Abstract
In Saccharomyces cerevisiae, genes placed near telomeres or the silent HML and HMR mating-type loci are transcriptionally repressed by a heterochromatin-like structure. We have generated nonreplicating DNA rings by recombination in vivo to examine the role of chromosomal context on transcriptional repression. Specifically, recombination at HMR was used to produce rings that lacked the E and I silencers. An altered level of DNA supercoiling was observed in these rings but not in comparable rings from derepressed loci. Our results indicate that a repressive chromatin structure persists in an extrachromosomal environment immediately following removal of the cis-acting control elements. Examination of both chromatin footprints and DNA sequence dependence revealed that changes in nucleosome number could account for the topology shifts. Upon continued cell growth, the differences in supercoiling were lost and transcriptional competence was restored. These results show that silencers are required for sustained persistence of repressive chromatin structure, even in the absence of DNA replication.
Keywords: Transcriptional repression and silencers, S. cerevisiae, mating-type locus, heterochromatin, DNA topology, recombination
The chromosomes of higher eukaryotes consist of both euchromatic and heterochromatic domains, and these regional variations in DNA packaging profoundly influence the transcriptional activity of genes. In Drosophila, for example, relocation of an active euchromatic gene near a heterochromatic domain results in a variegated pattern of gene expression; the gene is on in a subset of cells and off in others (1). The expression states are maintained stably over many generations because of the clonal propagation of heterochromatin. Switching between states occurs rarely.
Position effects also play an essential role in determining cell type in the yeast Saccharomyces cerevisiae. In this organism, haploid mating type is governed by the expression of either a- or α-specific genes at the MAT locus. Additional copies of the mating-type genes are stored at HML and HMR where they are transcriptionally repressed because of the combined action of cis-acting regulatory sequences, termed silencers, and trans-acting components (2). Silencers consist of different combinations of binding sites for Rap1p, Abf1p, and the origin recognition complex (ORC). Together, the DNA binding proteins recruit critical silencing factors encoded by the SIR genes, SIR1–4 (3), through a network of protein–protein contacts (4, 5).
Transcriptional silencing at HML and HMR is gene-independent and results from a heterochromatin-like structure that suppresses a number of physiological DNA transactions (2). The repressed chromatin is also refractory to probes of DNA accessibility (6), a property also observed at telomeres where the Sir2–4 proteins silence telomere-proximal genes (7). In addition, the amino-terminal tails of histones H3 and H4 are hypoacetylated at the silent mating-type loci (8). The deacetylated tails bind Sir3p and Sir4p directly in vitro (9), and crosslinking experiments have shown that Sir2p, Sir3p, and Sir4p span repressed chromosomal domains in vivo (10). Despite this extensive characterization, little is known about the detailed structure of silent chromatin and the mechanism by which transcriptional machinery is blocked.
An epigenetic pattern of gene expression has been observed in a subset of silencing pathway mutants. In a sir1 null strain, for example, repression of HML and HMR occurs in only a fraction of cells within a population and individual cells switch between states infrequently (11). Based on these and other data, Pillus and Rine (11) posited that transcriptional repression involves both establishment and maintenance processes. According to this view, Sir1p is required for efficient establishment of the silent state but not its maintenance. The establishment function of the protein is likely to be linked to its association with silencers, to which it is tethered (5), because mutations in silencers and in Rap1p illicit a similar response (12, 13). However, silencers also are involved in perpetuating the repressed state; the elimination of the elements results in reactivation of silent genes within a single generation (14).
In this report, we examine the role of chromosomal context in maintaining an altered chromatin structure at the silent HMR locus. We used site-specific recombination in vivo to produce nonreplicating DNA rings from different chromosomal positions. In SIR strains, rings from HMR initially maintained an altered DNA topology even though the excised DNA fragments lacked both E and I silencers. The altered supercoiling level was lost at later times, and transcriptional competence was eventually restored. Our results reveal new features of the chromatin structure at a repressed locus and show that silencers are required for long-term but not short-term persistence of repressive chromatin structure in DNA fragments removed from their normal chromosomal context.
MATERIALS AND METHODS
Plasmid Constructions.
