Abstract
Glyoxysomes are a subclass of peroxisomes involved in lipid mobilization. Two distinct peroxisomal targeting signals (PTSs), the C-terminal PTS1 and the N-terminal PTS2, are defined. Processing of the PTS2 on protein import is conserved in higher eukaryotes. The cleavage site typically contains a Cys at P1 or P2. We purified the glyoxysomal processing protease (GPP) from the fat-storing cotyledons of watermelon (Citrullus vulgaris) by column chromatography, preparative native isoelectric focusing, and 2D PAGE. The GPP appears in two forms, a 72-kDa monomer and a 144-kDa dimer, which are in equilibrium with one another. The equilibrium is shifted on Ca2+ removal toward the monomer and on Ca2+ addition toward the dimer. The monomer is a general degrading protease and is activated by denatured proteins. The dimer constitutes the processing protease because the substrate specificity proven for the monomer (Φ-Arg/Lys↓) is different from the processing substrate specificity (Cys-Xxx↓/Xxx-Cys↓) found with the mixture of monomer and dimer. The Arabidopsis genome analysis disclosed three proteases predicted to be in peroxisomes, a Deg-protease, a pitrilysin-like metallopeptidase, and a Lon-protease. Specific antibodies against the peroxisomal Deg-protease from Arabidopsis (Deg15) identify the watermelon GPP as a Deg15. A knockout mutation in the DEG15 gene of Arabidopsis (At1g28320) prevents processing of the glyoxysomal malate dehydrogenase precursor to the mature form. Thus, the GPP/Deg15 belongs to a group of trypsin-like serine proteases with Escherichia coli DegP as a prototype. Nevertheless, the GPP/Deg15 possesses specific characteristics and is therefore a new subgroup within the Deg proteases.
Keywords: Arabidopsis thaliana, Ca2+ signal, Citrullus vulgaris, monomer/dimer equilibrium
Numerous matrix enzymes have to be imported from the cytosol into peroxisomes, in plants especially into glyoxysomes for seed storage oil mobilization or into leaf peroxisomes for photorespiration. The majority of these enzymes are imported in their mature form and targeted by a C-terminal SKL designated peroxisomal targeting signal 1 (PTS1) (1). In a few matrix enzymes, the peroxisomal targeting signal 2 (PTS2) with the consensus RL-X5-HL is located in the N-terminal 30 to 50 amino acids of the protein (2). In plants, these are four enzymes of the glyoxylate cycle and β-oxidation of fatty acids: glyoxysomal malate dehydrogenase (gMDH), glyoxysomal citrate synthase (gCS), acyl-CoA oxidase, and thiolase. In mammals, three enzymes with a PTS2 have been identified: thiolase, alkyl-DHAP synthase, and phytanoyl-CoA hydroxylase. In higher eukaryotes, such as plants and mammals, the PTS2 is removed on import; a Cys is consistently found near the cleavage site [supporting information (SI) Table 2]. In lower eukaryotes, such as yeasts, a PTS2 is present in the N terminus of the mature subunit of thiolase and amine oxidase (2) (SI Table 2). The Cys in position P2 is required for processing the presequences of gCS and gMDH in pumpkin; deletion of the Cys prevents cleavage, and its exchange to Gly or Phe diminishes the processing efficiency to 20–30% of wild type (3, 4). Import or enzyme activity is not impeded by the absence of presequence removal.
In this paper, we present a procedure for purification and characterization of the glyoxysomal processing protease (GPP) from cotyledons of 3-day-old dark-grown watermelon seedlings (Citrullus vulgaris). The protein was identified with a fluorogenic substrate covering the cleavage sites of gMDH, gCS, and thiolase, and with an antibody raised against the Arabidopsis peroxisomal AtDeg15 protease. A T-DNA insertion mutant in the AtDEG15 gene of Arabidopsis (At1g28320) prevents processing of the higher molecular mass precursor of gMDH. The GPP from C. vulgaris appears in two forms, as a 72-kDa monomer and a 144-kDa dimer under reducing and denaturing conditions. They migrate with 55 kDa and 110 kDa under conditions preserving a native and folded protein conformation. They are in equilibrium with one another and could be detected with gelatine-activity gels. The equilibrium is shifted on Ca2+ removal toward the monomer and on Ca2+ addition toward the dimer. The dimeric GPP/Deg15 protease cleaves specifically substrates with Cys in the P1 or P2 position and constitutes the peroxisomal processing peptidase. The monomer cleaves denatured peroxisomal matrix proteins into small peptides; it also exhibits an unusual temperature optimum for eukaryotic proteases at 45°C and a pH optimum in the basic range.
Results
Partial Purification of the GPP.
Purified glyoxysomes with an isocitrate lyase activity of 0.14 units/ml were sonicated and filter-sterilized, and the membranes were removed by centrifugation. The resulting glyoxysomal extract was loaded onto a DEAE-anion exchange column. Protease activity of the starting material and eluate were analyzed with the fluorogenic peptide N-benzoyl-carbonyl-Phe-Arg-7-amino-4-methylcoumarin (Cbz-FR-AMC), yielding an eluate fraction containing 15 mg of protein, a recovery of 97% of the activity applied, and a purification factor of 12.3. A further 301-fold purification was achieved by Superdex75 gel filtration, hydroxyapatite chromatography, and MonoQ-anion exchange chromatography, but the losses resulted in a yield of no more than 5% (SI Table 3).
