Abstract
The development of kinase inhibitors is revolutionizing cancer treatment. Assessing the oncogenic potential of individual kinase activities and ensuring that a drug of interest acts by direct inhibition of its putative target kinase are clear priorities. We developed a genetic strategy to selectively inactivate the catalytic activity of kinases. This approach generates isogenic cells in which a given kinase gene is expressed but is devoid of enzymatic activity. As a model to test this approach, we chose the MET receptor, which is involved in multiple cancers and is the focus of several therapeutic efforts. The exon encoding the ATP-binding site of MET was deleted from the genome of colorectal, bladder, and endometrial cancer cells. The derivative isogenic cells expressed a kinase-inactive Met (MET-KD) and were completely unresponsive to its ligand hepatocyte growth factor (HGF), indicating the exclusivity of this ligand–receptor axis. The in vivo tumorigenic potential of MET-KD cells was reduced but could be partially restored by HGF, suggesting that concomitant targeting of the receptor and its ligand should be therapeutically exploited. A reportedly selective Met-kinase inhibitor (SU-11274) markedly affected the growth of MET-KD cancer cells, indicating this compound exerts its effects not only through the intended target. The genetic strategy presented here is not limited to kinase genes but could be broadly applicable to any drug/protein combination in which the target enzymatic domain is known.
Keywords: knockout, somatic knockout, targeted therapy, kinase inhibitor, invasive growth
Based on their relevance as therapeutic targets, kinase genes have been among the first to be systematically analyzed in human cancers (1, 2).
Establishing the contribution of the enzymatic activity of individual kinases to the oncogenic properties of cancer cells is crucial for the identification of therapeutic targets. Moreover, ensuring that a drug of interest exerts its effect by inhibiting its putative kinase target is difficult because of the homology among the active sites of kinases, and because diverse kinases are essential for cell growth.
In this study, we addressed these issues by developing a genetic strategy to permanently abrogate a specific protein function such as the catalytic activity of a kinase by stable modification of the corresponding genomic locus. To this end, we used somatic cell genetics to delete the exon coding for the ATP-binding site of a kinase gene in cancer cells. This strategy generates cells in which a given protein kinase is expressed but is catalytically inactive (Fig. 1A). This approach therefore resembles the chronic pharmacological treatment of cancer cells with a specific and selective kinase inhibitor.
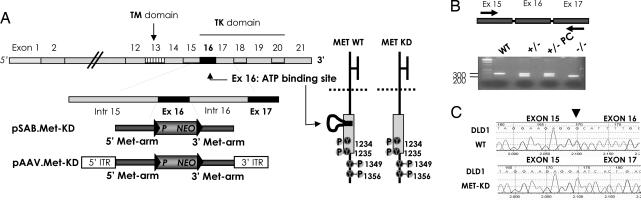
Fig. 1.
Targeted deletion of the ATP-binding site of the MET receptor in cancer cells. (A) The schematic structure of the Met genomic locus, the targeting vector, and the effects of the deletion on the MET protein are represented. (+/−), heterozygous; (−/−), homozygous. PC, post-CRE recombinase treatment. (B) Electrophoresis analysis of PCR products generated by using the cDNA obtained from WT and targeted (−/−) DLD1 cells using primers flanking exon 16 of the MET gene. The PCR strategy and the position of the primers are represented. (C) Sequence analysis of the MET transcript in WT and MET-KD cells. Upper and Lower represent the sequence of the MET cDNA obtained from WT and targeted cells, respectively. The arrow indicates the location of the boundary between exons.
As a model to test this approach, we chose the MET receptor, which is genetically altered in multiple cancers (3–7). Met is activated by its cognate ligand hepatocyte growth factor (HGF), thus triggering “invasive-growth,” a complex biological program resulting in proliferation, motility, and invasion (8). Deregulation of these activities leads to uncontrolled cell proliferation and metastasis. Targeting the MET receptor could therefore be a promising therapeutic approach (9). Accordingly, a number of pharmaceutical enterprises are developing Met inhibitors (10–12).