Cassettes for excision of mating-type sequences were constructed by inserting fragments of the HMRa locus from pLSD12 (D. Shore, University of Geneva) between a pair of tandemly repeated 58-bp RS sites (15) in plasmid pABX22 (M.R.G., unpublished results). The fragments along with flanking RS sites were then transferred back to pLSD12. This procedure yielded hmr∷rHMR, which contains RS sites at SnaBI and PvuI sites of HMR (outside E and I), and hmr∷rA(1.8), which contains RS sites at BspMI and BclI sites (within E and I). Δhmr∷rAΔp(1.6) was constructed by similar manipulations with the exception that overlap PCR was used to generate an hmr fragment that abutted the start codons of the divergent a1 and a2 genes. 523- and 735-bp LYS2 fragments were inserted downstream of the a2 gene in hmr∷rA(1.8) and Δhmr∷rAΔp(1.6), respectively, to generate hmr∷rA1A2 and Δhmr∷rA1A2Δp. To construct Δhmr∷rKWD50N, a 2.75-kilobase (kb) rKWD50N excision cassette from plasmid pKWD50N (16) was used to replace the XbaI-BglII fragment between the E and I silencers in pLSD12. Δhmr∷r50NY/Z was constructed by replacing a 275-bp NcoI-HpaI LYS2 fragment in Δhmr∷rKWD50N with a Y/Z junction-containing fragment of equal length (starting from the BglII site of HMR). The Δhmr∷r50NY/Z construction was then transferred to pRS414.
pUC18-Δlys2 was created by deleting the BglII-XhoI LYS2 fragment from pUC18-LYS2. The 2.75-kb rKWD50N excision cassette described above was inserted into the deletion site of pUC18-Δlys2 to generate Δlys2∷rKWD50N. A NotI site within this cassette was destroyed, introducing an additional 4 bp relative to Δhmr∷rKWD50N. pΔmat∷URA3 was constructed by replacing the BglII-BstBI region of the MATa locus in plasmid pDC283 (17) with the URA3 gene.
Strain Constructions.
The yeast strains used in this study were derived from either W303–1A or a W303–1A derivative, THC1, in which the mating-type genes of HMRa were replaced by URA3 (T.-H.C., unpublished results). All of the gene disruptions or replacements were performed by the one-step method (18) and confirmed by Southern hybridization or PCR. THC3 and THC13 were derived from THC1 and W303–1A, respectively, by disruption of SIR3 with psir3∷HIS3 (16). THC9, THC23, and THC24 were derived from THC3 by replacement of the chromosomal Δhmr∷URA3 with plasmids Δhmr∷rKWD50N, hmr∷rA1A2, and Δhmr∷rA1A2Δp, respectively. THC10, THC25, THC26, and YCL1 were derived from THC9, THC23, THC24, and YCL2, respectively, by regeneration of the chromosomal SIR3 using plasmid pAR3 (S. Holmes, Wesleyan University). THC16 and THC17 were derived from THC13 and W303–1A, respectively, by disruption of LYS2 with plasmid Δlys2∷rKWD50N. THC27 and THC28 were derived from THC10 and THC9, respectively, by disruption of LYS2 with plasmid pUC18-Δlys2. THC40 and THC41 were generated from THC23 and THC25, respectively, by disruption of MATa with Δmat∷URA3. YCL2 was derived from THC3 by replacement of the chromosomal Δhmr∷URA3 with hmr∷rHMR.
Formation of Intracellular DNA Rings and Analysis of Topoisomer Distributions.
Strains were transformed with the recombinase expression vector pHM153 (15) and pregrown at 30°C to mid-log phase in 50 ml of synthetic complete media lacking leucine and containing raffinose (2%). Recombinase expression was induced by the addition of galactose (Cf = 2%) for 60 min, unless specified otherwise. DNA rings were isolated by a spheroplasting procedure (16). Then, 25% of each DNA sample was electrophoresed for 18 hr at 1.5 V/cm in 0.8% agarose gels containing 2.0 μg/ml chloroquine diphosphate (Sigma). After transfer to Zeta Probe GT membrane (Bio-Rad), blots were hybridized with a randomly primed 735-bp LYS2 probe (beginning at StuI and spanning upstream sequences), unless specified otherwise. Topoisomer bands were quantified by phosphorimaging (Bio-Rad), and the centers of the topoisomer distributions were determined by the Gaussian method (19).
Analysis of Chromatin Structure by Micrococcal Nuclease (MNase) Digestion and Indirect End Labeling.
Footprinting by MNase was performed in NP-40 (Sigma) permeabilized spheroplasts essentially as described by Kent et al. (20) with the following exceptions. Ten times as many cells were used, and all of the buffer volumes were thus raised fivefold. Spheroplasting was accomplished by treatment with 1 ml of spheroplasting solution [20% Sorbitol, 0.3–0.5 mg/ml Zymolyase T100 (Seikagaku, Japan), and 0.5 mM β-mercaptoethanol] for 5 min at 30°C. MNase digests (5 min at 30°C) were terminated by the sequential addition of 125 μl of prewarmed 250 mM EDTA/5% SDS (65°C), 125 μl of 10% Triton X-100, and 25 μl RNase A (10 mg/ml). After a 30 min incubation at 37°C, 60 μl of Proteinase K (10 mg/ml) was added and the samples were incubated at 65°C overnight, extracted with phenol/chloroform twice, and precipitated. Sample were linearized with StuI, electrophoresed in 1% agarose, and transferred to nylon membrane. Blots were hybridized with the LYS2 probe described above. For naked DNA controls, DNA was isolated from spheroplasts, as described above, before MNase digestion.