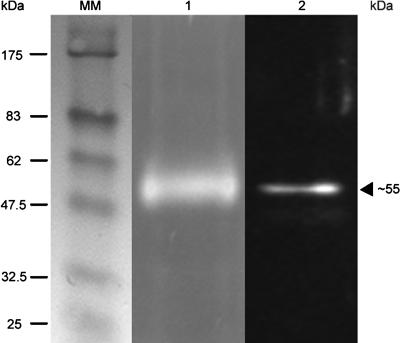
A synthetic peptide of 15 amino acids (ETTAAVHPGGSGGAV) close to the catalytic Ser (SI Fig. 7) of the AtDeg15 protease was used to prepare polyclonal antibodies. These antibodies labeled a glyoxysomal protease band of 55 kDa visualized through activity gel assay with Cbz-FR-AMC in a subsequent Western blot (Fig. 1, lanes 1 and 2). The 55-kDa band was visible in the protease fraction purified by DEAE anion exchange and benzamidine affinity chromatography and separated on a native 8% polyacrylamide gel [360,000 relative fluorescence units (RFUs)]. The same 55-kDa activity band was obtained by assaying the hydroxyapatite pool (data not shown). A Western blot of the same gel revealed the GPP to be immunologically identical to the AtDeg15 protease.
Fig. 1.
The watermelon GPP is immunologically identical to the AtDeg15 protease. The glyoxysome extract was purified by DEAE-anion exchange and p-aminobenzamidine affinity chromatography. The partially purified GPP was analyzed by native PAGE and incubation with the fluorogenic peptide Cbz-FR-AMC (lane 1), followed by Western blot analysis of the same gel and decoration with the α-Deg15 peptide antibody (lane 2).
Purification of GPP by DEAE-Anion Exchange Chromatography, Preparative Native Sephadex-Isoelectric Focusing (IEF), and Narrow pH Range 2D PAGE.
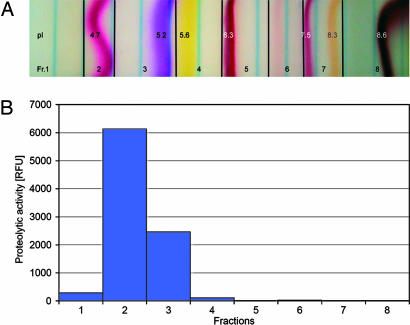
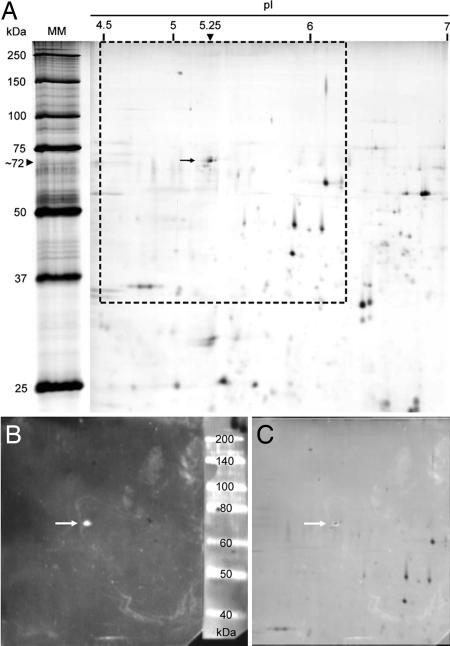
The partially purified GPP from the DEAE column was mixed with the Sephadex matrix, the immobilines pH range 3–10, and the colored pI markers and pipetted into the flat-bed. Separation in the electric field yielded a single-activity peak with Cbz-FR-AMC as a substrate with a pI of 4.7–5.2 (Fig. 2). For additional proof of identity of GPP with AtDeg15 and more precise determination of the molecular mass of the protein under reducing and denaturing conditions, 2D PAGE was performed (Fig. 3). The protease-containing fraction from the native Sephadex-IEF was resolved by IEF in an immobilized pH gradient (IPG) of 4–7 (IPG-IEF). For separation of the proteins according to molecular mass, duplicate IPG strips were mounted onto 13% SDS polyacrylamide gels and subjected to electrophoresis in the second dimension. One gel was stained with silver nitrate (Fig. 3A), and the other was decorated with α-AtDeg15 antibodies (Fig. 3B). The single spot identified by the antibody corresponded to a single spot in the 2D gel as shown by superimposition (Fig. 3C). The 2D PAGE revealed a pI of 5.25 for the GPP/AtDeg15; this pI is in accordance with the pI of 4.7–5.2 as estimated by the preparative native IEF, assuming that complete unfolding of the protease in the 2D PAGE might lead to a slight pI shift. Watermelon GPP had an apparent molecular mass of 72 kDa under reducing and denaturing conditions, whereas the GPP migrated at 55 kDa under conditions preserving a native and folded protein conformation (Fig. 1). This molecular mass of the watermelon GPP is in agreement with the glyoxysomal Deg15 protease in Arabidopsis (At1g28320), which has a calculated molecular mass of 76 kDa according to its amino acid sequence (5).