Results
To identify cellular models in which HGF-mediated Met activation could be assessed both at the biochemical and the biological level, we screened a panel of 15 human cancer cell lines. [supporting information (SI) Figs. 5 and 6]. From this panel, we selected four cell lines, including colorectal (HCT116 and DLD1), bladder (RT112), and endometrial (HEC1A) cancer cells in which the HGF-mediated Met activation could be readily and unambiguously measured. The catalytic activity of protein kinases relies on their ability to bind ATP and transfer a phosphate group to their targets. This process involves a highly conserved protein moiety known as the catalytic loop, which contains the ATP-binding site.
We devised a genetic strategy to generate cancer cells expressing a kinase-inactive Met receptor, hereafter referred to as MET-KD (MET kinase dead). This strategy implies the use of homologous recombination to generate an in-frame deletion of exon 16, which encodes the ATP-binding site (Fig. 1 and SI Fig. 7). A two-step genetic strategy was used to obtain first heterozygous and then homozygous clones, in which both alleles of the MET locus were correctly targeted (SI Fig. 7). Analysis of the RNA extracted from the targeted cells confirmed expression of a MET transcript with an in-frame deletion of exon 16 (Fig. 1 B and C). Multiple homozygous clones could be readily obtained in HCT116, DLD1 and HEC1A cells (Table 1). Interestingly, although multiple MET heterozygous clones could be obtained in RT112 cells, we were unable to generate RT112 cells in which both alleles of the MET exon 16 had been targeted. Instead, we repeatedly obtained retargeting of the first allele. Statistical analysis of the targeting frequency shows these results are significant (P < 0.004, Fisher's exact test) and likely suggest that homozygous deletion of the MET exon 16 yields nonviable RT112 cells (Table 1). This suggests that the kinase activity of the Met receptor may be required for the survival of this bladder cancer cells. The expression of the mutated receptor protein (MET-KD) was evaluated in the targeted cell lines. Fig. 2 shows that the deleted protein has a slightly lower molecular weight that is consistent with the deletion of the amino acid residues corresponding to the ATP-binding site. The Met protein is a glycosylated heterodimeric membrane receptor composed of an α and a β chain linked by disulfide bonds, which are generated by proteolitic cleavage of a single-chain precursor. In human cells, the Met protein is typically detected by immunoblotting in two forms: p145 Met, corresponding to the mature form of the receptor, and p170 Met, which is thought to correspond to the partially glycosylated single-chain precursor. Interestingly, we found that the ratio of the p170/p145-Met protein is increased in MET-KD cells (Fig. 2). This effect was particularly evident in late passages of the targeted cells. The increase in p170kD MET-KD suggests that deletion of the ATP-binding site could, at least in part, affect the post-translational processing of the receptor. Importantly, the MET-KD protein is correctly located at the cell surface as shown by cell-surface biotinylation experiments (Fig. 2C). Next we assessed whether MET-KD was capable of transducing the intracellular signaling cascade triggered by HGF, the Met-specific ligand. Biochemical experiments showed that deletion of the ATP-binding site results in a kinase-inactive Met receptor, which is incapable of autophosphorylation both in basal conditions and upon HGF stimulation (Fig. 2 A and B).
Table 1.
Allele-specific frequencies at which targeted clones were obtained in cancer cells
| Cell line | Allele A, % | Allele B, % |
|---|---|---|
| DLD1 | 4.5 (9/200) | 2 (8/400) |
| HCT116 | 3.25 (13/400) | 1 (3/300) |
| HEC1A | 1 (5/500) | 0.3 (1/300) |
| MDA-MB435S | 1 (6/600) | Not done |
| RT112 | 1 (4/400) | 0 (0/1,200, P < 0.004) |
The frequency at which heterozygous and homozygous clones were obtained and the name of the cell lines are indicated. In parentheses are indicated the numbers of positive clones with respect to those analyzed. Statistical significance of the probability of targeting the second allele vs. the retargeting of the first allele was determined by using Fisher's exact test.
