Abstract
The early phases of carcinogenesis resemble embryonic development, often involving the reexpression of embryonic mesenchymal genes. The NCI60 panel of human tumor cell lines can genetically be subdivided into two superclusters (SCs) that correspond to CD95 Type I and II cells. SC1 cells are characterized by a mesenchymal and SC2 cells by an epithelial gene signature, suggesting that SC1 cells represent less differentiated, advanced stages of cancer. miRNAs are small 20- to 22-nucleotide-long noncoding RNAs that inhibit gene expression at the posttranscriptional level. By performing miRNA expression analysis on 10 Type I and 10 Type II cells, we have determined that SC1 cells express low and SC2 cells high levels of the miRNA let-7, respectively, suggesting that let-7 is a marker for less advanced cancers. Expression of the let-7 target high-mobility group A2 (HMGA2), an early embryonic gene, but not of classical epithelial or mesenchymal markers such as E-cadherin or vimentin, inversely correlated with let-7 expression in SC1 and SC2 cells. Using ovarian cancer as a model, we demonstrate that expression of let-7 and HMGA2 is a better predictor of prognosis than classical markers such as E-cadherin, vimentin, and Snail. These data identify loss of let-7 expression as a marker for less differentiated cancer.
Keywords: HMGA2, miRNA, ovarian cancer, tumor progression, supercluster
It has been demonstrated that miRNAs act either as oncogenes (e.g., miR-155, miR-17–5p, and miR-21) (1, 2) or tumor suppressors (e.g., miR-15a, miR-16–1, and let-7) (3–7). Members of the ubiquitously expressed let-7/miR-98 family are expressed late in mammalian embryonic development (4–9), and in humans let-7 has been shown to act as a tumor suppressor for lung and colon cancer, in part through targeting of RAS (4–7). It is becoming increasingly evident that miRNAs play an important role in oncogenesis by regulating genes that are involved in transformation. Because there are ≈25,000 to 30,000 genes in the human genome, but only ≈400 miRNAs, there may be miRNA families that govern entire gene expression programs presumably by regulating a distinct pattern of differentiation. We previously identified two cell types that can be classified by the way they respond to stimulation of the death receptor, CD95 (Fas/APO-1) (12). We subsequently demonstrated that CD95-sensitive Type I (mitochondria-independent death) and Type II (mitochondria-dependent death) cells correspond to supercluster (SC)1 and SC2 cells, respectively [supporting information (SI) Fig. 7] (13, 14). To determine whether miRNAs could be responsible for the differences seen between SC1 and SC2 cells, we performed an miRNA gene array analysis. We report the identification of the let-7 family of miRNAs as being preferentially expressed in SC2 cells and identify high-mobility group A2 (HMGA2) as a direct let-7 target that is preferentially expressed in SC1 cells. Testing ovarian cancer (OC) as a model, we demonstrate that let-7 and HMGA2 serve as prognostic markers for this disease. We identify low let-7 and high HMGA2 expression as markers for advanced cancer.
Results and Discussion
Let-7 Genes Are Preferentially Expressed in Type II/SC2 Cells.
To determine whether miRNA could regulate the difference between Type I and II cells, we subjected 10 Type I/SC1 and 10 Type II/SC2 cell lines to an miRNA expression array analysis (Fig. 1A). Detected miRNAs were sorted based on their expression in the two groups. Four members of the let-7 family of miRNAs (let-7f, let-7d, miR-98, and let-7g) were preferentially expressed in SC2 cells (P ≤ 0.001). This result was confirmed by using quantitative real-time RT-PCR; all four let-7 miRNAs were expressed significantly higher in Type II cells than in Type I cells (Fig. 1B). By performing a nonparametric Wilcoxon two sample rank test, we determined that seven of the nine let-7 family members on the chip were significantly higher expressed in Type II cells, with let-7d and let-7g being the best differentiators (SI Fig. 8). These seven let-7 family members also clustered together in a hierarchical cluster analysis (SI Fig. 9).
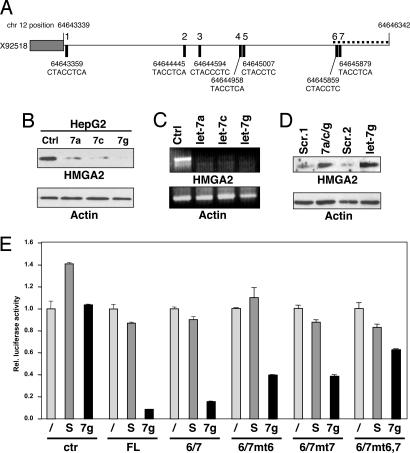
Fig. 1.