Northern Blots.
Aliquots of culture were harvested and preserved on dry ice until additional processing. Total RNA was extracted by the hot acid phenol procedure and electrophoresed in 1% agarose/formaldehyde gels (18). Nucleic acids were transferred to Zeta Probe membrane. Blots were hybridized sequentially with a randomly primed a1-specific probe (spanning the entire ORF as well as 3′-untranslated sequences) and a SNR17a(U3 snRNA)-specific probe (21).
RESULTS
Methodology.
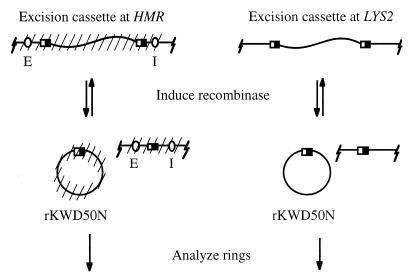
The R site-specific recombinase of yeast Zygosaccharomyces rouxii can excise and recircularize DNA fragments from appropriately designed constructs (15, 22). Inducible expression of the enzyme in living cells provides a means to separate DNA fragments from their original chromosomal positions (23). Furthermore, circularization of the excised fragments permits examination of the DNA topology of sequences that were formerly linear. Our initial experiments used an excision cassette consisting of a 2.4-kb LYS2 fragment flanked by recombinase target sites, termed RS sites (16). The cassette was integrated either at the HMR locus on chromosome III, replacing the transcriptionally repressed a1 and a2 genes, or within the nonrepressed LYS2 gene on chromosome II (Fig. 1). Excision from either location produced a nonreplicating ring of chromosomal DNA, designated rKWD50N, which lacked known promoters of transcription. Importantly, excision cassettes at HMR were positioned between the E and I silencers; recombination uncouples the regulatory elements from the ring to be studied.
Figure 1.
Methodology for production of extrachromosomal DNA rings. A fragment of internal coding sequence of the LYS2 gene was flanked by tandemly oriented RS sites (half-filled rectangles) and integrated at HMR or LYS2. Cross-hatches in the figure indicate the potential for Sir-mediated repression. Induction of the R recombinase gene creates a 2,465-bp extrachromosomal ring, rKWD50N.
Excision from HMR Yields DNA Rings with Altered DNA Topology.
In eukaryotes, DNA supercoiling is determined largely by the wrapping of the DNA double helix around histone cores. Each nucleosome constrains approximately one negative supercoil (24). Topoisomer distributions of intracellular DNA provide a sensitive probe of chromatin structure because nucleosome density and subtle structural changes in individual nucleosomes can affect DNA supercoiling (see ref. 25 for example). To test whether Sir-mediated silencing is accompanied by a change in supercoiling, we examined topoisomer distributions of ring rKWD50N from repressed and nonrepressed loci. Using a galactose-inducible recombinase expression plasmid (15), ≈80% of the excision cassette at HMR or LYS2 was circularized within 60 min in both SIR and sir3 strains (data not shown). The high rate and efficiency of excision indicates that recombination is not limited to a subpopulation of unrepresentative chromatin templates.
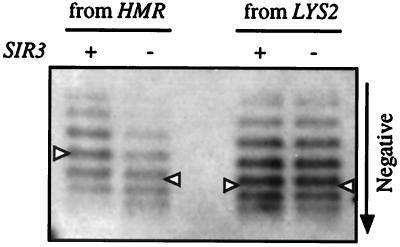
DNA topoisomers were resolved by electrophoresis in buffer containing chloroquine diphosphate such that more negatively supercoiled topoisomers migrated more rapidly. When excision occurred at the repressed HMR locus, the distribution of rKWD50N topoisomers was less negatively supercoiled, by approximately one turn, relative to the distribution in a sir3 strain (Fig. 2). Similar results were obtained in sir2 and sir4 mutants (data not shown). In contrast, when rKWD50N was excised from the LYS2 locus, the topology of the ring was not influenced by a sir3 mutation (Fig. 2). Furthermore, the superhelical density of the ring from LYS2 was comparable to the ring from the derepressed HMR locus (see Fig. 2 legend).
Figure 2.
DNA supercoiling of extrachromosomal rings. rKWD50N was produced by excision from either HMR in strains THC10 (SIR) and THC9 (sir3) or from LYS2 in strains THC17 (SIR) and THC16 (sir3). Topoisomers were resolved by electrophoresis in buffer containing chloroquine diphosphate. Autoradiograms were cropped to show DNA rings exclusively. Gaussian centers of the topoisomer distributions are marked with a triangle. The SIR3-dependent shift in linking number was 1.1 ± 0.1 for the ring from HMR but negligible for the ring from LYS2 (based on four independent measurements). The linking number difference between rings from LYS2 and from the derepressed HMR locus was also negligible. A slight increase in mobility in topoisomers from LYS2 arose from four additional bases in the excision cassette (see Materials and Methods).