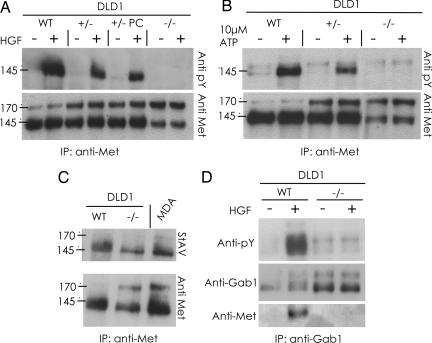
Fig. 2.
MET-KD is expressed on the surface of cancer cells but is catalytically and signaling inactive. (A) The Met protein was immunoprecipitated from lysates of the indicated cells, untreated or stimulated with HGF, and detected by immonoblotting with anti-MET and antiphosphotyrosine antibodies. (B) The Met protein was immunoprecipitated as in A from the indicated cells, and its ability to autophosphorylate on tyrosine residues was assessed. (C) The surface proteins of the indicated cells were labeled with biotin, the Met receptor was immunoprecipitated by using streptavidin (StAv) and revealed by Western blotting analysis. MDA cells were used as positive control. (D) The multidocking protein Gab1, which acts as the main MET phosphorylation substrate, was immunoprecipitated from the indicated cells in the presence and absence of HGF. The ability of the indicated Met proteins to bind and phosphorylate Gab1 was evaluated by Western blotting by using anti-MET and antiphosphotyrosine antibodies. (+/−), heterozygous; (−/−), homozygous; PC, post-CRE recombinase treatment; MDA, MDA 435S cancer cells; ATP, AdenosinTriPhosphate.
We also found that MET-KD is completely incapable of interacting with and phosphorylating the multidocking protein Gab1, which acts as the main phosphorylation substrate of this receptor (Fig. 2D). Furthermore, in contrast to its WT counterpart, MET-KD cannot activate either the MAPK or the AKT signaling pathways (SI Fig. 8). These results indicate that a catalytically active Met is critical to mediate the intracellular signaling initiated by HGF.
Next we assessed the effect of the genetic inactivation of Met kinase activity on the oncogenic properties of cancer cells in vitro. The proliferation of MET-KD cancer cells was comparable to that of their WT counterpart (Fig. 3A). In the absence of HGF, anchorage-independent growth of cell lacking a functional Met receptor was unaffected (Fig. 3B and SI Fig. 9B). These results were confirmed by using multiple independent MET- KD clones, thus excluding the potential effect of clonal variations. These data show that selective targeting of the Met catalytic activity does not have a profound effect on the proliferative potential of the targeted cancer cells in vitro.
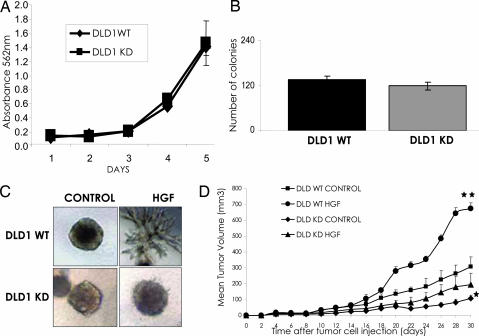
Fig. 3.
Biological and tumorigenic properties of cancer cells expressing a catalytically inactive MET receptor. (A) Proliferation of cancer cells lacking Met kinase activity. The indicated cells were seeded at the same concentration on day 0, and cell density was measured for 5 consecutive days. Each point represents the mean of triplicates; error bars, SD. (B) Anchorage-independent growth of cancer cells lacking Met kinase activity. The indicated cells were seeded in soft agar, and the colonies appearing after 15 days of incubation were counted. Each column represents the mean of triplicates; error bars, SD. (C) Measurement of the “invasive growth” potential of cancer cells lacking the Met kinase activity. The indicated cells were seeded in collagen and examined for their ability to form branched tubules in response to HGF stimulation. Representative pictures are presented. (D) Tumorigenic potential of cancer cells lacking the Met tyrosine activity. Cells were transduced with a control vector or with a lentivirus expressing HGF and were injected s.c. into the right posterior flank of 6-week-old immunodeficient nude mice. Tumor growth was measured for 30 days. Statistical significance was determined by two-tailed Student's t test. Mean ± SEM, n = 4.