Identification of let-7 as a marker for Type II/SC2 cells. (A) Gene array analysis of the expression of miRNAs in 10 Type I and 10 Type II cell lines. Seventy-nine of 287 human miRNAs on the chip were significantly expressed in the 20 cell lines (see Methods and SI Table 1). No single miRNA was found to be equally expressed in all cells, suggesting that miRNAs do not have general housekeeping functions in tumor cells. The expression of miRNAs between SC1 and SC2 cells was therefore calculated relative to the mean of the signal intensities of the 79 miRNAs that are expressed in the 20 tumor cells (SI Table 2). Shown are all miRNAs that were significantly expressed (see Methods) and sorted according to P values from t tests of expression differences between Type I or II cell lines. Four miRNAs with a highly significant P value of <0.001 are boxed with a stippled line. (B) Relative fold difference of let-7d, mir-98, let-7a/f, and let-7g expression between the 10 Type I and 10 Type II cancer cell lines measured by real-time PCR. Data were normalized by determining the ratio of let-7 expression to that of the small nuclear RNA U6 as described before (33). The let-7/U6 ratios were plotted relative to the ratio found in HepG2 cells. HeLa and HepG2 cells were chosen as positive or negative controls for let-7 expression, respectively. We determined that the probe for let-7f does not discriminate between let-7f and let-7a (data not shown). (C) Analysis of the expression of let-7d in 59 of the NCI60 cells. (Left) Expression of let-7d in the 22 Type I (blue) and Type II (red) cells previously identified among the NCI60 cells (13) is shown. (Left) Expression in the SC1 (blue) and SC2 (red) cell lines among the 59 NCI60 cells is shown. The asterisk marks an outlier in the SC1 group. Without this one value, the P value changed to 0.0012 (two sample t test). In this independent performed analysis, we compared the expression of let-7d relative to the mean of expression of a very similar set of 65 miRNAs that was used to analyze the miRNA chip data (SI Table 2).
To determine whether the results with a subset of NCI60 cells also applied to all of the NCI60 cells, we interrogated a data set of miRNA expression that was recently generated with 59 of the NCI60 cells (15). Let-7d showed the highest significance of preferential expression in the Type II and SC2 cells, correlating well with our own analysis (Fig. 1C and data not shown). This analysis confirms that Type I cells represent SC1 cells and Type II cells represent SC2 cells. Although there was overlap between the groups, let-7d expression discriminates between the two SCs more effectively than any of the previously tested mRNAs (13).
Identification of HMGA2 as a Direct Target for Let-7 in Human Cancer Cell Lines.
To identify genes that are up-regulated in SC1 cells, we searched for putative let-7 targets employing four of the most widely used programs to predict miRNA targets (TargetScan, TargetScanS, PicTar, and miRNAviewer) (16). All four algorithms predicted HMGA2 as the number one target (SI Fig. 10), consistent with recent reports identifying HMGA2 as a let-7 target (10, 11). The 3′-UTR of human HMGA2 contains seven putative let-7 complementary sites (LCSs) (Fig. 2A). To test whether introduction of let-7 could reduce expression of HMGA2 in a human cancer cell line that expresses low let-7, we transfected HepG2 cells that have high HMGA2 expression with three different let-7 precursor miRNAs: let-7a, let-7c, and let-7g. HMGA2 expression was substantially reduced on introduction of the let-7 miRNAs (Fig. 2B). Introduction of exogenous let-7a, let-7c, or let-7g caused degradation of HMGA2 mRNA (Fig. 2C). To determine whether expression of HMGA2 in cancer cells may be suppressed by endogenous levels of let-7 found in SC2 cells, we transfected the high let-7-expressing HeLa cells with inhibitors of let-7 (Fig. 2D). Both a mixture of inhibitors specific for let-7a, let-7c, and let-7g and the let-7g inhibitor alone caused induction of HMGA2 protein expression, indicating that it is let-7 expression that prevents HMGA2 expression in these cells.
Fig. 2.
