Abstract
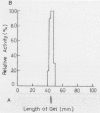
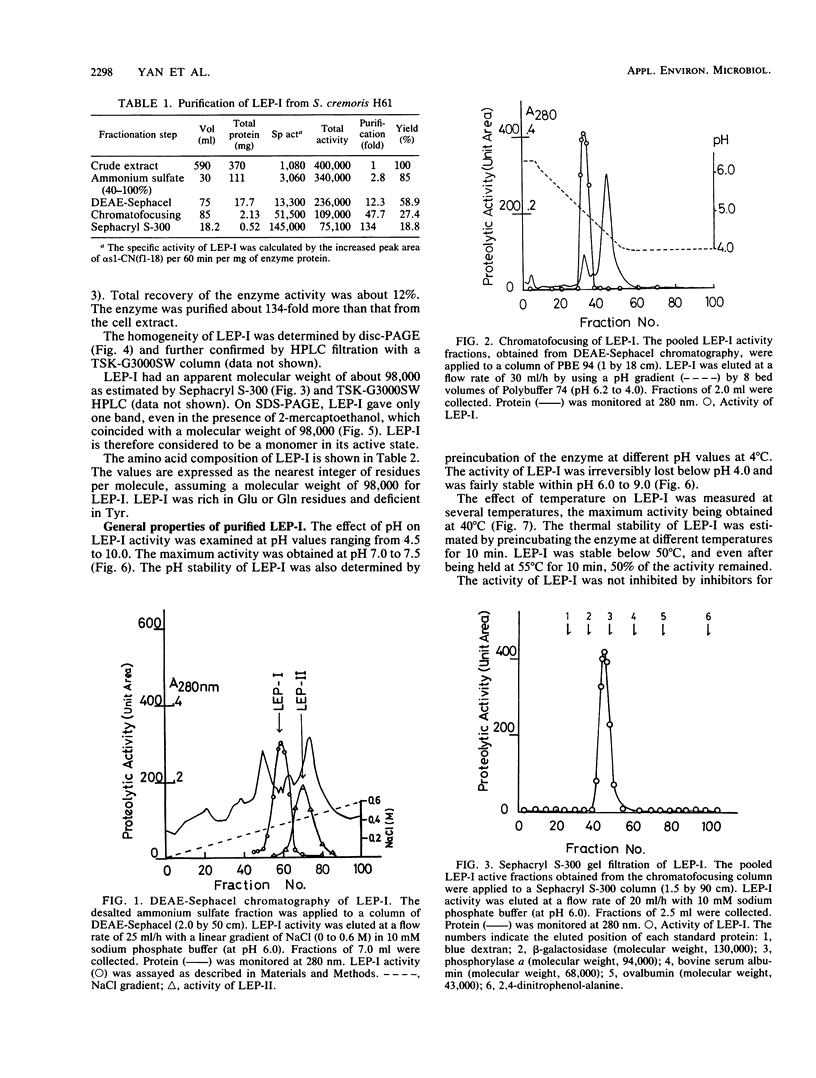
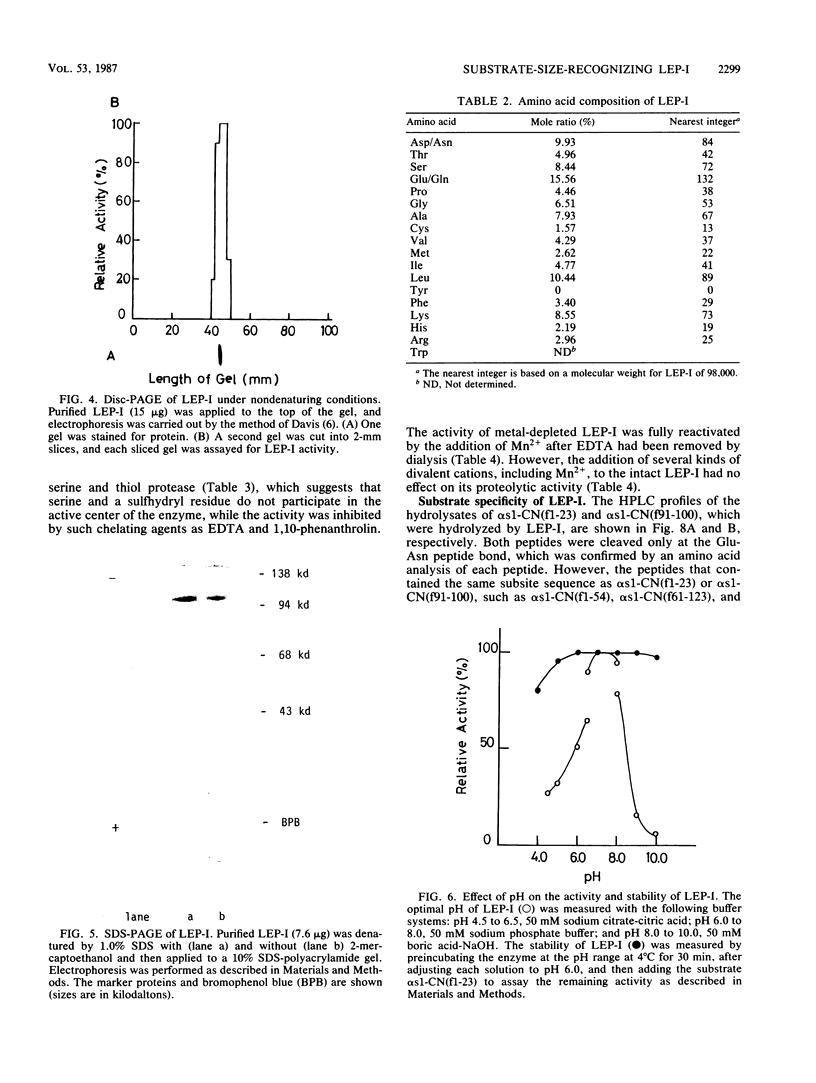
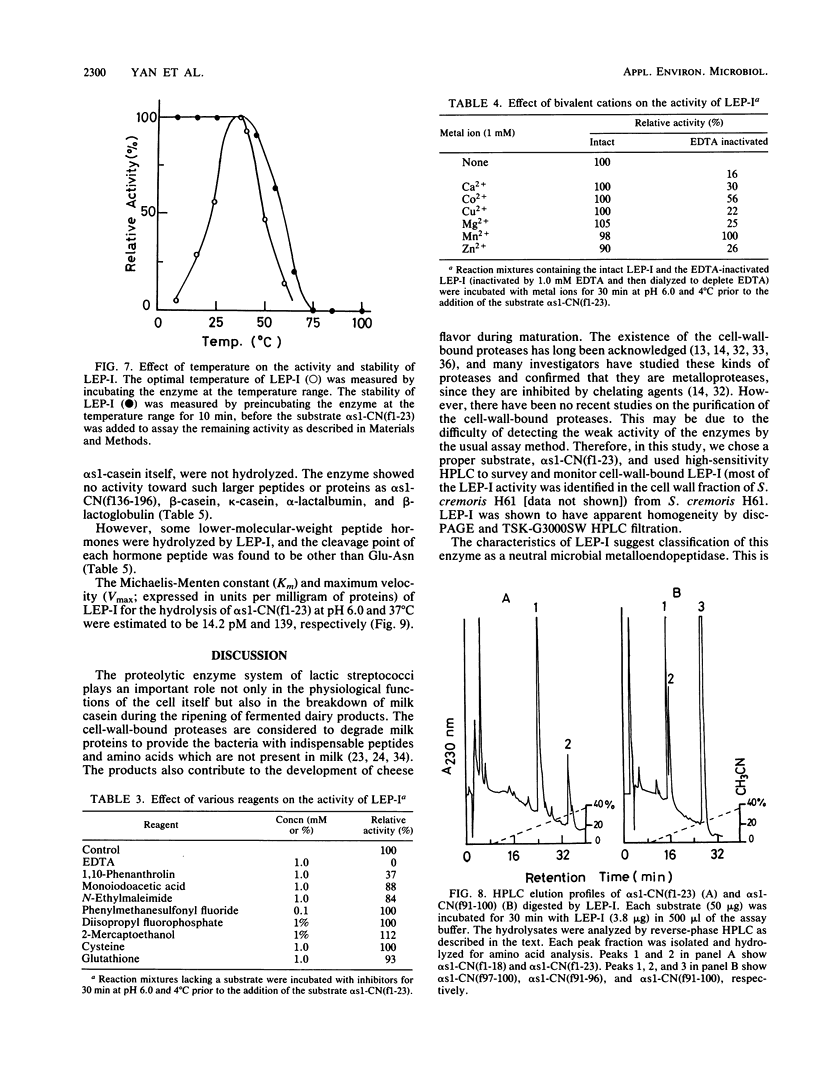
During the ripening of Gouda-type cheese, two kinds of endopeptidases were found to participate in the degradation of αs1-CN(f1-23), a specific product from αs1-casein hydrolyzed by chymosin. One of the endopeptidases, lactic acid bacteria endopeptidase (LEP-II), which can recognize the size of its substrates, has already been purified and characterized (T. R. Yan, N. Azuma, S. Kaminogawa, and K. Yamauchi, Eur. J. Biochem. 163:259-265, 1987). The other endopeptidase, LEP-I, was purified to homogeneity by conventional chromatographic techniques from Streptococcus cremoris H61. The enzyme appeared to be monomeric, with an apparent molecular weight of 98,000, and its isoelectric point was 5.1. For the hydrolysis of αs1-CN(f1-23), the enzyme had an optimum pH and temperature of 7.0 to 7.5 and 40°C, respectively. Its activity was inhibited by such chelating agents as EDTA and 1,10-phenanthrolin, and it could be fully reactivated by Mn2+. Inhibitors specific for serine and thiol proteases had no effect on the protease activity. The enzyme showed a high affinity toward the Glu-Asn peptide bond of αs1-CN(f1-23) and αs1-CN(f91-100) but showed no hydrolysis activity toward αs1-CN(f1-52), αs1-CN(61-122), αs1-CN(136-196), αs1-casein, β-casein, κ-casein, α-lactalbumin, and β-lactoglobulin. The Km and Vmax of LEP-I for αs1-CN(f1-23) were 14.2 pM and 139 U, respectively.