Abstract
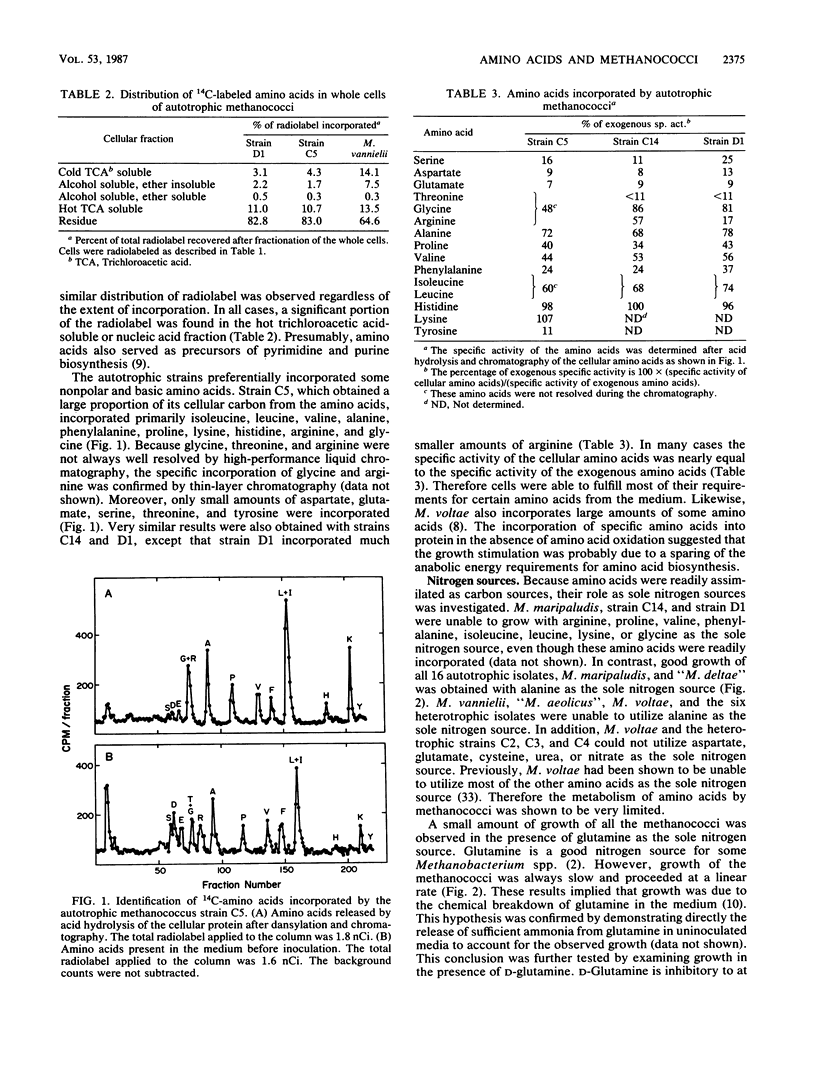
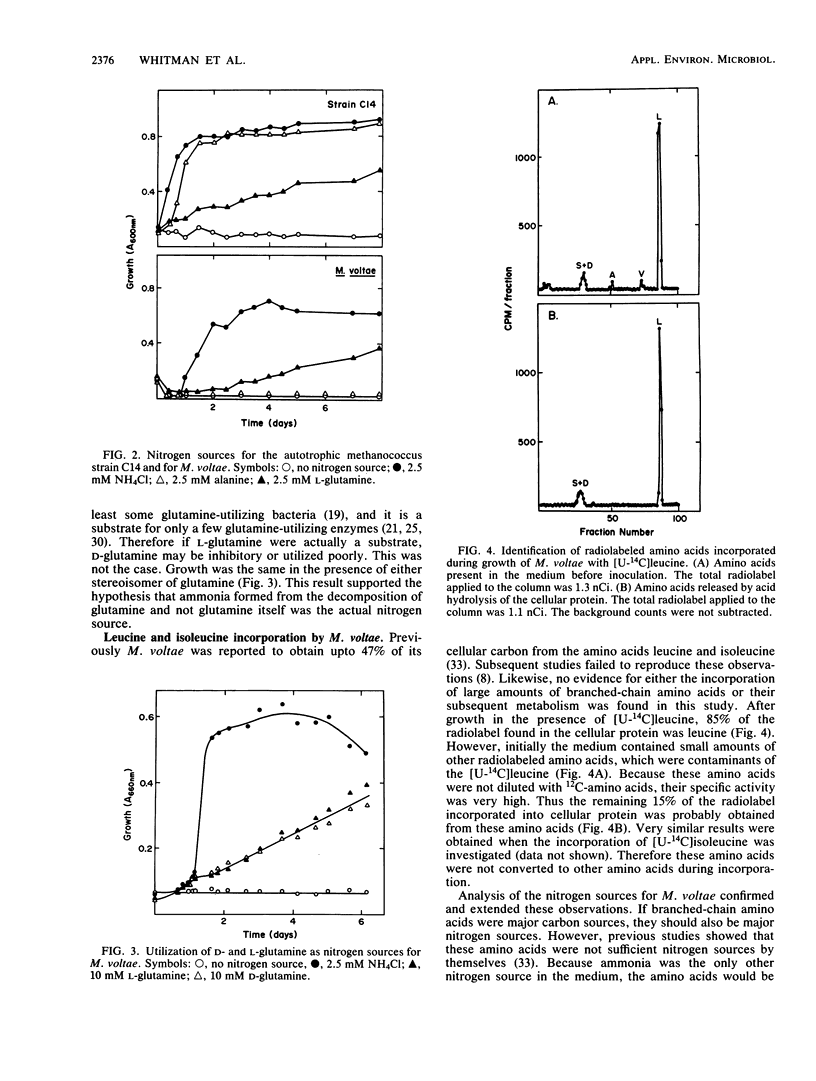
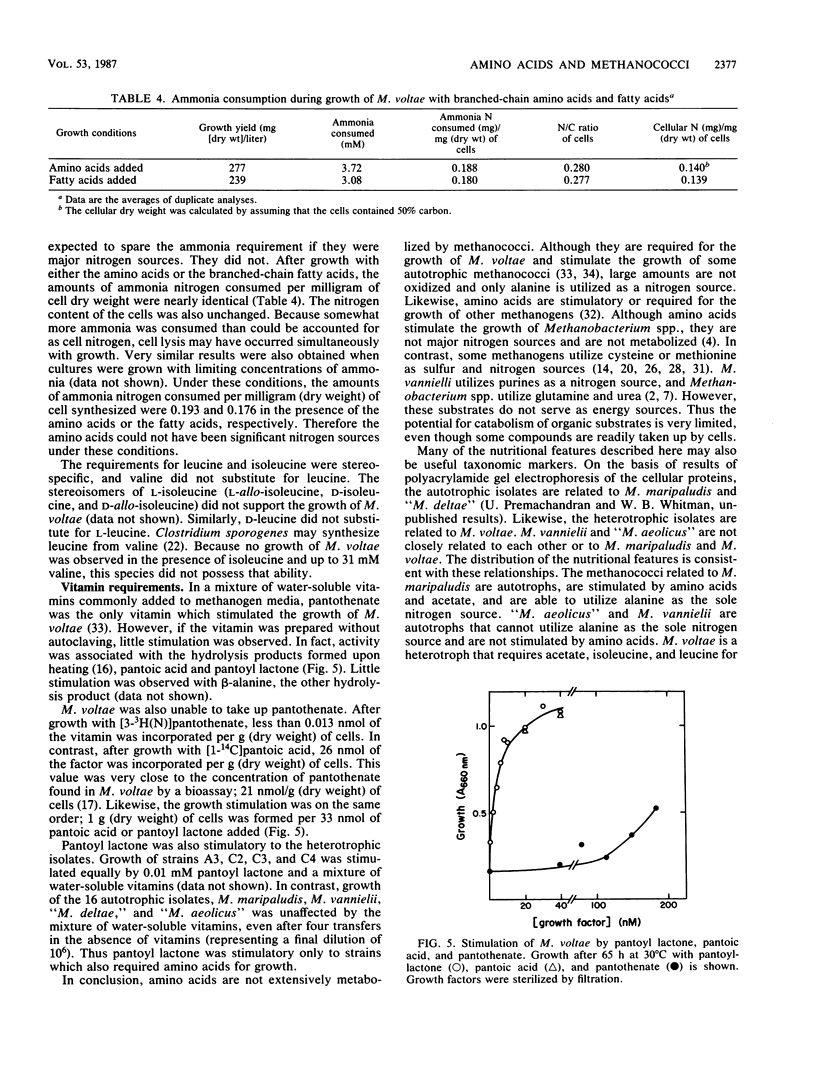
In this study we found that autotrophic methanococci similar to Methanococcus maripaludis obtained up to 57% of their cellular carbon from exogenous amino acids. About 85% of the incorporation was into protein. Primarily nonpolar and basic amino acids and glycine were incorporated; only small amounts of acidic and some polar amino acids were taken up. An additional 10% of the incorporation was into the nucleic acid fraction. Because little 14CO2 was formed from the 14C-amino acids, little metabolism of the amino acids occurred. Therefore the growth stimulation by amino acids was probably due to the sparing of anabolic energy requirements. Of the amino acids incorporated, only alanine was also a sole nitrogen source for these methanococci. In contrast, Methanococcus vannielii and “Methanococcus aeolicus” are autotrophic methanococci which did not incorporate amino acids and did not utilize alanine as a sole nitrogen source. Although glutamine served as a sole nitrogen source for the autotrophic methanococci and Methanococcus voltae, a heterotrophic methanococcus, growth was due to chemical deamination in the medium. M. voltae requires leucine and isoleucine for growth. However, these amino acids were not significant nitrogen sources, and alanine was not a sole nitrogen source for the growth of M. voltae. The branched-chain amino acids were not extensively metabolized by M. voltae. Pantoyl lactone and pantoic acid were readily incorporated by M. voltae. The intact vitamin pantothenate was neither stimulatory to growth nor incorporated. In conclusion, although amino acids and vitamins are nutritionally important to both autotrophic and heterotrophic methanococci, generally they are not subject to extensive catabolism.