Identification of HMGA2 as a direct target of let-7. (A) Schematic of the 3′UTR at the human HMGA2 genomic locus located on chromosome 12. The gray box depicts the 3′-end of the ORF, whereas the horizontal line depicts the 3′-UTR spanning ≈3 kb. Solid rectangles indicate the precise locations with nucleotide positions on chromosome 12 of the seven putative let-7 binding sites and the seed match to the let-7 family of miRNAs. The dotted line located at the distal end of the 3′-UTR represents the fragment of the 3′-UTR containing the sixth and seventh putative let-7 sites that was used for the Renilla reporter experiments in Fig. 2E. (B) HMGA2 protein expression is negatively regulated by let-7. HepG2 cells were transiently transfected with let-7a, let-7c, let-7g, or control precursor miRNA. β-actin was detected to demonstrate equal loading. Ctrl, scrambled control. (C) HMGA2 RNA is degraded with increased levels of let-7. HepG2 cells were transfected with miRNAs as in Fig. 2B, and after 72 h mRNA for HMGA2 was detected by RT-PCR. β-actin is shown as a control. (D) Induction of HMGA2 protein through inhibiting let-7. HeLa cells were transfected either with a mixture of let-7a, let-7c, and let-7g (total of 180 pmol) or with let-7d (60 pmol) inhibitor alone. Scr, scrambled oligonucleotide as controls at two concentrations; Scr.1, 180 pmol; Scr.2, 60 pmol. (E) HMGA2 is posttranscriptionally regulated through the 3′-UTR by let-7. Renilla luciferase reporter assays were performed with reporter plasmid psiCHECK (ctr), psiCHECK-HMGA2 full-length 3′-UTR (FL), or psiCHECK-HMGA2 3′-UTR 6/7 (6/7), together with either 1 pmol of premiR scrambled (S) or premiR let-7g (7g). PsiCHECK-HMGA2 3′-UTR 6/7mt6 (6/7mt6), psiCHECK-HMGA2 3′-UTR 6/7mt7 (6/7mt7), or psiCHECK-HMGA2 3′-UTR 6/7mt6,7 (6/7mt6,7), which harbor mutations in the seed matches in either LCS6, LCS7, or both, were also used in the luciferase assay experiment. Renilla luciferase activity was normalized to the internally controlled firefly luciferase activity.
It was recently demonstrated that five of the seven LCSs in the 3′UTR of human HMGA2 are functional (10). The two distal LCSs, 6 and 7, are identical in the 3′UTR of humans and mice, with LCS6 showing the highest degree of identity with let-7 family members (SI Fig. 11). To determine whether targeting these two LCSs was sufficient to suppress expression of HMGA2, we cloned a fragment of the 3′UTR containing LCS6/7 into a luciferase reporter plasmid and determined how transfection of let-7 affects luciferase activity (Fig. 2E). Both let-7d and let-7g caused significant repression of luciferase activity of full-length 3′UTR as well as the 6/7 fragment (Fig. 2E and data not shown). Mutation of either one of the two seed matches in the 6/7 fragment caused a moderate reduction of the ability of let-7 to suppress luciferase activity. When both LCS in the 6/7 fragment were mutated, let-7g could not suppress luciferase activity anymore when compared to the effect on control plasmid (Fig. 2E). These data suggest that both distal LCSs in the HMGA2 3′UTR are targets for let-7, in contrast to a recent report (10), and they are sufficient to regulate HMGA2 expression.
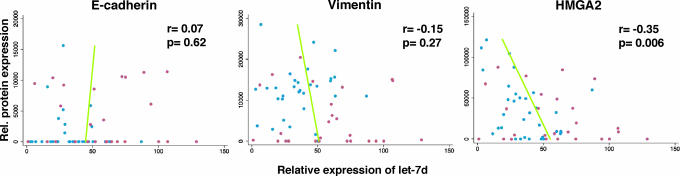
It has recently been shown that tumor cells express lower levels of miRNAs than primary tissues (17), raising the question of whether the differences in let-7 expression between SC1 and SC2 tumor cell lines are functionally significant and whether the level of let-7 detected in these cells could control expression of HMGA2. We therefore quantified the amount of HMGA2 protein in the NCI60 cells and compared its expression with the let-7 expression in every cell line. Because SC1 cells express a mesenchymal gene signature and SC2 cells an epithelial gene signature, we also determined protein expression of E-cadherin (an epithelial marker) and vimentin (a mesenchymal marker) in the two cell types (Fig. 3 and SI Fig. 12). This analysis revealed a significant inverse correlation between the expression of let-7d, let-7g, and let-7f and HMGA2 in individual cell lines (Fig. 3 and SI Fig. 13). We did not find such a correlation between the expression of let-7 and E-cadherin or vimentin (Fig. 3 and SI Fig. 13). The inverse ratio of expression of let-7 and HMGA2 characterizes the difference between SC1 and SC2 cells and could therefore be an indicator for more or less advanced human cancer.