The supercoiling shift was not caused by restricted access of topoisomerases to silent chromatin. We found that the supercoil densities of silent and nonsilent rings increased rapidly upon an upshift in cell culture temperature, consistent with topoisomerase action in response to thermal unwinding of DNA (ref. 26 and T.-H.C., unpublished results). The relative difference in the topology of rings in SIR and sir3 strains, however, was unaltered by the temperature jump (23°C to 38°C).
Three conclusions can be drawn from these observations. First, Sir-mediated effects on DNA topology occur in the vicinity of a silenced locus. Second, the altered DNA supercoiling of silent chromatin is not caused by a deficiency in topoisomerase action. Third, the structural differences detected by this approach persist in DNA rings despite uncoupling from the E and I silencers. Thus, identical DNA sequences are packaged differently at repressed and nonrepressed loci, and this difference persists on their removal from a chromosomal context.
Chromatin Footprints of rKWD50N.
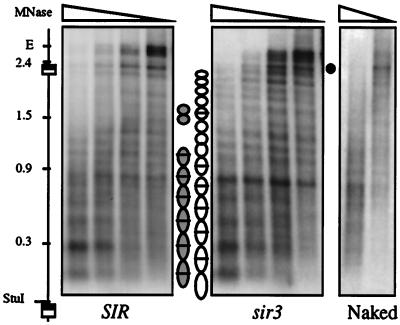
One simple explanation for the observed shift in DNA topology is that repressed chromatin contains fewer nucleosomes. To analyze the influence of SIR-mediated repression on nucleosome placement, we generated MNase footprints of the chromosomal excision cassette at HMR in spheroplasted cells. Cleavage sites were mapped by indirect end-labeling (43). Fig. 3 shows that the unexcised rKWD50N template yields similar although nonidentical digestion patterns in repressed and derepressed strains. Comparable results were obtained from digestion of the excised rings (data not shown). Regions of protection spanning ≈165 bp are indicative of positioned nucleosomes and are marked with ovals. Internucleosomal bands exist at some locations and these are marked with bars. It seems unlikely that the bands correspond to disrupted or “split” nucleosomes because they were not observed in footprints of the native HMR locus (ref. 27 and T.-H.C., unpublished results). Rather, the pattern of bands suggests that there are two possible settings for nucleosomes in the SIR strain (either shaded ovals or open ovals) whereas only one setting is used in the sir3 strain (open ovals). Gain of a supercoil upon derepression, according to one interpretation, is caused by the presence of an additional nucleosome, on average, in the derepressed setting. An alternative explanation holds that the Sir-mediated topology shift is caused by a change nucleosome conformation, not a change in nucleosome number (see Discussion). The data presently available cannot differentiate between these two possibilities.
Figure 3.
MNase footprinting of chromatin templates. Spheroplasts were made from strains THC27 (SIR) and THC28 (sir3), and DNA was digested in situ with MNase (see Materials and Methods). The indirect end-labeling probe, a LYS2 fragment, hybridizes within the rKWD50N excision cassette adjacent to the StuI site (these strains lack the LYS2 gene). The positions of HMR E, RS sites, and size markers (in kb) are denoted. (•) identifies a single hypersensitive site outside the excision cassette that appears upon derepression. Digestion of purified chromosomal DNA is also shown (marked Naked). Units of MNase/ml used in each lane: (Left and Center) 160, 80, 40, and 20; (Right) 10 and 5.
The Role of DNA Sequence in Sir-Mediated Shifts in DNA Topology.
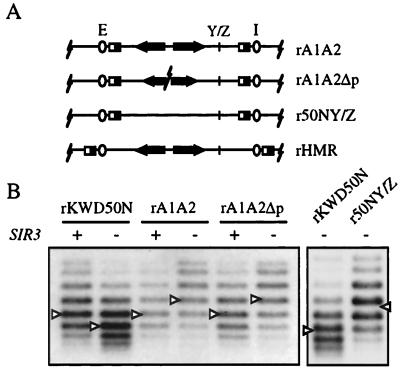
Next, we asked whether the Sir-mediated supercoiling shift at HMR was dependent on the sequence of the excised fragment. Two additional excision cassettes were constructed from fragments of the native HMR locus. The first yields ring rA1A2, containing both the a1 and a2 mating-type genes and the second yields ring rA1A2Δp, bearing the same genes with their promoters deleted (Fig. 4A). When excised from HMR, each ring possessed a different level of supercoiling in a SIR strain than in a sir3 strain (Fig. 4B). Upon repression, rA1A2 and rA1A2Δp gained a supercoil whereas rKWD50N lost a supercoil. Nevertheless, rKWD50N and the two new rings bore comparable DNA topoisomer distributions in the repressed case. These results indicated that Sir-mediated changes in the rings yielded similar repressed chromatin structures.