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhatnagar L., Jain M. K., Aubert J. P., Zeikus J. G. Comparison of assimilatory organic nitrogen, sulfur, and carbon sources for growth of methanobacterium species. Appl Environ Microbiol. 1984 Oct;48(4):785–790. doi: 10.1128/aem.48.4.785-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANEY A. L., MARBACH E. P. Modified reagents for determination of urea and ammonia. Clin Chem. 1962 Apr;8:130–132. [PubMed] [Google Scholar]
- DeMoll E., Tsai L. Utilization of purines or pyrimidines as the sole nitrogen source by Methanococcus vannielii. J Bacteriol. 1986 Aug;167(2):681–684. doi: 10.1128/jb.167.2.681-684.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiel I., Jarrell K. F., Sprott G. D. Amino acid biosynthesis and sodium-dependent transport in Methanococcus voltae, as revealed by 13C NMR. Eur J Biochem. 1985 Jun 3;149(2):437–444. doi: 10.1111/j.1432-1033.1985.tb08944.x. [DOI] [PubMed] [Google Scholar]
- Ekiel I., Smith I. C., Sprott G. D. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J Bacteriol. 1983 Oct;156(1):316–326. doi: 10.1128/jb.156.1.316-326.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippe H., Caspari D., Fiebig K., Gottschalk G. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci U S A. 1979 Jan;76(1):494–498. doi: 10.1073/pnas.76.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenealy W. R., Thompson T. E., Schubert K. R., Zeikus J. G. Ammonia assimilation and synthesis of alanine, aspartate, and glutamate in Methanosarcina barkeri and Methanobacterium thermoautotrophicum. J Bacteriol. 