Abstract
Methylglyoxal is an α-ketoaldehyde and dicarbonyl formed in cells as a side product of normal metabolism. Endogenously produced dicarbonyls, such as methylglyoxal, are involved in numerous pathogenic processes in vivo, including carcinogenesis and advanced glycation end-product formation; advanced glycation end-products are contributors to the pathophysiology of aging and chronic diabetes. Despite recent advances in understanding of the systemic effects of methylglyoxal, the full significance of this compound remains unknown. Herein we provide evidence that the majority of the methylglyoxal present in vivo is bound to biological ligands. The basis for our finding is an experimental approach that provides a measure of the bound methylglyoxal present in living systems, in this instance Chinese hamster ovary cells; with our approach, as much as 310 μM methylglyoxal was detected, 100- to 1,000-fold more than observed previously in biological systems. Several artifacts were considered before concluding that the methylglyoxal was associated with cellular structures, including phosphate elimination from triose phosphates, carbohydrate degradation under the assay conditions, and interference from the derivatizing agent used as part of the assay procedure. A major source of the recovered methylglyoxal is most probably modified cellular proteins. With methylglyoxal at about 300 μM, 0.02% of cellular amino acid residues could be modified. As few as one or two “hits” with methylglyoxal per protein molecule have previously been reported to be sufficient to cause protein endocytosis and subsequent degradation. Thus, 5–10% of cellular proteins may be modified to physiologically significant levels.
Methylglyoxal is an α-ketoaldehyde and dicarbonyl that has been found in all mammalian tissues and cell lines for which it has been assayed (1). Recent research indicates that biogenic dicarbonyls, such as methylglyoxal, are produced during normal cell metabolism and from degradation reactions, such as glucose autoxidation and lipid peroxidation (2), and may be involved in a variety of detrimental processes in vivo, including advanced glycation (glycosylation) end-product (AGE) formation with proteins (3) and DNA modification (4, 5). Methylglyoxal metabolism has been linked to certain complications of diabetes mellitus, possibly mediated through increased blood levels of methylglyoxal evident in diabetics under hyperglycemic conditions (6). Reaction with human serum albumin results in the formation of AGE-like compounds and is believed to be an important signal for the degradation of senescent proteins (7–9). In addition, methylglyoxal can modify unprotected plasmid DNA and cause gene mutation and abnormal gene expression at physiological concentrations (5). The major source of methylglyoxal is nonenzymatic phosphate elimination from glycolytic pathway intermediates (10). In animal cells the principal route for methylglyoxal catabolism is the glyoxalase pathway, which consists of two enzymes, glyoxalase I (EC 4.4.1.5, lactoylglutathione lyase) and glyoxalase II (EC 3.1.2.6, hydroxyacylglutathione hydrolase). This pathway converts methylglyoxal to d-lactate via S-d-lactoylglutathione (10); reduced glutathione is a required cofactor.
Despite advances in understanding the systemic effects of biogenic methylglyoxal, much remains unknown. In large part, this is because that intracellular methylglyoxal exists as a complex mixture of free methylglyoxal and methylglyoxal bound in adduct form (7). Further complicating the issue is that methylglyoxal interacts both reversibly and irreversibly with cellular structures—reversible and irreversible being functional definitions based on whether bound methylglyoxal can be considered in dynamic equilibrium with free methylglyoxal under assay conditions. Irreversibly bound methylglyoxal is stable even under the harshest of assay conditions and can be measured by isolating and quantifying the various irreversible adducts, as in the study of Nagaraj et al. (11), for example, where imidazoline, an adduct formed between lysine residues and methylglyoxal, was quantified in human serum proteins after acid hydrolysis. Reversibly bound methylglyoxal is difficult to measure with the same techniques because the adducts formed are inherently unstable. This instability is a known source of error in the standard assay for free methylglyoxal (12–14). We hypothesize that it will also allow the direct quantification of reversibly bound methylglyoxal and have developed another experimental approach that uses a diaminobenzene derivative, also found in the standard assay, as a thermodynamic trap or sequestering agent to strip bound methylglyoxal from cellular macromolecules. These macromolecules are removed early in the standard assay procedure as part of sample deproteinization with perchloric acid (PCA). Addition of PCA is important for extinguishing metabolic activity and preventing formation of methylglyoxal from glycolytic pathway intermediates, but we believe that the immediate removal of precipitated macromolecules excludes much of the reversibly bound methylglyoxal from quantification.