The hallmark of MET biological activities is known as “invasive growth” (9, 13, 14). This process integrates multiple cellular outputs, including proliferation, motility, apoptosis, and invasion, that are deregulated in many human cancers. “Invasive growth” can be measured by growing Met-expressing cells in a 3D collagen matrix in the presence of HGF. In these conditions, activation of the HGF/Met signaling triggers the formation of typical multicellular branched structures (Fig. 3C). Although cancer cells carrying WT MET could be instructed by HGF stimulation to perform the “invasive growth program,” MET-KD cells were completely unresponsive (Fig. 3C).
Next we took advantage of the WT and MET-KD cells to assess the role of the catalytic activity of Met in mediating the antiapoptotic properties of HGF. When challenged with 5-FU treatment, the MET-KD cells were unable to elicit an antiapoptotic response upon HGF stimulation (SI Fig. 9A). Previous work suggested that Met-mediated prevention of apoptosis could be independent from its catalytic activity (15). Our data clearly indicate that the mechanisms by which Met triggers the antiapoptotic response are also strictly reliant on its kinase activity. Overall, these results indicate that the kinase activity of the Met receptor is required to mediate the biological effects of HGF. They also unequivocally prove the exclusivity of this ligand–receptor axis. This information has obvious implications for the therapeutic targeting of the HGF-Met ligand–receptor pair.
We then evaluated the contribution of the kinase activity of the Met receptor to the tumorigenic properties of human cancer cells in vivo. The oncogenic potential of WT and MET-KD cells was evaluated by s.c. injection into immunodeficient mice. The data showed that cells harboring MET-KD had a reduced tumorigenic potential (Fig. 3D). These results were reproducible in independent experiments and in another cancer cell line (HCT116), in which the kinase activity of the Met receptor had also been targeted (SI Fig. 6 and SI Fig. 10).
Growth factors play a crucial role in promoting tumor growth, by acting not only on cancer cells but also on the surrounding normal parenchyma. We reasoned that cells lacking MET kinase activity could represent an ideal model system to dissect the relative contribution of the HGF to tumor growth. An HGF autocrine loop was established by lentiviral transduction in both WT and MET-KD cancer cells, and their tumorigenic potential was assessed in mouse models. Intriguingly, HGF-expressing MET-KD cells showed a marked and reproducible increase in their tumorigenic potential with respect to the MET-KD cells not expressing HGF (Fig. 3D). This effect was independent of the activity of HGF on the tumor cells, because the latter express a functionally inactive Met. Similar results were obtained in another cancer cell line (HCT116; see SI Fig. 10). Interestingly, we found there was a direct correlation between vessel density and the size of the tumors, suggesting that HGF was promoting its tumorigenic response by increasing the angiogenic potential of tumors (SI Fig. 11). We conclude that the Met receptor plays a dual role in tumor progression: on one side, it is involved in the control of the growth of cancer cells, and on the other, Met and its ligand HGF promote tumor progression by acting on surrounding noncancerous cells, most likely by triggering angiogenesis. These results underscore the importance of assessing the effect of a drug on its target protein in the appropriate tumor environment. Furthermore, they suggest that therapies aimed at interfering with the Met–HGF axis will be more effective if they concomitantly target both the receptor and its ligand.
The most exciting new developments in cancer revolve around the use of kinase inhibitors to treat patients with genetic alterations that affect kinase activity. Because of the homology among the active sites of kinases, and because diverse kinases are essential for cell growth, it is very difficult to ensure that a drug of interest interferes with any biological property directly by inhibiting its putative target.