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ASCHAFFENBURG R., DREWRY J. Improved method for the preparation of crystalline beta-lactoglobulin and alpha-lactalbumin from cow's milk. Biochem J. 1957 Feb;65(2):273–277. doi: 10.1042/bj0650273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carles C., Ribadeau Dumas B. Kinetics of the action of chymosin (rennin) on a peptide bond of bovine alpha s1-casein. Comparison of the behaviour of this substrate with that of beta- and kappa o-caseins. FEBS Lett. 1985 Jun 17;185(2):282–286. doi: 10.1016/0014-5793(85)80923-6. [DOI] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- DELFOUR A., JOLLES J., ALAIS C., JOLLES P. CASEINO-GLYCOPEPTIDES: CHARACTERIZATION OF A METHIONINE RESIDUE AND OF THE N-TERMINAL SEQUENCE. Biochem Biophys Res Commun. 1965 May 3;19:452–455. doi: 10.1016/0006-291x(65)90145-2. [DOI] [PubMed] [Google Scholar]
- Eggimann B., Bachmann M. Purification and Partial Characterization of an Aminopeptidase from Lactobacillus lactis. Appl Environ Microbiol. 1980 Nov;40(5):876–882. doi: 10.1128/aem.40.5.876-882.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Soda M., Desmazeaud M. J., Bergère J. L. Peptide hydrolases of Lactobacillus casei: isolation and general properties of various peptidase activities. J Dairy Res. 1978 Oct;45(3):445–455. doi: 10.1017/s0022029900016666. [DOI] [PubMed] [Google Scholar]
- Exterkate F. A., de Veer G. J. Partial Isolation and Degradation of Caseins by Cell Wall Proteinase(s) of Streptococcus cremoris HP. Appl Environ Microbiol. 1985 Feb;49(2):328–332. doi: 10.1128/aem.49.2.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROSS E., WITKOP B. Nonenzymatic cleavage of peptide bonds: the methionine residues in bovine pancreatic ribonuclease. J Biol Chem. 1962 Jun;237:1856–1860. [PubMed] [Google Scholar]