Fig. 2.
Localization of the watermelon GPP activity in the preparative native Sephadex-IEF. (A) Separation of partially purified GPP in the electric field with colored pI markers and immobilines from pH 3–10. (B) A single peak for protease activity with the fluorogenic substrate Cbz-FR-AMC was obtained at pI 4.7–5.2.
Fig. 3.
Purification and identification of the watermelon GPP. (A) The glyoxysome extract purified by DEAE chromatography and native Sephadex-IEF was resolved by 2D PAGE with an immobilized pH gradient and SDS/PAGE and stained with silver nitrate. (B) A parallel gel was blotted (framed area in A) and decorated with α-Deg15 antibodies. (C) Superimposition of A (framed area) and B identify a single protein spot with a pI of 5.25 and an Mr of 72 kDa.
Characteristics of the GPP/Deg15 Enzyme.
A test of the proteolytic activity in the pH range from 4.5 to 9.0 revealed a linear increase with low activity at pH 4.5 to high activity at pH 9.0; in phosphate buffer, the enzyme activity increased sigmoidally from low to high pH, reaching its maximum at pH 8–9 (SI Fig. 8 A and B). The temperature optimum of the enzyme for Cbz-FR-AMC was 45°C. Temperature dependence increased from 2,500 RFUs at 25°C to 6,500 RFUs at 45°C (SI Fig. 9). Several protease inhibitors were tested on the DEAE and Sephadex-IEF eluates (SI Fig. 10). GPP was inhibited by antipain, leupeptin, and N-p-tosyl-l-Lys chloromethylketone, which inhibit Cys proteases such as papain and trypsin-like Ser proteases. The lack of inhibition by the chymotrypsin inhibitors chymostatin and pepstatin A, as well as the serine proteases inhibitor PMSF, was noteworthy. An important characteristic was an increased proteolytic activity on incubation with EDTA and cystatin.
GPP Exists as a Monomer and a Dimer with Different Substrate Specificities.
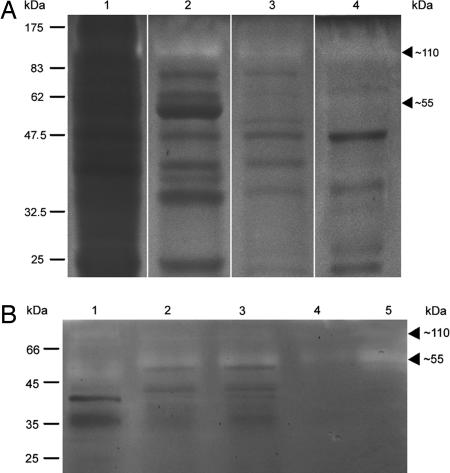
Separation of activity from the DEAE column, Sephadex75 gel filtration, hydroxyapatite, and MonoQ column by 8% native PAGE with 0.05% gelatine and incubation of the gel at 30°C identified protease activity at ≈110 kDa, with little or no activity at 55 kDa (Fig. 4A, lanes 1–4). Thus, a 110-kDa protease activity coeluted over four different chromatography columns with the 55-kDa protease that cleaves fluorogenic Cbz-FR-AMC, which was used for monitoring GPP activity during purification. This outcome raised the possibility that the 55-kDa and 110-kDa protease activity could result from a monomer and a dimer of the same protein. High concentrations of GPP activity separated at room temperature on a native polyacrylamide gel and elicited an extremely fluorescent signal at the 55-kDa position when incubated with Cbz-FR-AMC, whereas no cleavage of the fluorogenic peptide occurred by the 110-kDa form (data not shown). On incubation of the gelatine activity gel at 45°C, the glyoxysome extract, the DEAE eluate pool, and the native Sephadex-IEF eluate all exhibited protease activity at the 55-kDa level and only a weak activity at 110 kDa (Fig. 4B, lanes 1, 2, and 5). With Cbz-FR-AMC as a substrate, the optimal temperature for GPP in vitro was 45°C (SI Fig. 9).
Fig. 4.
The GPP/Deg15 exists as an equilibrium of monomers and dimers. The GPP was partially purified and proteolytic activity was localized by native PAGE in a gel with 0.05% gelatine after incubation at 30°C or 45°C. (A) At 30°C, analysis of the activity pools from DEAE (lane 1), Sephadex75 (lane 2), hydroxyapatite (lane 3), and MonoQ chromatography (lane 4) revealed a strong proteolytic activity at 110 kDa (lanes 1–4) and a barely visible activity at 55 kDa (lane 4). (B) At 45°C, analysis of the glyoxysome extract (lane 1), DEAE pool (lane 2), and Sephadex-IEF eluate (lane 5) revealed a strong activity at 55 kDa and a weaker activity at 110 kDa. Fractionation of the DEAE pool by ultrafiltration into proteins <100 kDa (lane 3) and >100 kDa (lane 4) likewise indicated activity at both positions. This result indicates interconversion among monomers and dimers.