Fig. 3.
Analysis of the protein expression levels of HMGA2, E-cadherin, and vimentin, and correlation with let-7d expression in 59 NCI60 cell lines. Protein expression was determined by Western blotting (see SI Fig. 12). Expression levels in SC1 cells are shown as blue dots and SC2 cells as red dots. Predicted let-7d values from a univariate linear regression are plotted against protein expression. Pearson correlation coefficients (r) and P values (p) are reported.
Let-7 Regulates HMGA2 Expression in OC and Predicts Disease Progression.
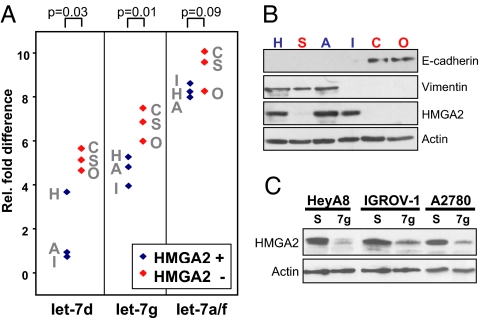
Now that we established that endogenous let-7 levels found in SC2 cells can control HMGA2 expression, we sought to test the significance of these findings for human cancer progression. Therefore, we examined the expression of let-7 at different stages of OC. OC develops from a single layer of cells on the surface of the ovary and progresses to an aggressive, dedifferentiated cancer expressing mesenchymal markers (18, 19). Recently, it was shown that genomic alterations affecting the expression of let-7 are found in OC (20). In view of our findings, we tested whether the correlation between let-7 and HMGA2 held for six randomly selected epithelial OC lines (Fig. 4 A and B). Three of the tested cell lines expressed low amounts of let-7, whereas three expressed high levels (Fig. 4A). Only the three cell lines expressing low let-7 expressed HMGA2 (Fig. 4B), and introduction of let-7d or let-7g into these cell lines abrogated HMGA2 expression, demonstrating that let-7 can target HMGA2 in OC cells (Fig. 4C and data not shown).
Fig. 4.
HMGA2 in OC cell lines is under control of let-7. (A) Relative fold difference of let-7d, let-7g, and let-7a/f miRNA expression in six randomly chosen OC cell lines measured by real-time PCR and normalized to U6 expression. A, A2780; C, CAOV3; H, HeyA8; I, IGROV-I; O, OVCAR-5; S, SKOV3ip. Two sample t test results are shown. (B) Western blot analysis of the same cells analyzed in A. Let-7hi/HMGA2lo cells in A and B are labeled in red, let-7lo/HMGA2hi cells in blue. β-actin was detected to demonstrate equal loading. (C) The three HMGA2-expressing cells were transfected with let-7g, and HMGA2 protein amount was determined by Western blotting. β-actin was detected to demonstrate equal loading. S, scrambled control.
Next we determined the expression level of HMGA2 in tissues from patients with OC (Fig. 5A). HMGA2 was undetectable in normal ovarian surface epithelium or benign ovarian tumors, but expressed in patients with OC (Fig. 5A). Although normal appearing ovarian epithelium did not express HMGA2 (Fig. 5B Inset), tumor cells in the early stage represented by in situ cancers (see Inset) clearly expressed the HMGA2 protein, and its expression was further increased in full-blown carcinoma (C in Fig. 5B). These data suggest that HMGA2 expression is up-regulated early on transformation of ovarian epithelial cells.
Fig. 5.
HMGA2 expression is increased early in OC progression. (A) Immunohistochemistry for HMGA2 of representative normal (Left Upper) and tumor tissues (Right Upper and Lower) from OC patients. (Magnification: tumors, ×200; ovary, ×400.) Arrowhead points to the surface epithelium of a normal ovary in a patient with a benign tumor. (B) Immunohistochemical detection of HMGA2 in tumor from a patient with advanced OC. C, carcinoma; O, ovary.