- Hill R. D., Lahav E., Givol D. A rennin-sensitive bond in alpha-s1 B-casein. J Dairy Res. 1974 Feb;41(1):147–153. doi: 10.1017/s0022029900015028. [DOI] [PubMed] [Google Scholar]
- Law B. A., Kolstad J. Proteolytic systems in lactic acid bacteria. Antonie Van Leeuwenhoek. 1983 Sep;49(3):225–245. doi: 10.1007/BF00399500. [DOI] [PubMed] [Google Scholar]
- Ohmiya K., Sato Y. Purification and Properties of Intracellular Proteinase from Streptococcus cremoris. Appl Microbiol. 1975 Nov;30(5):738–745. doi: 10.1128/am.30.5.738-745.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. H., Morris H. A., Castberg H. B., McKay L. L. Hydrolysis of milk proteins by bacteria used in cheese making. J Agric Food Chem. 1976 Nov-Dec;24(6):1106–1113. doi: 10.1021/jf60208a026. [DOI] [PubMed] [Google Scholar]
- Sullivan J. J., Jago G. R., Mou L. Pptidase activities in group N streptococci. J Dairy Res. 1975 Feb;42(1):147–155. doi: 10.1017/s002202990001517x. [DOI] [PubMed] [Google Scholar]
- Thomas T. D., Jarvis B. D., Skipper N. A. Localization of proteinase(s) near the cell surface of Streptococcus lactis. J Bacteriol. 1974 May;118(2):329–333. doi: 10.1128/jb.118.2.329-333.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Yan T. R., Azuma N., Kaminogawa S., Yamauchi K. Purification and characterization of a novel metalloendopeptidase from Streptococcus cremoris H61. A metalloendopeptidase that recognizes the size of its substrate. Eur J Biochem. 1987 Mar 2;163(2):259–265. doi: 10.1111/j.1432-1033.1987.tb10796.x. [DOI] [PubMed] [Google Scholar]