The GPP dimer and monomer seem to be in equilibrium because the 110-kDa form can be generated from the 55-kDa form. During Superdex75 gel filtration, the dimeric 110-kDa form was in the void volume and did not react with Cbz-FR-AMC. This result can explain the loss of 40% of the loaded activity during Superdex75 gel filtration (SI Table 3). However, the proteolytically active fraction eluting at 55 kDa from the Superdex75 column, detectable with Cbz-FR-AMC, contained the gelatin-hydrolyzing 110-kDa form (Fig. 4A, lane 2). Furthermore, when the DEAE eluate was fractionated by ultrafiltration into <100 kDa and >100 kDa, both fractions contained monomeric as well as dimeric proteolytic activity, although the 55-kDa monomer should be absent from the >100-kDa fraction (Fig. 4B, lanes 3 and 4).
Specificity of the Sites Cleaved by Monomeric and Dimeric GPP.
Four fluorogenic peptides covering the cleavage sites of gCS, gMDH, thiolase, and a negative control peptide, in which the Cys in P1 position of thiolase was substituted by Gly, were synthesized (SI Table 4). In all, 10 nmol of peptide was incubated for 3 h at 24°C with GPP (67,000 RFUs) that had been purified by DEAE chromatography, native Sephadex-IEF, and removal of peptides by 5-kDa ultrafiltration. The cleavage products were sequenced by N-terminal sequence analysis and mass spectrometry (Table 1). The gCS peptide was cleaved in the vicinity of Cys, but not at the authentic site with Cys at P2. The gMDH peptide was cleaved at four positions, including next to the predicted Arg at P1 with Cys at P2. The thiolase peptide was cleaved at two positions, one being the predicted one adjacent to Cys. The negative control peptide with the substituted Cys was resistant to cleavage. The furin substrate was cleaved after Arg (P1) and Lys (P2). The GPP protein was present as a mixture of a monomer and a dimer. The monomer, but not the dimer, cleaves the fluorogenic Cbz-FR-AMC peptide. We interpret the nonauthentic cleavages to be due to the activity of monomers and the authentic cleavages to be performed by dimers.
Table 1.
Cleavage by GPP of fluorogenic peptides containing the cleavage sites of glyoxysomal higher molecular weight precursor proteins
| Fluorogenic peptide | Sequence | Cleavage products |
|---|---|---|
| gCS | Abz-EAHCV†SAQ‡TMY*D | Abz-EAH … + TMY*D |
| gMDH | Abz-R‡ANCR†‡AK‡G‡GAY*D | Abz-RANCR + ANCR + AKG + GGAY*D + GAY*D |
| Thiolase | Abz-SASVC†‡AA‡GDSY*D | Abz-SASV … + AAGDS … + GDSY*D |
| Control | Abz-SASVG†AAGDSY*D | None |
| Furin substrate | Abz-RVKR‡GLAY*D | Abz-RVKR + GLAY*D |
Y*, (NO2)Y, m-nitro-tyrosine as quencher (acceptor); Abz, anthralinic acid as fluorogene (donor; excitation 320, emission 420 nm).
†Predicted cleavage site.
‡Cleavage sites as identified by N-terminal sequence analysis and mass spectroscopy of the cleavage products.
GPP Degrades Glyoxysomal Matrix Proteins at 45°C.
The partially purified protein pool eluted from the DEAE column contains many glyoxysomal matrix proteins in addition to GPP. A 24-h digestion of the pool at 30°C displays many protein bands on SDS/PAGE. However, an incubation for 24 h at 45°C reduced the number of protein bands dramatically (SI Fig. 11 A and B). We interpret this result to show that the monomer degrades proteins by cleaving Φ-R/K↓ with Φ identifying hydrophobic and apolar residues. The limited endoproteolysis by GPP of higher molecular weight precursor proteins is carried out by the dimer at lower temperatures, implying two different proteolytic specificities for the two forms of the enzyme.
Ca2+ Ions Promote Dimerization of GPP.
The DEAE eluate was concentrated by ultrafiltration. After incubating with 6.25 mM EDTA for 10 min, the proteolytic activity was increased by 35%. A similar result was obtained in the course of inhibitor studies with 10 mM EDTA final concentration, which increased the proteolytic activity 2.5-fold (SI Fig. 10). No significant change was observed by adding 25 mM Ca2+ ions to the DEAE eluate. A 10-min incubation with EDTA, followed by the addition of 25 mM Ca2+ for 10 min, diminished the proteolytic activity to 20%, as measured with the substrate for the monomer Cbz-FR-AMC. Apparently dimer formation requires Ca2+ ions. A pretreatment with EDTA might remove other divalent cations from the binding site.
A direct comparison of proteolytic activity of the DEAE eluate toward the Cbz-FR-AMC substrate in 20 mM Tris·HCl buffer (pH 8.5) with that in 50 mM Na-phosphate buffer (pH 8.5) revealed a 2-fold higher activity in phosphate than in Tris buffer (SI Fig. 12). The effect of phosphate buffer is probably because of the precipitation of Ca3(PO4)2, thus pushing the equilibrium toward the monomer. Formation of the dimer on addition of Ca2+ ions might explain the 57% loss of activity against the Cbz-FR-AMC monomer substrate during hydroxyapatite chromatography (SI Table 3) because this ceramic material contains Ca2+, phosphate, and hydroxide ions.
A Knockout Mutation in the DEG15 Gene of Arabidopsis Prevents Processing of pre-gMDH.