Figure 4.
Sequence dependence of DNA supercoiling for rings excised from HMR. (A) Excision cassettes based on HMR sequences. rA1A2 contains the a1 and a2 genes. rA1A2Δp contains the same genes but lacks the promoters. r50NY/Z is a derivative of rKWD50N that contains the Y/Z junction. rHMR contains the entire HMR locus. All rings, with the exception of rHMR, are 2,465 bp and contain a fragment of LYS2 (Materials and Methods). (B) Topoisomer analysis. rA1A2 from strains THC25 (SIR) and THC23 (sir3), rA1A2Δp from strains THC26 (SIR) and THC24 (sir3), and rKWD50N from THC10 (SIR) and THC9 (sir3) were examined as described in Fig. 2. r50NY/Z was excised from a modified hmr locus on a CEN vector (pRS414) in strain THC13 (sir3). The linking number shift was 1.0 for rKWD50N, −1.2 for rA1A2, and −1.0 for rA1A2Δp. Additional trials of the experiment gave similar results.
To sustain comparable levels of supercoiling in the repressed state, each ring must also contain the same number of nucleosomes. In isogenic sir3 mutants, however, rKWD50N was more negatively supercoiled than both rA1A2 and rA1A2Δp by approximately two turns (Fig. 4B). The difference cannot be attributed to transcription of ring-borne genes because rA1A2Δp lacks the a1 and a2 promoters. Rather, the difference is most easily explained by rA1A2 and rA1A2Δp containing fewer nucleosomes in the derepressed state. Previously, chromatin footprints of HMR showed that nuclease hypersensitive sites, indicative of disrupted or displaced nucleosomes, appeared at a small number of positions upon derepression (27). Indeed, incorporating one of these sites, the Y/Z junction, into rKWD50N changed its DNA topology. The new ring r50NY/Z contained 1–2 fewer supercoils than rKWD50N in a sir3 mutant (Fig. 4).
Altered DNA Topology and Transcriptional Silencing do not Persist Indefinitely.
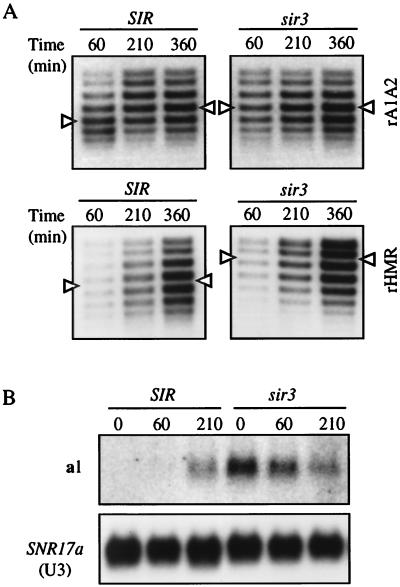
In our initial experiments, few if any cells would have traversed a single cell cycle during the short 60-min excision period (Fig. 2). To test whether the DNA supercoiling shifts of silent DNA persist indefinitely, we examined the topoisomer distributions of rings from HMR at later times. Cells were harvested at 210 and 360 min after the addition of galactose, time points which correspond to one and two doublings in cell number after an initial 60-min excision interval. Upon extended growth, the supercoiling shift of rA1A2 was lost; topoisomer distributions of the ring were similar in both SIR and sir3 strains by 210 min (Fig. 5A). Similar results were obtained for rKWD50N, which became more negatively supercoiled at later times (data not shown). These results demonstrated that Sir-mediated alterations in chromatin structure persisted for only a limited period after uncoupling from silencers and displacement from the chromosome. In contrast, a ring that contains silencers, rHMR (Fig. 4A), maintained its Sir-mediated supercoiling shift throughout the experiment (Fig. 5A). This suggests that silencers and not chromosomal continuity are required for long-term persistence of an altered DNA topology. In agreement, centromeric plasmids that contain the entire HMR locus also maintain a Sir-dependent supercoiling shift and support transcriptional repression (28).
Figure 5.
Structural and functional fate of DNA rings from HMR. (A) Topoisomer distributions of rings at extended intervals after galactose induction (Time 0). rA1A2 (Top) was from strain THC25 (SIR) and THC23 (sir3). rHMR (Bottom) was from YCL1 (SIR) and YCL2 (sir3). rHMR was visualized by an HMR probe. (B) a1 mRNA determined by Northern blot analysis. Experiment was performed in strains THC41 (SIR) and THC40 (sir3). Total RNA was isolated from cells at timed intervals after induction of the R recombinase. Twenty micrograms of nucleic acid were loaded in each lane. a1 mRNA and SNR17a (U3) were visualized sequentially with gene-specific probes. SNR17a serves as a loading control.
