427. doi: 10.1016/s0167-5699(97)01112-2. [DOI] [PubMed] [Google Scholar]
- 8.Brauweiler A, Tamir I, Dal Porto J, Benschop RJ, Helgason CD, Humphries RK, Freed JH, Cambier JC. J Exp Med. 2000;191:1545–1554. doi: 10.1084/jem.191.9.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.March ME, Ravichandran K. Semin Immunol. 2002;14:37–47. doi: 10.1006/smim.2001.0340. [DOI] [PubMed] [Google Scholar]
- 10.Bolland S, Pearse RN, Kurosaki T, Ravetch JV. Immunity. 1998;8:509–516. doi: 10.1016/s1074-7613(00)80555-5. [DOI] [PubMed] [Google Scholar]
- 11.Scharenberg AM, El-Hillal O, Fruman DA, Beitz LO, Li Z, Lin S, Gout I, Cantley LC, Rawlings DJ, Kinet JP. EMBO J. 1998;17:1961–1972. doi: 10.1093/emboj/17.7.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carver DJ, Aman MJ, Ravichandran KS. Blood. 2000;96:1449–1456. [PubMed] [Google Scholar]
- 13.Galandrini R, Tassi I, Mattia G, Lenti L, Piccoli M, Frati L, Santoni A. Blood. 2002;100:4581–4589. doi: 10.1182/blood-2002-04-1058. [DOI] [PubMed] [Google Scholar]
- 14.Okada H, Bolland S, Hashimoto A, Kurosaki M, Kabuyama Y, Iino M, Ravetch JV, Kurosaki T. J Immunol. 1998;161:5129–5132. [PubMed] [Google Scholar]
- 15.Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM, Borowski A, Jirik F, Krystal G, Humphries RK. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgason CD, Kalberer CP, Damen JE, Chappel SM, Pineault N, Krystal G, Humphries RK. J Exp Med. 2000;191:781–794. doi: 10.1084/jem.191.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson MC, Guinamard R, Bolland S, Sankala M, Steinman RM, Ravetch JV. J Exp Med. 2003;198:333–340. doi: 10.1084/jem.20030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rauh MJ, Ho V, Pereira C, Sham A, Sly LM, Lam V, Huxham L, Minchinton AI, Mui A, Krystal G. Immunity. 2005;23:361–374. doi: 10.1016/j.immuni.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Edmunds C, Parry RV, Burgess SJ, Reaves B, Ward SG. Eur J Immunol. 1999;29:3507–3515. doi: 10.1002/(SICI)1521-4141(199911)29:11<3507::AID-IMMU3507>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Freeburn RW, Wright KL, Burgess SJ, Astoul E, Cantrell DA, Ward SG. J Immunol. 2002;169:5441–5450. doi: 10.4049/jimmunol.169.10.5441. [DOI] [PubMed] [Google Scholar]
- 21.Tomlinson MG, Heath VL, Turck CW, Watson SP, Weiss A. J Biol Chem. 2004;279:55089–55096. doi: 10.1074/jbc.M408141200. [DOI] [PubMed] [Google Scholar]
- 22.Dong S, Corre B, Foulon E, Dufour E, Veillette A, Acuto O, Michel F. J Exp Med. 2006;203:2509–2518. doi: 10.1084/jem.20060650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashiwada M, Cattoretti G, McKeag L, Rouse T, Showalter BM, Al-Alem U, Niki M, Pandolfi PP, Field EH, Rothman PB. J Immunol. 2006;176:3958–3965. doi: 10.4049/jimmunol.176.7.3958. [DOI] [PubMed] [Google Scholar]
- 24.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 25.Yamane H, Zhu J, Paul WE. J Exp Med. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O'Shea JJ. Proc Natl Acad Sci USA. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita T, Reis LF, Watanabe N, Kimura Y, Taniguchi T, Vilcek J. Proc Natl Acad Sci USA. 1989;86:9936–9940. doi: 10.1073/pnas.86.24.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park IK, Shultz LD, Letterio JJ, Gorham JD. J Immunol. 2005;175:5666–5674. doi: 10.4049/jimmunol.175.9.5666. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Proc Natl Acad Sci USA. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Proc Natl Acad Sci USA. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 32.Naramura M, Kole HK, Hu RJ, Gu H. Proc Natl Acad Sci USA. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenderfer SE, Ke B, Hollmann TJ, Wetsel RA, Lan HY, Braun MC. J Am Soc Nephrol. 2005;16:3572–3582. doi: 10.1681/ASN.2005040373. [DOI] [PubMed] [Google Scholar]
- 34.Sandler NG, Mentink-Kane MM, Cheever AW, Wynn TA. J Immunol. 2003;171:3655–3667. doi: 10.4049/jimmunol.171.7.3655. [DOI] [PubMed] [Google Scholar]
- 35.Mentink-Kane MM, Cheever AW, Thompson RW, Hari DM, Kabatereine NB, Vennervald BJ, Ouma JH, Mwatha JK, Jones FM, Donaldson DD, et al. Proc Natl Acad Sci USA. 2004;101:586–590. doi: 10.1073/pnas.0305064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.