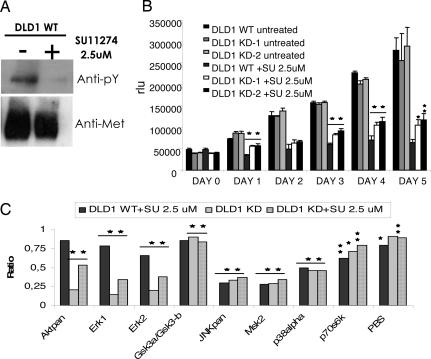
Clearly, in vitro systems for testing specificity cannot provide definitive answers to this question. Similarly, systems in which kinases are expressed exogenously and are not physiologically regulated can provide misleading answers. The strategy we have developed allows the genetic inactivation of a specific catalytic activity and should therefore overcome these limitations. To test this hypothesis, we used SU11274, a Met kinase inhibitor thought to specifically inhibit the kinase activity of Met (12). Any effect achieved by SU11274 on MET-KD cells should be associated with the inhibition of cellular activities independent from the enzymatic activity of the receptor. In accordance with previous reports when used at 2.5 μM concentration, SU11274 almost completely inhibited Met kinase activity in DLD1 colorectal cancer cells (Fig. 4A). We then measured the effect of 2.5 μM SU11274 on the proliferation of parental cells and on two independently obtained MET-KD DLD1 clones. SU11274 affected the growth of WT cells but also markedly impaired the proliferation of cells carrying a kinase-inactive Met, indicating this compound affects one or more Met-independent cellular functions (Fig. 4B). To begin the identification of the “non-Met” targets of SU11274, we measured activation of multiple signaling pathways by using a high-throughput phospho-specific antibodies platform (the Human Phospho-MAPK Array kit, R&D Systems, Minneapolis, MN). Parental and MET-KD cells were treated with SU11274, and cell signaling was assessed in the corresponding cell lysates. These experiments showed that the Met kinase activity modulates a number of key signaling molecules in cancer cells. For example, in MET-KD cells, the phosphorylation status of JNK, MSK, p38α, and p70s6K was markedly reduced both by the Met kinase inhibitor and by genetic inactivation of the Met kinase. Conversely GSK3α/GSK3β phosphorylation is relatively unaffected in both systems (Fig. 4C). Interestingly, these experiments also showed that SU11274 treatment affects a number of signaling pathways in cells carrying a kinase-inactive Met. For example, we found that SU11274 can modulate AKT and ERK phosphorylation in a Met-independent fashion (Fig. 4C). This suggests that SU11274 may inhibit or activate central signaling switches possibly by interfering with the activity of presently unknown kinases acting upstream to AKT and ERK.
Fig. 4.
Biological and biochemical effects of the MET inhibitor SU11274 in cancer cells lacking the MET kinase activity. (A) The ability of SU11274 to inhibit the kinase activity of the MET receptor was evaluated on WT DLD1 cells. Cells were treated for 16 h with 2.5-μM SU11274 and then lysed. The Met protein was immunoprecipitated and analyzed by Western blotting by using the specified antibodies. (B) Cell lines were treated as indicated, and the cell number was evaluated by using an ATP-based assay. Each column represents the mean of triplicates; error bars, SD. RLU, relative light units. (C) Modulation of intracellular signaling pathways by the MET enzymatic activity and by SU11274. The indicated cells were incubated for 16 h in the absence and presence of 2.5 μM SU11274. Phosphorylation of intracellular signaling molecules was assessed by using the Human Phospho-MAPK Array Kit. The columns represent the result of the densitometric analysis of the dot images corresponding to the phosphorylation status of individual proteins. The numbers are referred to the untreated DLD1 WT cells that were given an arbitrary value of 1. Statistical significance was determined by two-tailed Student's t test by using WT untreated cells as reference.
Discussion
The pharmacological modulation of proteins with enzymatic activity (such as kinases) is revolutionizing medicine (16). Establishing the contribution of individual enzymatic activities to the biology of human cells is essential for the identification of therapeutic targets. Ensuring that a drug of interest exerts its effect by inhibiting its putative target is also critical.