Given our results that loss of let-7 induces HMGA2 expression and identifies a mesenchymal cancer subtype (similar to SC1 cells), we sought to determine whether expression of HMGA2 is associated with patient prognosis. A human tissue microarray containing samples from primary tumors of 100 patients with International Federation of Gynecology and Obstetrics (FIGO) stages II-IV advanced OC was stained for HMGA2 protein expression. High expression of HMGA2 correlated significantly with an adverse prognosis both for progression-free (Fig. 6A) and overall survival (SI Fig. 14) of the patients. No significant difference was found in the expression of HMGA2 between primary tumors and corresponding omental or peritoneal metastases from the same patients (P = 0.5909). These data indicate that HMGA2 expression is a marker for early tumor progression and not for metastasis.
Fig. 6.
HMGA2 and let-7d expression inversely correlate with survival of OC patients. (A) Progression-free survival of 100 OC patients depending on the staining intensity for HMGA2. Primary tumors from 100 patients (with FIGO stage II–IV) on a tissue array were stained for HMGA2. Splitting the group at the median resulted in 0 < 132 and 1 > 132. Only samples with >5% HMGA2-positive cells were included. The patient group is described in SI Table 3. (B) Expression of let-7d in RNA samples extracted from tumor tissue with low <100 (n = 8) or high >160 (n = 9) HMGA2 staining. Fold difference relative to the sample with the lowest let-7d expression (set to 1) is shown. Expression of let-7d was normalized to U6. P value is the result of two sample t test. (C) Progression-free survival by the ratio of HMGA2 intensity to let-7 ΔCT (splitting at the median: 0 < 54 and 1 > 54 high) including 53 subjects with FIGO stage II, III, or IV and HMGA 2% positive >5%. Note that two let-7 ΔCT values <0 were given a threshold value of 0.20.
HMGA2 was shown to be up-regulated in advanced cancers, and its expression correlates with poor prognosis (21–25). Because let-7 regulates the expression of HMGA2, we investigated whether let-7 is down-regulated in OC tissue from patients with an adverse prognosis. RNA was extracted from tumor tissues with low (<100) staining and compared to tumor tissues with high (>160) staining for HMGA2 from patients analyzed in Fig. 6A. A significantly higher expression of let-7d was detected in the patient group with low HMGA2 intensity (Fig. 6B). In contrast, no significant correlation between the expression of E-cadherin or vimentin and progression-free or overall survival was found (SI Fig. 14). We also did not find a correlation between the expression of Snail and either progression-free survival or let-7d expression (SI Fig. 15). Finally, a combined score of let-7d and HMGA2 expression was also able to separate the patients into two groups (Fig. 6C): The estimate of 5-year progression-free survival in the group with a lower HMGA2/let-7 ratio was 39.7% (95% confidence interval: 21.4–57.6%). The estimate of 5-year progression-free survival in the group with a higher HMGA2/let-7 ratio was 9.6% (95% confidence interval: 1.8–25.4%).
Our data suggest that human tumors can be divided into two major subtypes, the let-7hi- and let-7lo-expressing tumor cells. The separation of tumor cells into two such SCs may not be restricted to the NCI60 cells. A similar separation into two major clusters was observed in unsupervised hierarchical cluster analyses of cDNAs in cell lines or primary tumor samples from patients with breast (26), ovarian (27), and other cancers (28). In all these cases, in addition to tumor- and class-specific gene clusters, two SCs emerged from the analyses. The differences in gene expression between the SCs were larger than any differences between tumor types in these analyses. Recently, the analysis of miRNA expression among different normal tissues also resulted in the separation of tissues into two similar sized SCs (29). Interestingly, the miRNA that best allowed the separation of these two SCs was let-7a, suggesting that the concept of two SCs separated by their expression levels of let-7 may not be limited to tumor cells.