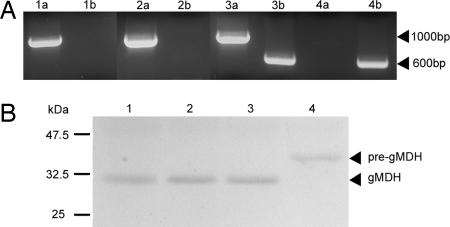
SALK-line 007184 contains a T-DNA insertion in intron 4 of the DEG15 gene (At1g28320; 13 exons). Genomic DNA from 10-day-old segregating plants was isolated. Two primers specific for the wild-type gene amplified a 1,000-bp product, whereas the gene-specific primer with the T-DNA primer gave a product of 600 bp. Genomic DNA of the heterozygous plants gave both amplification products, whereas genomic DNA of the homozygous plant yielded only the 600-bp fragment (Fig. 5A). These mutants had no obvious altered plant phenotype. A protein extract was isolated from homozygous mutant plants and separated by SDS/12.5% PAGE. Immunoblot with α-gMDH antibodies showed that processing of pre-gMDH to gMDH no longer occurred in the homozygous mutant plant, whereas the heterozygous and wild-type plants processed the pre-gMDH to the mature enzyme (Fig. 5B).
Fig. 5.
Analysis of knockout T-DNA insertion mutation in the DEG15 gene (At1g28320) of Arabidopsis. (A) PCR amplificates from wild-type and mutant plant DNA. Primers specific for the wild-type gene amplify a 1,000-bp fragment, whereas the gene-specific primer with the T-DNA gives a 600-bp product. PCR products are as follows: lanes 1 a and b, wild-type Columbia; lanes 2 a and b, wild type from segregating population; lanes 3 a and b, heterozygous mutant plant; lanes 4 a and b, homozygous mutant plant. (B) Western blots of protein extracts from wild-type and mutant plants decorated with antibodies specific for glyoxysomal malate dehydrogenase. Lane 1, wild-type control; lane 2, wild type; lane 3, heterozygous plant of segregating population; lane 4, homozygous mutant fails to process pre-gMDH to its mature form.
In addition to Deg15, analysis of the Arabidopsis genome disclosed two peroxisomal proteases with a PTS1 that may be possible candidates for a peroxisomal processing protease (5): a Zn-metallo peptidase with the pitrylisin family typical inversed HXXEH-Zn binding motif, also called insulinase (At2g4170); and a Lon-protease, a serine protease with the catalytic diad Ser-Lys (At5g47040). Knockout mutants with T-DNA insertions have been identified for both candidates: for At2g4170 (26 exons), the SALK-line 023917 with the T-DNA insertion in the 21 exon; and for At5g47040 (17 exons), the SALK-line 043857 with the T-DNA insertion in exon 17. Homozygous mutants were characterized in a segregating population. Protein extracts from both homozygous mutants processed the pre-gMDH to its mature form. The proteins were separated by SDS/12.5% PAGE, and decoration with α-gMDH antibodies consistently yielded the processed mature gMDH (data not shown). Thus, both the Zn-metallopeptidase and the Lon-protease are not involved in the processing of glyoxysomal higher molecular weight precursor.
Discussion
In contrast to the majority of peroxisomal matrix enzymes targeted with the PTS1, a smaller number employ the PTS2 (2). Removal of the PTS2-containing presequence in higher eukaryotes by the peroxisomal processing protease takes place at a Cys at P1 or P2. In the latter case, P1 is occupied by Arg, Val, or Thr (SI Table 2). Processing of the PTS2 is not necessary for enzyme activity in higher eukaryotes (6, 7); it does not take place in lower eukaryotes such as yeasts (8, 9), and pre-gMDH from higher plants is enzymatically active in the heterologous host H. polymorpha (10). An Arabidopsis peroxisomal protease (GPP) of the Deg/HtrA family of ATP-independent serine endopeptidases was purified from watermelon. GPP occurs in monomeric and dimeric forms; dimerization requires Ca2+ ions. Evidence is presented that the dimer carries out the specific cleavages needed to remove the presequence, whereas the monomer attacks sites that can degrade the removed presequences and matrix enzymes that are misfolded or no longer needed during development. The protein extracted from a T-DNA insertion mutant in Arabidopsis deg15 is not able to process the presequence. Of the three known peroxisomal Arabidopsis proteases (5), only the AtDeg15 protease can process presequences; the Zn-metallopeptidase (11) and the Lon-protease†† are unable to remove the presequence.