1982 Jun;150(3):1357–1365. doi: 10.1128/jb.150.3.1357-1365.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiene R. P., Oremland R. S., Catena A., Miller L. G., Capone D. G. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl Environ Microbiol. 1986 Nov;52(5):1037–1045. doi: 10.1128/aem.52.5.1037-1045.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H. L., Jr, Dyar R. E., Wilken D. R. Ketopantoyl lactone and ketopantoic acid reductases. Characterization of the reactions and purification of two forms of ketopantoyl lactone reductase. J Biol Chem. 1974 Aug 10;249(15):4689–4695. [PubMed] [Google Scholar]
- Leigh J. A. Levels of water-soluble vitamins in methanogenic and non-methanogenic bacteria. Appl Environ Microbiol. 1983 Mar;45(3):800–803. doi: 10.1128/aem.45.3.800-803.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccubbin A. E., Hodson R. E. Mineralization of detrital lignocelluloses by salt marsh sediment microflora. Appl Environ Microbiol. 1980 Oct;40(4):735–740. doi: 10.1128/aem.40.4.735-740.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P. S., Hong J. S. Genetics of the glutamine transport system in Escherichia coli. J Bacteriol. 1981 Sep;147(3):805–819. doi: 10.1128/jb.147.3.805-819.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder T. K., Nishio N., Fukuzaki S., Nagai S. Effect of Sulfur-Containing Compounds on Growth of Methanosarcina barkeri in Defined Medium. Appl Environ Microbiol. 1986 Oct;52(4):617–622. doi: 10.1128/aem.52.4.617-622.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticello D. J., Costilow R. N. Interconversion of valine and leucine by Clostridium sporogenes. J Bacteriol. 