The two tumor types that are characterized by the expression of let-7 go beyond the conventional definition of epithelial and mesenchymal cells. The gene expression profiles corresponding to SC1 and SC2 cells, as described by Ross et al. (14), do not always yield morphological features. Many of the established “epithelial” cell lines among the NCI60 cells, such as PC3, Hs578T, BT-549, U251, A549, H226, ACHN, CAKI-1, OVCAR-5, IGROV-I, EKVX, and H460, separated into SC1 in the hierarchical cluster analysis (SI Fig. 7). In addition, many lymphoid cell lines that cannot be viewed as being derived from epithelial tissues clustered together with the epithelial group. However, because the gene signature of SC2 cells overall has an epithelial character, classical definitions of epithelial cells may not be applicable anymore. It appears that whole gene expression patterns can be regulated by miRNAs, and let-7 is a good candidate to define this “epithelial” gene signature. Consequently, let-7 was found to be superior in differentiating SC2 from SC1 cells to established markers such as E-cadherin or vimentin.
One of the models used to explain carcinogenesis suggests that, during cancer progression, a number of embryonic genes that are not expressed in adult tissues are reexpressed, causing tumors to dedifferentiate and become more mobile and invasive in a manner similar to embryonic cells (30). Because tumor cells with a mesenchymal gene profile represent more advanced stages of cancer, let-7 down-regulation could be seen as part of the process of tumor progression. Recently, it was demonstrated that impaired miRNA processing accelerates oncogenic transformation (31). Altering let-7 expression levels caused a change in tumorigenicity in a number of cancer cell lines (31). These data were taken to support the finding that a global reduction of miRNAs increases oncogenic potential of cancer cells. Our work now focuses on the loss of members of the let-7 family as the major determinant of cancer progression because no other miRNA was allowed to differentiate SC1 from SC2 cells as effectively (see SI Table 1). We propose that one of the functions of let-7 is to maintain differentiated states by suppressing the expression of genes that are expressed in dedifferentiated tissues, such as HMGA2.
Methods
miRNA Chip Analysis.
miRNA was enriched from cells by using the mirVana miRNA Isolation Kit (Ambion, Austin, TX). Size distribution of enriched miRNAs was confirmed by using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Probes were labeled by attaching a 20- to 50-nucleotide tail of a mixture of ATP and amine-modified ATP to the 3′ end of miRNA using Escherichia coli poly(A) polymerase. The amine-modified miRNA was then coupled to Cy5 dye. The Cy5-labeled miRNA was cleaned and hybridized at 42°C for 16 h to the mirVana miRNA bioarray. After a wash and drying step, the array was scanned at a PMT setting of 600 with 100% power and 5-μm resolution with the GenePix pro 4.1 software in the GenePix 4000B microarray scanner (Axon Instruments, Union City, CA). To determine relative expression levels of miRNA in different tumor cells, four miRNAs (miR103, miR107, miR93, and miR191) were selected on the chip that had roughly similar expression levels in all cell lines. The expression of all miRNAs was normalized to the mean of these four invariant miRNAs. The sum of the signal intensity of all 20 cell lines was calculated. There were 87 human miRNAs with cumulative signal intensity >1,000 that were used for further analysis (with the highest signal of 326,169 for miR21). Fold change was calculated, and a two sample t test was conducted to test for differences in the expression between the two cell types. miRNAs were sorted by P value. This preliminary analysis identified four let-7 family members as being preferentially expressed in Type II cells with a significant P value. To refine this analysis, we recalculated the signals by normalizing all values to the mean of all significantly expressed human miRNAs (not including let-7 family members) (see SI Table 2). All signals were divided by the mean of these miRNAs. Negative values were set to zero. When all samples were added again for each row (miRNA), the highest value was 143.31 (for miR-21) and the lowest was 0.58 (for miR-132). The eight miRNAs below a value of 1.0 were deleted. Significance analysis of the remaining 79 significantly expressed miRNA was performed by using a two-tailed t test with unequal variance. Samples were sorted according to P value, and a hierarchical cluster analysis was performed by using Dchip and default settings. Additionally, a Wilcoxon rank test was conducted to test whether there was a difference in let-7 expression for each of the nine let-7 miRNAs between the two cell types (SI Fig. 8).