DNA replication of the rings represents a likely step during which repressive chromatin structure could be remodeled. However, two lines of evidence suggest that the extrachromosomal rings do not replicate. First, the rA1A2 excision cassette was constructed from sequences that possess no ARS activity (29). Second, and more importantly, the copy number of the excised ring remained constant over a 5-hr period of growth whereas chromosomal DNA content quadrupled (data not shown). Thus, we conclude that loss of the Sir-mediated shift in DNA topology does not require DNA replication-coupled remodeling of the rings.
Lastly, we examined whether the altered DNA supercoiling of rings from HMR was correlated with transcriptional repression. For this purpose, we used northern blot analysis to measure the steady–state levels of a1 mRNA from rA1A2. Before recombination, a1 mRNA was absent in the SIR strain and present in the isogenic sir3 strain (Fig. 5B, lanes 1 and 4). This result indicates that incorporation of RS sites at HMR does not disrupt normal gene regulation. After a 60-min recombinase induction, a1 mRNA was barely detectable in the SIR strain. By the 210-min time point, however, the level of a1 transcript was comparable in SIR and sir3 strains. The time-dependent diminution of transcript in the sir3 strain can be explained, in part, by the dilution of the nonreplicating template in growing culture. These results demonstrated that transcriptional repression of HMR, along with altered DNA supercoiling, does not persist indefinitely in extrachromosomal rings that lack silencers. Loss of silent chromatin could be governed by either a time-dependent decay process or a cell–cycle-mediated event.
DISCUSSION
Silencing is Associated with an Altered DNA Path in Chromatin.
Saccharomyces lacks the cytological features of constitutively condensed heterochromatin found in higher eukaryotes. Heterochromatin-like domains thus have been identified by their ability to repress transcription and to limit access to DNA modifying enzymes (2). Here, we show that DNA supercoiling is also a sensitive probe of silenced chromatin in yeast. Importantly, the altered superhelical density of rings excised from HMR indicates that the packaging of repressed DNA is different; silencing does not arise from simply encasing standard chromatin in an impenetrable sheath of silencing factors.
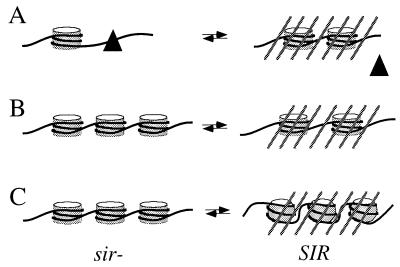
The simplest model to account for Sir-mediated changes in DNA supercoiling is that silencing alters the number of nucleosomes on a template. According to this model, the preferred settings and stabilities of nucleosomes may differ in repressed and derepressed states. For example, rings rA1A2 and rA1A2Δp contain the Y/Z junction, a site which is cut by nucleases at all mating-type loci in derepressed strains but only at MAT in repressed strains (27). Regulated cleavage of the site at MAT by the endogenous Ho endonuclease initiates mating-type interconversion (17). A Y/Z junction binding protein, YZbp, was recently purified from yeast (30), and it was proposed that the factor facilitates cleavage by maintaining an open chromatin conformation (27, 30). The loss of one supercoil at HMR upon derepression is consistent with YZbp, or some other factor, occluding or unwrapping nucleosomes (Fig. 6A).
Figure 6.
Models for Sir-mediated transitions that can alter DNA topology. (A) Silencing favors nucleosome formation at the expense of binding sequence-specific binding factors (triangle). Negative supercoils are gained upon repression. (B) Silencing favors an altered nucleosome distribution, which can dictate a different total number of nucleosomes. Negative supercoils are either lost (as depicted here) or gained upon repression. (C) Silencing promotes a conformational change in all nucleosomes within a repressed region, which leads to a loss of negative supercoils upon repression (based on rKWD50N). Cross-hatched lines denote repressed chromatin.
Unlike rA1A2 and rA1A2Δp, rKWD50N gains a supercoil upon derepression, as do other rings composed of non-HMR yeast DNA (Y.-C.L., unpublished results). Ascribing the shift to a change in nucleosome number is difficult because chromatin footprints of rKWD50N do not provide clear evidence of such an event. However, the footprints are consistent with the presence of two preferred settings for nucleosomes with one strongly favored in the derepressed state and either one used in the repressed state. The supercoiling of rKWD50N can be explained if the derepressed setting permits binding of additional histone octamers (Fig. 6B).