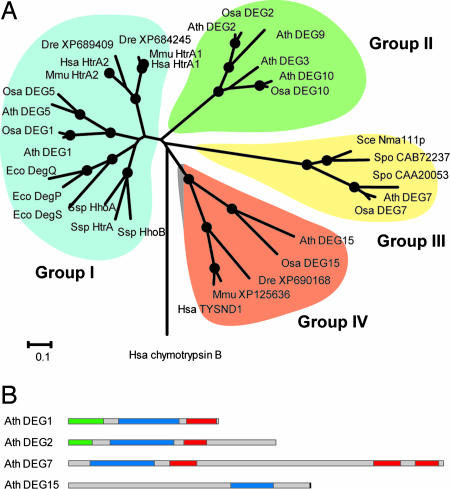
Deg15 belongs to the Deg/HtrA proteases, which form trimeric and hexameric complexes (12). In addition to the trypsin-like domain, most of them contain one to three PDZ domains located toward the C terminus of the polypeptide chain. Crystallographic analyses of the trimers and hexamers suggest that the PDZ domains play a role in substrate recognition and activation of the protease domain (13). Sixteen genes coding for putative Deg/HtrA proteases have been identified in the genome of Arabidopsis, of which six are predicted to target to mitochondria and seven to chloroplasts (14–16). DEG15 (At1g28320) is the only gene coding for a peroxisomal protein. Paralogues of DEG15 exist in various eukaryotic organisms (SI Table 5). These proteases contain the C-terminal PTS1 (17) and lack a PDZ domain (Fig. 6). The human and mouse homologues are annotated as trypsin domain-containing domain 1 (TYSND1) and are classified as members of the S1A subfamily of trypsin-like serine proteases (18). Similarity analysis of the protease domains of known and putative Deg/HtrA proteases shows that this family is divided into four distinct groups (Fig. 6A). This distribution of the Deg/HtrA proteases into four groups is reflected by their respective protein domain structures, rather than their distribution across different species (Fig. 6B). Group I includes the canonical Deg/HtrA proteases from bacteria and eukaryotes, which carry one (e.g., HsaHtrA2) or two (e.g., EcoDegP/HtrA) PDZ domains at the C terminus of the protease domain (12, 19). The only exception is the Deg5 protease from plants, which does not contain a PDZ domain. Analysis of the thylakoid lumen proteome of Arabidopsis found the AtDeg5 to be a truncated version of a Deg protease (20–22), which suggests that Deg/HtrA proteases can function without containing a PDZ domain. This model is supported by the fact that bacterial DegS protease is fully functional without its PDZ domain (23). Outer membrane porin peptide signals initiate the envelope stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Group II includes mostly plant Deg proteases such as AtDeg2 (16, 24), which contain a putative PDZ domain and an elongated C terminus lacking clear similarity to any known protein. All Deg proteases with a chloroplast or mitochondrial targeting sequence are located in group I or II. Group III includes all Deg/HtrA proteases found in fungi, such as the nuclear-localized Nma111p protease of S. cerevisiae (25), and some plant proteases. The putative proteases of this group are about twice as long as Deg proteases of groups I and II and possess three PDZ domains, one adjacent to the C terminus of the protease domain and two at the C terminus of the protein. Because the PTS2 in yeast enzymes investigated is not cleaved off on import, it is interesting to note that no DEG15 paralogue has been identified in fungi. Group IV is formed by AtDEG15 and related enzymes from other organisms. The proteins of this group carry the protease domain more toward the C terminus than other Deg proteases and, as mentioned, do not contain recognizable PDZ domains. The diverging domain structure of group IV Deg proteases, their placement in the phylogenetic tree, and their capacity to remove the PTS2-containing presequence suggest that this group might be a subfamily of the Deg/HtrA proteases.
Fig. 6.
Phylogenetic analysis and domain structure of Deg proteases. (A) The neighbor-joining phylogenetic tree of the Deg protease domains. Black dots indicate a support by 80% or more of 10,000 bootstrap replicates. National Center for Biotechnology Information protein accession numbers are given for unnamed proteins. (B) Domain structures of representative Deg/HtrA proteases from the four distinct groups. Blue, trypsin-like protease domains; red, PDZ domains; green, chloroplast transit peptides; black, SKL signal. The following abbreviations are used: Ath, Arabidopsis thaliana; Bsu, Bacillus subtilis; Dme, Drosophila melanogaster; Eco, Escherichia coli; Hsa, Homo sapiens; Mmu, Mus musculus; Osa, Oryza sativa; Sce, Saccharomyces cerevisiae; Spo, Schizosaccharomyces pombe; Ssp, Synechocystis sp. PCC 6803.
The 709-amino acid-long protein sequence of AtDeg15 has a distinct primary structure. The protease domain with 240 amino acids is located at positions 375–614. The N- and C-terminal regions have no obvious homology to other proteins. The primary structure is, however, conserved in the Deg15 protein of rice (SI Fig. 13). The protease domains of AtDeg15 and OsDeg15 share 45% identical amino acids, compared with 40% identity in the complete sequence. In the two plant Deg15 proteases (SI Fig. 7), 67 amino acids are inserted after the catalytic His, which is located in the short helix B in DegP (13). Because the loop is located in the vicinity of the catalytic pocket, it may determine access to the active center. The loop may be involved in electrostatic interactions due to the presence of many polar and charged amino acids.
It is not unusual for Deg proteases to alternate in two distinct forms. E.coli DegP alternates between proteolytic and chaperone functions (12), whereas E.coli DegS changes between a catalytically active and inactive form (26). Deg15 may exist as monomers and dimers exhibiting different substrate specificities.