Although RNAi-mediated gene knockdown can be used to address these issues, this technology has drawbacks. These include the incomplete down-regulation of the target protein and the well known off-target effects. Furthermore, many proteins, including kinases and phosphatases, have multiple independent functions associated to specific domains. Clearly, the RNAi approach does not allow abrogation of a specific function/domain (such as the catalytic activity of a kinase) while leaving the corresponding protein (and the associated additional functions) expressed in the target cells.
In this work, we address these issues by developing an innovative genetic strategy to permanently abrogate a specific protein function by stable modification of the corresponding genomic locus. This approach generates human cells in which a given enzyme is expressed but is catalytically inactive. This strategy therefore closely resembles the chronic pharmacological treatment of the target enzymatic domain with a specific and selective inhibitor. This approach was applied here to the tyrosine kinase receptor Met but can be used to assess the specific contribution of any given kinase gene to the oncogenic properties of cancer cells. Our strategy also allows for unequivocal establishment of whether a kinase inhibitor exerts its effects through the intended target protein.
The genetic approach presented here is not limited to kinase genes but could be broadly applicable to any drug/protein combination in which the target enzymatic domain is known. We also envisage the use of cells carrying genetically inactivated kinase alleles for synthetic lethality screening. For example, compound libraries could be screened against WT and MET-KD cells to identify drugs inhibiting oncogenic signaling pathways acting synergistically or in cooperation with the Met receptor. These drugs could then be used in combination with the Met kinase inhibitors currently being developed.
Materials and Methods
Cell Culture and Reagents.
Cell lines were purchased from American Type Culture Collection (Bethesda, MD) and cultured following standard procedures. Recombinant HGF was obtained from culture supernatant of Sf9 cells infected with the baculovirus vector containing the full-size human HGF. The lentiviral vector expressing HGF and the infection and transduction procedure have been described (17, 18). Geneticin (G418) was purchased from Gibco (Carlsbad, CA). SU11274, PP2, LY294002, and PD98059 were purchased from Calbiochem (San Diego, CA).
Gene Targeting.
The experimental procedures used to delete exon 16 from the genome of cancer cells are described in SI Methods and SI Fig. 7.
RT-PCR Analysis.
RNA was extracted from the indicated cell lines by using the TRIzol Reagent from Invitrogen (Life Technology, Carlsbad, CA). Retrotranscription was carried out with Moloney murine leukemia virus reverse-transcriptase RNase H minus (Promega, Madison, WI) and oligodeoxythymidine nucleotide, following the manufacturer's instructions. PCR conditions and primers for cloning are described in SI Methods. All primers were synthesized by Invitrogen.
Protein Analysis.
Immunoprecipitation and Western blotting were performed as described (19). In particular, the Met protein was immunoprecipitated with the monoclonal antibody DQ13 (20) and detected by Western blotting with DL21 monoclonal antibody (Upstate Biotechnology, Lake Placid, NY). Tyrosine phosphorylation was detected with the monoclonal antibody clone 4G10 (Upstate Biotechnology). Other primary antibodies used for immunoblotting were: anti-AKT, antiphospho-AKT S473, anti-MAPK, and antiphospho-MAPK were all purchased from Cell Signaling (Cell Signaling Technology, Danvers, MA); the antiactin antibody was purchased from Sigma (Sigma–Aldrich, St. Louis, MO). The biotinylation assay was performed as described (21). Phosphoprotein detection was performed by using the human phospho-MAPK Array kit (R&D Systems), according to the manufacturer's instructions. Image analysis was performed with MetaMorph 6.1 software (Universal Imaging, Downingtown, PA).
In Vitro Autophosphorylation Assay.
The Met receptor was immunoprecipitated with anti-Met antibodies from cell lysates in the absence of sodium orthovanadate to allow dephosphorylation. After extensive washing, immunoprecipitates were subjected to autophosphorylation in kinase buffer (50 mM Hepes; pH 7.5 150 mM NaCl; 12.5 mM MgCl2) in the presence of 10 μM ATP. The reaction was carried out for 10 min at room temperature, and then samples were washed and analyzed by 8% SDS/PAGE.