Real-Time PCR.
microRNA was enriched from cells by using the mirVana miRNA Isolation Kit (Ambion). microRNA and U6-specific cDNA was generated from between 10 and 20 ng of enriched or FFPET RNA by using the TaqMan MicroRNA RT Kit and the RNA-specific RT primer from the TaqMan micro RNA Assays (Applied Biosystems, Foster City, CA). miRNA levels were also measured by using the miRNA-specific TaqMan MGB probe included with the MicroRNA Assays on a 7,500 quantitative real-time PCR machine and SDS software (Applied Biosystems). miRNA abundance was normalized relative to U6. For the analysis of let-7 in 59 NCI60 cells, data sets containing the expression levels of let-7d, let-7g, let-7f, miR-98, and miR-63 control miRNAs obtained by real-time PCR using total RNA (15) were provided by Mark Israel (Dartmouth Medical School, Lebanon, NH). The expression level of let-7d relative to the mean of 63 control miRNAs (see SI Table 2) was determined for each cell line. Snail gene expression was quantified in ovarian tumor tissues by using cDNA generated from 100 ng RNA extracted from formalin-fixed paraffin-embedded tissue (FFPET) by using the High-Capacity cDNA RT Kit (Applied Biosystems) and TaqMan Gene Expression Assays for Snai1 (Mm00441533_g1; Applied Biosystems) and human β-gluco ronidase (4333767T; Applied Biosystems) as an endogenous control. Relative levels of miRNA or mRNA gene expression were calculated by using the 2−ΔΔCT method.
RNA Extraction from FFPET.
RNA was extracted from FFPET using the MasterPureRNA Purification Kit (Epicenter Technologies, Madison, WI) according to the manufacturer's instructions with the following modifications: Punched-out plugs (1.5 × 1.5 mm) of a paraffin block were deparaffinized in 1 ml of ACS grade Xylene (Fisher, Pittsburgh, PA) and incubated at 65°C for 10 min, followed by a 100% ACS grade ethanol (Sigma–Aldrich, St. Louis, MO) wash. Samples were then incubated with proteinase K at 65°C overnight while shaking. After the isopropanol precipitation step, samples were incubated with DNase I for 30 min at 37°C to ensure the removal of all contaminating genomic DNA. RNA samples were resuspended in nuclease-free water after the final precipitation steps, and quality and quantity were assessed by using the ND-1000 spectrophotometer (Nanodrop, Wilmington, DE). The paraffin plugs were enriched for tumor tissue by microscope control by using H&E-stained sections of the same sample as guidance.
Constructs and Mutagenesis.
The human HMGA2 3′-UTR (GenBank accession no. X92518, bases 1645–4218) was PCR-amplified from pBAC clone RP11–1025D9 with primers 5′-AAAACTCGAGGCCAACGTTCGATTTCTACCT-3′ and 5′- AAAAGCGGCCGCTACTGTTCCATTGGCCACAA-3′ that contain NotI and XhoI restriction site overhangs, respectively. This PCR product, which contains seven putative let-7 binding sites, was cloned into the psiCHECK-2 vector (Promega, Madison, WI) immediately downstream of the Renilla luciferase reporter gene and named psiCHECK-HMGA2 3′-UTR. A DNA stretch containing LCS6 and 7 (4023–4218) was PCR-amplified with primers 5′-AAAACTCGAGGGATGGGCCTTTTAGAAACC-3′ and 5′-AAAAGCGGCCGCTACTGTTCCATTGGCCACAA-3′ that contain NotI and XhoI restriction site overhangs, respectively, and cloned into psiCHECK generating psiCHECK-HMGA2 3′-UTR 6/7. Site-directed mutagenesis was performed by using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) to change three nucleotides in the second, fourth, and sixth positions in the seed matches of LCS6 and 7. The seed sequence of LCS6 CTACCTC was substituted by CAAGCAC, and LCS7 was changed from TACCTCA to TTCGTGA. The mutations were generated in the psiCHECK-HMGA2 3′-UTR 6/7 construct and named psiCHECK-HMGA2 3′-UTR 6/7mt6 and pSICHECK-HMGA2 3′UTR 6/7mt7 for a single-seed match mutation and psiCHECK- HMGA2 3′-UTR 6/7mt6/7 for double-seed match mutations.
Transfection and Luciferase Assay.
The 293TN cells (2.5 × 104) were seeded in each well of a 48-well plate 1 day before transfection. Cells were transfected with either psiCHECK, psiCHECK-HMGA2 3′-UTR, psiCHECK-HMGA2 3′-UTR 6/7, or psiCHECK-HMGA2 3′-UTR 6/7 mutants together with either PremiR let-7g or negative control #1 precursor miRNA (Ambion) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After 48 h, cells were lysed and measured for luciferase activity according to the manufacturer's instructions (Promega). All experiments were performed in triplicates and normalized to the activity of the Renilla luciferase gene that is contained within the psiCHECK-2 vector as an internal control.