1982 Nov;152(2):946–949. doi: 10.1128/jb.152.2.946-949.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Olsen G. J., Woese C. R. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986 May 9;45(3):325–326. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- Rivard C. J., Henson J. M., Thomas M. V., Smith P. H. Isolation and Characterization of Methanomicrobium paynteri sp. nov., a Mesophilic Methanogen Isolated from Marine Sediments. Appl Environ Microbiol. 1983 Aug;46(2):484–490. doi: 10.1128/aem.46.2.484-490.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K., Thomm M., Laminet A., Laue F. G., Kessler C., Stetter K. O., Schmitt R. Three new restriction endonucleases MaeI, MaeII and MaeIII from Methanococcus aeolicus. Nucleic Acids Res. 1984 Mar 26;12(6):2619–2628. doi: 10.1093/nar/12.6.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger A., Wuhrmann K. Influence of sulfide compounds on the metabolism of Methanobacterium strain AZ. Arch Microbiol. 1977 Oct 24;115(1):13–17. doi: 10.1007/BF00427839. [DOI] [PubMed] [Google Scholar]
- Whitman W. B., Ankwanda E., Wolfe R. S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982 Mar;149(3):852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F. Growth of methanogenic bacteria in pure culture with 2-propanol and other alcohols as hydrogen donors. Appl Environ Microbiol. 1986 May;51(5):1056–1062. doi: 10.1128/aem.51.5.1056-1062.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmeier V. T., Porterfield S. P., Hendrich C. E. Quantitation of Dns-amino acids from body tissues and fluids using high-performance liquid chromatography. J Chromatogr. 1982 Sep 10;231(2):410–417. doi: 10.1016/s0378-4347(00)81865-4. [DOI] [PubMed] [Google Scholar]