Recently, Tysnd1 was identified as the mammalian peroxisomal processing protease responsible for the removal of the PTS2 peptide and was classified as a cysteine endopeptidase based on inhibitor studies (27). Sequence analysis, however, shows that the highly conserved catalytic triad His-Asp-Ser in the serine protease domain is present in Tysnd1 (SI Fig. 7), whereas no conserved Cys is detectable. Mutation analysis of recombinant Arabidopsis Deg15 further confirmed that predicted catalytic His, Asp, and Ser are essential for catalysis (28). The Tysnd1 processing activity using pre-thiolase as a substrate was completely abolished with the cysteine protease inhibitor N-ethylmaleimide (27). In the light of our data, we suggest that N-ethylmaleimide inhibited the dimer formation of Tysnd1, thus abolishing processing of the specific substrate pre-thiolase. The observed partial inhibition of Tysnd1 by metal chelators removing Ca2+ ions supports this interpretation. In both cases, the presumed dimer formation of Tysnd1 would be inhibited.
Structural investigations are required to determine which alternative protein interaction domains GPP/Deg15 uses. Further studies are necessary to understand how the two functions of the enzyme (i.e., limited proteolysis and proteolysis of denatured proteins) are accomplished.
In conclusion, a peroxisomal protease that cleaves PTS2-carrying presequences has been purified to a single protein that immunologically interacted with specific antibodies recognizing the Arabidopsis peroxisomal Deg15 endoprotease. Knockout of the Deg15 protein eliminates cleavage of the presequences of the peroxisomal matrix enzymes. This Deg protein lacks PDZ domains and forms dimers in the presence of Ca2+ ions. Monomers and dimers have different substrate specificities.
Methods
Isolation of Glyoxysomes on a Sucrose (Suc) Gradient for Purification of the GPP.
Watermelon (C. vulgaris) was grown in the dark at 30°C for 3 days, and glyoxysomes were isolated. For details, see SI Methods.
GPP Activity Assay.
Preliminary experiments using processing of 35S-labeled pre-gMDH as an assay to monitor GPP activity found activity in the 120 mM NaCl eluate of a DEAE-anion exchange column. To simplify the GPP assay, fluorogenic peptides covering the cleavage sites of the higher molecular weight precursor proteins of gMDH, gCS, and thiolase were synthesized; the thiolase peptide with the Cys in P1 exchanged to Gly was used as a control (SI Table 4). The fluorogenic peptide anthranilic acid-RVKRGLA-m-nitro-tyrosine-d, a substrate for the eukaryotic Ca2+-dependent proprotein convertase, was also used for characterization. Because of the low yield and stability of the fluorescent signal of anthranilic acid upon cleavage, these peptides were not suitable for monitoring the GPP purification by screening diluted protease-containing fractions. However, the peptides were highly suitable for functional analysis of the purified and concentrated GPP. The peptide Cbz-FR-AMC, which is a substrate for many serine and cysteine proteases, was used to follow the purification. For fluorogenic peptide synthesis and assay details, see SI Methods.
Purification of GPP.
Pure glyoxysomes obtained from 90 Suc gradients with an isocitrate lyase activity of 0.14 unit/ml were diluted 1:10 with DEAE-buffer A, sonicated, centrifuged, and filtrated. The resulting glyoxysome extract was purified on a DEAE-anion exchange column followed by a Superdex75 column, a hydroxyapatite column, and a MonoQ column. In a second approach, the glyoxysome extract was purified on a DEAE column, followed by a p-aminobenzamidine affinity column. Peak activity fractions were pooled and used for activity gels and Western blot analysis. In a final approach, the purification of GPP was achieved by DEAE chromatography and sample prefractionation with preparative Sephadex isoelectric focusing, followed by narrow pH range 2D PAGE (29). The gels were either silver stained or used for Western blot analysis with α-AtDeg15 antibodies. The DEAE eluate pool was also subdivided by ultrafiltration in fractions <100 kDa and >100 kDa for analysis by gelatin activity polyacrylamide gels. For details, see SI Methods.
Biochemical Procedures.
Protease activity was analyzed by nondenaturing (native) PAGE, with SDS only in the running buffer; gelatine was added at a final concentration of 0.05% to the resolving gel. The gels were run at 4°C overnight, equilibrated with buffer, and incubated at either 30°C or 45°C overnight, followed by Coomassie blue staining. Alternatively, native PAGE without gelatine was performed, followed by equilibration with buffer, incubation with the fluorogenic substrate Cbz-FR-AMC, and recorded with a Nikon Coolpix4500 (Tokyo, Japan). Protease inhibitor studies, SDS/PAGE, and immunoblotting using the polyclonal primary rabbit α-Deg15 peptide or the α-gMDH antibody were performed. Anti-Deg15 antibodies were raised against the synthetic peptide ETTAAVHPGGSGGAV comprising 15 amino acids close to the catalytic Ser of the AtDeg15 protease. Anti-gMDH antibodies were raised against the overexpressed protein. Immunoreactive proteins were detected either with horseradish peroxidase-conjugated α-rabbit IgG, visualized by using chemiluminescent substrate, or with alkaline phosphate-conjugated α-rabbit IgG using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color reaction. For details, see SI Methods.
Bioinformatics.
Similarity searches against public protein sequence databases were performed as implemented at the web pages of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/blast) and the Institute for Genomic Research (www.tigr.org) using default parameters. For details, see SI Methods.
Supplementary Material
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft Grants Gi154/9-4, Gi154/9-5, (to C.G.), and Ad92/8–2 (to I.A.).