If silencing does not change the number of nucleosomes in rKWD50N then the supercoiling shift must reflect a conformational change in those nucleosomes present, as depicted in Fig. 6C. At least two types of transitions can be envisioned. Silencing could alter the three-dimensional path or helical repeat of DNA within nucleosomes, and small differences in each nucleosome could contribute additively to a change in supercoiling. Alternatively, silencing could alter interactions between nucleosomes (31). This second type of transition might not perturb nucleosome structure directly, but it would impact the DNA topology of internucleosomal linker DNA. Either of the transitions could be facilitated by interactions of the Sir proteins with histones (9) or by the altered acetylation pattern of histone tails in repressed regions (32). The direct influence of histone acetylation on DNA supercoiling, however, is still controversial (33, 34).
Short-Term vs. Long-Term Persistence of Repressive Chromatin Structure in the Absence of Silencers.
An altered chromatin structure persisted in silenced DNA immediately after excision from the chromosome and unlinkage from silencers. At extended times, however, the repressive DNA packaging was lost and the silenced genes were transcribed (Fig. 5). Thus, long-term preservation of silent chromatin requires silencers in cis. Our results confirm and extend earlier work of others. In DNA modification experiments with isolated nuclei, removal of silencers by endonucleolytic cleavage did not increase the accessibility of restriction endonuclease sites within HMR (6). Similarly, when a sole silencer was deleted from a chromosome by in vivo recombination, the adjacent mating-type genes were not derepressed, as long as cells were held in G1 (14). Upon release from growth arrest, however, expression occurred within a single generation, indicating that repression was not permanent. Taken together, these observations indicate that cell growth is required for reactivation of gene expression and topological reorganization. According to this view, the role of silencers may be to reinforce or reestablish silencing in the face of a specific physiological challenge during the cell cycle.
Global changes in chromosome structure during cell cycle progression could promote disassembly or decay of repressive chromatin. In this regard, DNA replication during S phase represents the most logical remodeling process. However, we show that nonreplicating DNA rings lose conformational distinctions and undergo changes in expression state. Therefore, replication of the extrachromosomal templates per se is not required for derepression. Inactivation of Sir3p, a constituent of silent chromatin, also leads to derepression without DNA replication (35).
Mitotic compaction of DNA represents another cell cycle-mediated event that could lead to dissociation of factors necessary for sustained repression. In human cells, for instance, nearly all sequence-specific transcription factors are displaced from highly condensed mitotic chromosomes every cell cycle (36). Two experimental observations suggest that maintenance of repression is compromised at or near this point in yeast. First, HMR is partially derepressed at the G2/M border when silencer binding proteins, Orc2p and Orc5p, are inactivated (37). Second, silencing of a telomere-proximal URA3 gene can be overcome exclusively at the G2/M border by overexpression of its cognate transcriptional regulator (38). Programmed degradation of a critical silencing components at this stage in the cell cycle could also account for these results.
Finally, excision of sequences from HMR may permit relocalization of DNA fragments from a nuclear compartment that supports transcriptional repression to those that do not. There is evidence in yeast that Sir proteins are sequestered at telomeres and that efficient transcriptional repression afforded by the silent mating-type loci relies, in part, on their proximity to chromosome ends (39). Potentially, extrachromosomal rings without silencers are reactivated because they escape from localized pools of silencing factors.
Persistence of Silencing in Other Organisms.
In female mammals, a cis-acting silencer-like element termed the X-inactivation center is required for establishment of repression of one of the two X chromosomes. Surprisingly, repression persists through successive generations after deletion of the element, indicating that the X-inactivation center is not required for inheritance of the silent state (40). A reasonable explanation is that structural features of the inactive chromosome are self-templating, even if the X-inactivation center is missing. The persistence of repressed chromatin also has been examined with in vivo recombination in Drosophila. In this case, transposed chromosomal elements that were subject to heterochromatic repression were excised (41). Similar to our results with yeast, the elements became transcriptionally active upon excision. Strikingly, reactivation occurred in cells that were mitotically quiescent and, therefore, did not require active growth as in Saccharomyces (14). Structural changes were shown to occur during cell-cycle progression, primarily during the G1 to G2/M interval.
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATa HMLα HMRa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein, Columbia University |
| THC1 | W303-1A Δhmr∷URA3 | This study |
| THC3 | W303-1A Δhmr∷URA3 Δsir3∷HIS3 | “ ” |
| THC9 | W303-1A Δhmr::rKWD50N Δsir3::HIS3 | “ ” |
| THC10 | W303-1A Δhmr∷rKWD50N | “ ” |
| THC13 | W303-1A Δsir3∷HIS3 | “ ” |
| THC16 | W303-1A Δlys2∷rKWD50N Δsir3∷HIS3 | “ ” |
| THC17 | W303-1A Δlys2∷rKWD50N | “ ” |
| THC23 | W303-1A hmr∷rA1A2 Δsir3∷HIS3 | “ ” |
| THC24 | W303-1A Δhmr∷rA1A2Δp Δsir3∷HIS3 | “ ” |
| THC25 | W303-1A hmr∷rA1A2 | “ ” |
| THC26 | W303-1A Δhmr∷rA1A2Δp | “ ” |
| THC27 | W303-1A Δhmr∷rKWD50N Δlys2 | “ ” |
| THC28 | W303-1A Δhmr∷rKWD50N Δlys2 Δsir3∷HIS3 | “ ” |
| THC40 | W303-1A hmr∷rA1A2 Δmat∷URA3 Δsir3∷HIS3 | “ ” |
| THC41 | W303-1A hmr∷rA1A2 Δmat∷URA3 | “ ” |
| YCL1 | W303-1A hmr∷rHMR | “ ” |
| YCL2 | W303-1A hmr∷rHMR Δsir3∷HIS3 | “ ” |
Acknowledgments
We thank D. Shore and S. Holmes for providing plasmids and A. Gabriel, R. Kamakaka, and R. Morris for critical reading of the manuscript. This work was funded by the National Institutes of Health (GM 51402). M.R.G. is an American Cancer Society Junior Faculty Research Professor (JFRA-507).