Analysis of OC Tumor Microarray.
Tissue blocks from 107 patients with FIGO stages II–IV advanced OC or peritoneal cancer who had undergone tumor debulking by a Gynecologic Oncologist at the Section of Gynecologic Oncology, University of Chicago, between 1994 and 2004 were selected for the study after Institutional Review Board approval was obtained. A tumor microarray was assembled, and clinicopathological parameters collected as described (32). Satisfactory immunohistochemical staining of HMGA2 was obtained in 100 patients (see SI Table 3 for details).
Additional Details.
Additional methods are described in SI Text.
Supplementary Material
Acknowledgments
We thank X. Li for help with the miRNA array analysis, S. Holbeck for providing the NCI60 cell lines, C. Burge for critically reading the manuscript, and M. Israel for providing real-time PCR data on the NCI60 cells. This work was supported by the Ovarian Cancer Research Fund (Liz Tilberis Scholars Program), National Institutes of Health Grants R01 CA111882 (to E.L.) and R01 GM61712 (to M.E.P), and University of Chicago Women's Board Fellowship (S.M.P.).
Abbreviations
- FIGO
International Federation of Gynecology and Obstetrics
- FFPET
formalin-fixed paraffin-embedded tissue
- HMGA2
high-mobility group A2
- LCS
let-7 complementary site
- SC
supercluster
- OC
ovarian cancer.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704372104/DC1.
References
- 1.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akao Y, Nakagawa Y, Naoe T. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 5.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 6.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 7.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 10.Lee YS, Dutta A. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayr C, Hemann MT, Bartel DP. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, Debatin KM, Krammer PH, Peter ME. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Algeciras-Schimnich A, Pietras EM, Barnhart BC, Legembre P, Vijayan S, Holbeck SL, Peter ME. Proc Natl Acad Sci USA. 2003;100:11445–11450. doi: 10.1073/pnas.2034995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 15.Gaur AB, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel M. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 16.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 18.Auersperg N, Pan J, Grove BD, Peterson T, Fisher J, Maines-Bandiera S, Somasiri A, Roskelley CD. Proc Natl Acad Sci USA. 1999;96:6249–6254. doi: 10.1073/pnas.96.11.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosano L, Spinella F, Di Castro V, Nicotra MR, Dedhar S, de Herreros AG, Natali PG, Bagnato A. Cancer Res. 2005;65:11649–11657. doi: 10.1158/0008-5472.CAN-05-2123. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giannini G, Kim CJ, Di Marcotullio L, Manfioletti G, Cardinali B, Cerignoli F, Ristori E, Zani M, Frati L, Screpanti I, Guilino A. Br J Cancer. 2000;83:1503–1509. doi: 10.1054/bjoc.2000.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abe N, Watanabe T, Suzuki Y, Matsumoto N, Masaki T, Mori T, Sugiyama M, Chiappetta G, Fusco A, Atomi Y. Br J Cancer. 2003;89:2104–2109. doi: 10.1038/sj.bjc.6601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiappetta G, Bandiera A, Berlingieri MT, Visconti R, Manfioletti G, Battista S, Martinez-Tello FJ, Santoro M, Giancotti V, Fusco A. Oncogene. 1995;10:1307–1314. [PubMed] [Google Scholar]
- 24.Miyazawa J, Mitoro A, Kawashiri S, Chada KK, Imai K. Cancer Res. 2004;64:2024–2029. doi: 10.1158/0008-5472.can-03-1855. [DOI] [PubMed] [Google Scholar]
- 25.Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, Salo J, Karjalainen A, Knuutila S, Anttila S. J Pathol. 2006;209:206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- 26.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 27.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S, et al. Mol Biol Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alizadeh AA, Ross DT, Perou CM, van de Rijn M. J Pathol. 2001;195:41–52. doi: 10.1002/path.889. [DOI] [PubMed] [Google Scholar]
- 29.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiery JP, Sleeman JP. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 31.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 32.Sawada K, Radjabi AR, Shinomiya N, Kistner E, Kenny H, Becker AR, Turkyilmaz MA, Salgia R, Yamada SD, Vande Woude GF, et al. Cancer Res. 2007;67:1670–1679. doi: 10.1158/0008-5472.CAN-06-1147. [DOI] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Jiang J, Liu Q, Yang L. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.