Cell-Based Assays.
Branching morphogenesis was evaluated by culturing cells in a collagen matrix, as described in ref. 22. Anchorage-independent growth was assessed in soft agar, as described (23). Growing or serum-starved cells were stimulated with recombinant HGF (30 or 60 ng/ml) for 15 min, as indicated in Results. SU11274 was diluted and used to treat cells as described (12). For proliferation assays, cells were seeded at equal density in 96-wells Costar microtiter plate on day 0, and cell number was measured for 5 consecutive days by using the ATPlite 1 step kit (PerkinElmer, Milan, Italy). For the survival assay, the cells were seeded at equal density in 96-well Costar microtiter plate. After treatment for 48 h with 80 μg/ml 5-fluoruracil, cell viability was measured by a luminescence ATP assay (ATPlite 1 step kit, PerkinElmer). All luminescence measurements were recorded by the DTX 880-Multimode plate reader (Beckman–Coulter, Fullerton, CA).
Animal Studies.
For ex vivo tumorigenesis assay, 5 × 106 cells transduced with a control or an HGF lentiviral vector were injected s.c. into the right posterior flank of 6-week-old immunodeficient nu−/− female mice on a Swiss CD1 background (Charles River Breeding Laboratories, Calco, Lecco, Italy). Tumor appearance was evaluated every 2 days by caliper measurement. Tumor volume was calculated by using the formula V × 4/3 π × (d/2)2 × D/2, where d is the minor tumor axis, and D is the major tumor axis. A mass of 15 mm3, corresponding approximately to the initial volume occupied by injected cells, was chosen as the threshold for tumor positivity. Tumors were weighed at the end of the observation period. All animal procedures were approved by the Ethical Commission of the University of Torino, Italy, and by the Italian Ministry of Health.
Statistics.
Results are given as the mean ± SD. or SEM where indicated. Statistical analyses were performed by the two-tailed Fisher's exact test or the two-tailed Student's t test by using the Instat program (GraphPad; GraphPad Software, Inc., San Diego, CA). Differences of means were considered significant at a level of 0.05.
Supplementary Material
Acknowledgments
We are grateful to Dr. Ben Park for helpful insights and critical suggestions on the use of the adeno-associated virus approach to achieve homologous recombination in human cells. We thank Patrizia Gasparini for help with the FISH analysis, Andrea Bertotti and Paolo Michieli for suggestions on data analysis, Christopher Torrance and Federica Di Nicolantonio for critical reading of the manuscript, and Catherine Tighe for final manuscript editing. This work was supported by grants from the Italian Association for Cancer Research (AIRC); the Italian Ministry of Health, Regione Piemonte; the Italian Ministry of University and Research; European Union FP-6 Grant MCSCs 037297; and the Association for International Cancer Research (AICR-UK).
Abbreviations
- HGF
hepatocyte growth factor
- MET-KD
MET kinase dead.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703205104/DC1.