ABBREVIATIONS
- MNase
micrococcal nuclease
- kb
kilobase
Note
After this work was completed, a similar study of the structural fate of silent chromatin at HML was published (42). Structural changes were shown to occur during cell-cycle progression, primarily during the G1 to G2/M interval.
References
- 1.Elgin S C R. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 2.Loo S, Rine J. Ann Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 3.Rine J, Hershkowitz I. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moretti P, Freeman K, Coodly L, Shore D. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 5.Triolo T, Sternglanz R. Nature (London) 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 6.Loo S, Rine J. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- 7.Gottschling D E. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 9.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 10.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 11.Pillus L, Rine J. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 12.Mahoney D J, Marquardt R, Shei G-J, Rose A B, Broach J R. Genes Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- 13.Sussel L, Vannier D, Shore D. Mol Cell Biol. 1993;13:3919–3928. doi: 10.1128/mcb.13.7.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes S G, Broach J R. Genes Dev. 1996;10:1021–1032. doi: 10.1101/gad.10.8.1021. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzaki H, Nakajima R, Nishiyama J, Araki H, Oshima Y. J Bacteriol. 1990;172:610–618. doi: 10.1128/jb.172.2.610-618.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirabella A, Gartenberg M R. EMBO J. 1997;16:523–533. doi: 10.1093/emboj/16.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostriken R, Strathern J N, Klar A J S, Hicks J B, Heffron F. Cell. 1983;35:167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1993. [Google Scholar]
- 19.Depew D E, Wang J C. Proc Natl Acad Sci USA. 1975;72:4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent N A, Bird L E, Mellor J. Nucleic Acids Res. 1993;21:4653–4654. doi: 10.1093/nar/21.19.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myslinski E, Segault V, Branlant C. Science. 1990;247:1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- 22.Gartenberg M R, Wang J C. Proc Natl Acad Sci USA. 1993;90:10514–10518. doi: 10.1073/pnas.90.22.10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghuraman M K, Brewer B J, Fangman W L. Science. 1997;276:806–809. doi: 10.1126/science.276.5313.806. [DOI] [PubMed] [Google Scholar]
- 24.Simpson R T, Thoma F, Brubaker J M. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 25.Roth S Y, Shimizu M, Johnson L, Grunstein M, Simpson R T. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- 26.Saavedra R A, Huberman J A. Cell. 1986;45:65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- 27.Nasmyth K. Cell. 1982;30:567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- 28.Abraham J, Feldman J, Nasmyth K A, Strathern J N, Klar A J S, Broach J R, Hicks J B. Cold Spring Harbor Symp Quant Biol. 1982;47:989–998. doi: 10.1101/sqb.1983.047.01.113. [DOI] [PubMed] [Google Scholar]
- 29.Abraham J, Nasmyth K, Strathern J N, Klar A J S, Hicks J B. J Mol Biol. 1984;176:307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang R, Jin Y, Norris D. Mol Cell Biol. 1997;17:770–777. doi: 10.1128/mcb.17.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 32.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 33.Lutter L C, Judis L, Paretti R F. Mol Cell Biol. 1992;12:5004–5014. doi: 10.1128/mcb.12.11.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norton V G, Imai B S, Yau P, Bradbury E M. Cell. 1989;57:449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- 35.Miller A M, Nasmyth K A. Nature (London) 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Balbas M A, Dey A, Rabindran S K, Ozato K, Wu C. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 37.Fox C A, Loo S, Dillin A, Rine J. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- 38.Aparicio O M, Gottschling D E. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 39.Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser S M. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- 40.Brown C J, Willard H F. Nature (London) 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 41.Ahmad K, Golic K G. Genetics. 1996;144:657–670. doi: 10.1093/genetics/144.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bi X, Broach J R. Mol Cell Biol. 1997;17:7077–7087. doi: 10.1128/mcb.17.12.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C. Nature (London) 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]