Abbreviations
- Cbz-FR-AMC
N-benzoyl-carbonyl-Phe-Arg-7-amino-4-methylcoumarin
- gCS
glyoxysomal citrate synthase
- gMDH
glyoxysomal malate dehydrogenase
- GPP
glyoxysomal processing protease
- IEF
isoelectric focusing
- PTS1
peroxisomal targeting signal 1
- PTS2
peroxisomal targeting signal 2
- RFU
relative fluorescence unit.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704733104/DC1.
De Walque, S., Fransen, M., Mannaerts, G. P., Van Veldhoven, P. P., Poster presentation, International Symposium “Peroxisomal Disorders and Regulation of Genes,” Sept. 23–25, 2002, Ghent, Belgium.
References
- 1.Keller GA, Krisans S, Gould SJ, Sommer JM, Wang CC, Schliebs W, Kunau W, Brody S, Subramani S. J Cell Biol. 1991;114:893–904. doi: 10.1083/jcb.114.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flynn CR, Mullen RT, Trelease RN. Plant J. 1998;16:709–720. doi: 10.1046/j.1365-313x.1998.00344.x. [DOI] [PubMed] [Google Scholar]
- 3.Kato A, Hayashi M, Kondo M, Nishimura M. Plant Cell. 1996;8:1601–1611. doi: 10.1105/tpc.8.9.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato A, Takeda-Yoshikawa Y, Hayashi M, Kondo M, Hara-Nishimura I, Nishimura M. Plant Cell Physiol. 1998;39:186–195. doi: 10.1093/oxfordjournals.pcp.a029356. [DOI] [PubMed] [Google Scholar]
- 5.Reumann S, Ma C, Lemke S, Babujee L. Plant Physiol. 2004;136:2587–2608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gietl C, Seidel C, Svendsen I. Biochim Biophys Acta. 1996;1274:48–58. doi: 10.1016/0005-2728(96)00009-6. [DOI] [PubMed] [Google Scholar]
- 7.Cox B, Chit MM, Weaver T, Gietl C, Bailey J, Bell E, Banaszak L. FEBS J. 2005;272:643–654. doi: 10.1111/j.1742-4658.2004.04475.x. [DOI] [PubMed] [Google Scholar]
- 8.Mathieu M, Zeelen J, Pauptit RA, Erdmann R, Kunau WH, Wierenga RK. Structure (London) 1994;2:797–808. doi: 10.1016/s0969-2126(94)00081-6. [DOI] [PubMed] [Google Scholar]
- 9.Faber KN, Haima P, Gietl C, Harder W, Ab G, Veenhuis M. Proc Natl Acad Sci USA. 1994;91:12985–12989. doi: 10.1073/pnas.91.26.12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietl C, Faber KN, Van der Klei IJ, Veenhuis M. Proc Natl Acad Sci USA. 1994;91:3151–3155. doi: 10.1073/pnas.91.8.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Authier F, Bergeron JJM, Ou WJ, Rachubinski RA, Posner BI, Walton PA. Proc Natl Acad Sci USA. 1995;92:3859–3863. doi: 10.1073/pnas.92.9.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clausen T, Southan C, Ehrmann M. Mol Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 13.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 14.Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K, Clarke AK. Plant Physiol. 2001;125:1912–1918. doi: 10.1104/pp.125.4.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokolenko A, Pojidaeva E, Zinchenko V, Glaser VM, Herrmann RG, Shestakov SV. Curr Genet. 2002;41:291–310. doi: 10.1007/s00294-002-0309-8. [DOI] [PubMed] [Google Scholar]
- 16.Huesgen PF, Schuhmann H, Adamska I. Physiol Plant. 2005;123:413–420. [Google Scholar]
- 17.Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. J Mol Biol. 2003;328:581–592. doi: 10.1016/s0022-2836(03)00319-x. [DOI] [PubMed] [Google Scholar]
- 18.Rawlings ND, Tolle DP, Barrett AJ. Nucleic Acid Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pallen MJ, Wren BW. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 20.Adam Z, Clarke AK. Trends Plants Sci. 2002;7:451–456. doi: 10.1016/s1360-1385(02)02326-9. [DOI] [PubMed] [Google Scholar]
- 21.Peltier JB, Emanuelson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, van Heijne G, van Wijk KJ. Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert M, Petersson UA, Haas BJ, Funk C, Schroder WP, Kieselbach T. J Bio Chem. 2002;277:8354–8365. doi: 10.1074/jbc.M108575200. [DOI] [PubMed] [Google Scholar]
- 23.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 24.Hauβühl K, Andersson B, Adamska I. EMBO J. 2001;20:713–722. doi: 10.1093/emboj/20.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fahrenkrog B, Sauder U, Aebi U. J Cell Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- 26.Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 27.Kurochkin IV, Mizuno Y, Konagaya A, Sakaki Y, Schönbach C, Okazaki Y. EMBO J. 2007;26:835–845. doi: 10.1038/sj.emboj.7601525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuhmann H, Huesgen PF, Gietl C, Adamska I. J Biol Chem. 2007 in press. [Google Scholar]
- 29.Görg A, Boguth G, Köpf A, Reil G, Parlar H, Weiss W. Proteomics. 2002;2:1652–1657. doi: 10.1002/1615-9861(200212)2:12<1652::AID-PROT1652>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.