References
- 1.Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, et al. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 2.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, Holcomb T, Pujara K, Stinson J, Fu L, et al. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 4.Smolen GA, Muir B, Mohapatra G, Barmettler A, Kim WJ, Rivera MN, Haserlat SM, Okimoto RA, Kwak E, Dahiya S, et al. Cancer Res. 2006;66:3452–3455. doi: 10.1158/0008-5472.CAN-05-4181. [DOI] [PubMed] [Google Scholar]
- 5.Jagadeeswaran R, Ma PC, Seiwert TY, Jagadeeswaran S, Zumba O, Nallasura V, Ahmed S, Filiberti R, Paganuzzi M, Puntoni R, et al. Cancer Res. 2006;66:352–361. doi: 10.1158/0008-5472.CAN-04-4567. [DOI] [PubMed] [Google Scholar]
- 6.Olivero M, Valente G, Bardelli A, Longati P, Ferrero N, Cracco C, Terrone C, Rocca-Rossetti S, Comoglio PM, Di Renzo MF. Int J Cancer. 1999;82:640–643. doi: 10.1002/(sici)1097-0215(19990827)82:5<640::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Di Renzo MF, Olivero M, Martone T, Maffe A, Maggiora P, Stefani AD, Valente G, Giordano S, Cortesina G, Comoglio PM. Oncogene. 2000;19:1547–1555. doi: 10.1038/sj.onc.1203455. [DOI] [PubMed] [Google Scholar]
- 8.Trusolino L, Comoglio PM. Nat Rev Cancer. 2002;2:289–300. doi: 10.1038/nrc779. [DOI] [PubMed] [Google Scholar]
- 9.Peruzzi B, Bottaro DP. Clin Cancer Res. 2006;12:3657–3660. doi: 10.1158/1078-0432.CCR-06-0818. [DOI] [PubMed] [Google Scholar]
- 10.Ma PC, Jagadeeswaran R, Jagadeesh S, Tretiakova MS, Nallasura V, Fox EA, Hansen M, Schaefer E, Naoki K, Lader A, et al. Cancer Res. 2005;65:1479–1488. doi: 10.1158/0008-5472.CAN-04-2650. [DOI] [PubMed] [Google Scholar]
- 11.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, Chen J, Wang X, Ruslim L, Blake R, et al. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 12.Berthou S, Aebersold DM, Schmidt LS, Stroka D, Heigl C, Streit B, Stalder D, Gruber G, Liang C, Howlett AR, et al. Oncogene. 2004;23:5387–5393. doi: 10.1038/sj.onc.1207691. [DOI] [PubMed] [Google Scholar]
- 13.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Nat Rev Cancer. 2003;4:915–925. [Google Scholar]
- 14.Boccaccio C, Comoglio PM. Nat Rev Cancer. 2006;6:637–645. doi: 10.1038/nrc1912. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, DeFrances MC, Dai Y, Pediaditakis P, Johnson C, Bell A, Michalopoulos GK, Zarnegar R. Mol Cell. 2002;9:411–421. doi: 10.1016/s1097-2765(02)00439-2. [DOI] [PubMed] [Google Scholar]
- 16.Imai K, Takaoka A. Nat Rev Cancer. 2006;6:714–727. doi: 10.1038/nrc1913. [DOI] [PubMed] [Google Scholar]
- 17.Mazzone M, Basilico C, Cavassa S, Pennacchietti S, Risio M, Naldini L, Comoglio PM, Michieli P. J Clin Invest. 2004;114:1418–1432. doi: 10.1172/JCI22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vigna E, Naldini L. J Gene Med. 2000;2:308–316. doi: 10.1002/1521-2254(200009/10)2:5<308::AID-JGM131>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Bardelli A, Basile ML, Audero E, Giordano S, Wennstrom S, Menard S, Comoglio PM, Ponzetto C. Oncogene. 1999;18:1139–1146. doi: 10.1038/sj.onc.1202607. [DOI] [PubMed] [Google Scholar]
- 20.Ruco LP, Ranalli T, Marzullo A, Bianco P, Prat M, Comoglio PM, Baroni CD. J Pathol. 1996;180:266–270. doi: 10.1002/(SICI)1096-9896(199611)180:3<266::AID-PATH658>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 21.Crepaldi T, Pollack AL, Prat M, Zborek A, Mostov K, Comoglio PM. J Cell Biol. 1994;125:313–320. doi: 10.1083/jcb.125.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medico E, Mongiovi AM, Huff J, Jelinek MA, Follenzi A, Gaudino G, Parsons JT, Comoglio PM. Mol Biol Cell. 1996;7:495–504. doi: 10.1091/mbc.7.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giordano S, Corso S, Conrotto P, Artigiani S, Gilestro G, Barberis D, Tamagnone L, Comoglio PM. Nat Cell Biol. 2002;4:720–724. doi: 10.1038/ncb843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.