In this report, we describe application of our approach to Chinese hamster ovary (CHO) cells grown in culture. From these studies, it is apparent that biological systems can contain astonishing levels of methylglyoxal, indeed, several orders of magnitude higher than previously observed. Several potential artifacts were considered before concluding that the detected methylglyoxal had originally been associated with cellular structures. These artifacts fall into two categories—those caused when methylglyoxal is created from cell materials as a result of the assay procedure and those caused by the coelution of artifact peaks with 2-methylquinoxaline (2-MQ); 2-MQ is the o-phenylenediamine (o-PD) derivative of methylglyoxal, the quantification of which by HPLC forms the basis for the methylglyoxal assays used herein. Our results represent evidence that the majority of methylglyoxal in vivo is bound to biological ligands. This is significant because methylglyoxal has often been dismissed on basis of measurements of free methylglyoxal, which is typically present at very low levels. Our data indicate that methylglyoxal is a chemical modifier of proteins in CHO cells grown in culture and possibly in other mammalian systems, also.
MATERIALS AND METHODS
Chemicals and Solutions.
All chemicals were of reagent grade. PBS (pH 7.3), methylglyoxal, d- and l- methotrexate, 2-MQ, 5-methylquinoxaline (5-MQ), and o-PD stock solutions were made as described (11, 14).
Cell Culture.
CHO cells were grown in Iscove’s modified Dulbecco’s medium or MEM (GIBCO/BRL/Life Technologies) as described (15).
Methylglyoxal Assays.
Cells were incubated in MEM containing 1% fetal bovine serum, 100 mM glucose, and 5 mM glutamine for 12 h before harvest. For the standard assay (free methylglyoxal), samples were analyzed as described (14). For our assay, cell extracts were prepared as described (16); PCA was added to a final concentration of 0.45 M; 1,250 pmol of 5-MQ was added as an internal standard, and various amounts of 0.1% o-PD were added as a derivatizing agent. The samples were derivatized at 20°C for 24 h before centrifugation (12,000 × g, 10 min) and removal of the PCA precipitate. This was followed by sample concentration and analysis for 2-MQ content by HPLC (14). To account for methylglyoxal formation from nucleic acid degradation, calf thymus DNA was treated with 0.5 M PCA (16), but with sample preparation modified to approximate the various sample preparation conditions.
Methylglyoxal Recovery in PCA.
All the cell samples were separated into aliquots from a common pool. Noncontrol samples were prepared and assayed for methylglyoxal with our assay but with various periods of derivatization. Control samples were prepared and assayed similarily, but o-PD was not added after PCA addition. The samples were incubated for the required length of time, the PCA precipitate was removed by centrifugation (12,000 × g, 10 min), and the supernatant was passed through a C18 solid-phase extraction cartridge (Waters Sep-Pak tC18 plus cartridge, Millipore) that had been prepared by flushing with 6–8 ml of acetonitrile and 6–8 ml of 10 mM KH2PO4 (pH 2.5). o-PD was added for 2 h before sample concentration and HPLC analysis. Methylglyoxal formation from nucleic acid degradation was accounted for as described above.
Methylglyoxal Recovery in Acetic Acid.
All cell samples were separated into aliquots from a common pool. Noncontrol and control samples were prepared as described for the PCA time course experiment, but glacial acetic acid, to a final concentration of 0.1 M, was substituted for PCA. For the noncontrol samples, PCA, to a final concentration of 0.45 M, was added as a deproteinizing agent after derivatization, and the samples were incubated on ice for 10 min and centrifuged (12,000 × g, 10 min) to remove the PCA-precipitated material. Samples were concentrated and analyzed by HPLC (14). The control samples were deproteinized by PCA at each point and extracted by using solid-phase extraction as described above. Samples were then derivatized with o-PD for 1 h before sample concentration and HPLC analysis. In both cases, nucleic acid degradation during the PCA deproteinization step was accounted for as described above.
Effect of Increasing o-PD Concentrations.
Samples were prepared as for our assay procedure but were derivatized with 50, 100, 200, 500, 1,000, 2,000, or 5,000 μM o-PD. 2,3-Butanedione was detected as 2,3-dimethylquinoxaline (16). The 2,3-dimethylquinoxaline/5-MQ peak area ratios were adjusted to account for an interfering peak that coelutes with 2,3-dimethylquinoxaline. A ratio of 0.025 was used for an addition of 250 nmol of o-PD and 2.50 nmol of 5-MQ to the original sample. This ratio was scaled, as necessary, for the different o-PD concentrations used.
Interaction of Methylglyoxal with Reduced Glutathione.
Reduced glutathione (2 mM, final concentration) and methylglyoxal (20 μM, final concentration) were reacted in PBS for 1 min at 20°C.
Recovery of bound methylglyoxal.
PCA (0.5 M, final concentration) and o-PD (175 μM, final concentration) were added to the reaction mixture, which was analyzed by HPLC as described above. Controls were methylglyoxal and o-PD without reduced glutathione, reduced glutathione and o-PD without methylglyoxal, and methylglyoxal and reduced glutathione without o-PD.
Measurement of free methylglyoxal.
PCA (0.5 M, final concentration) was added to the reaction mixture. Samples were taken over a 47-h period. Each sample was passed through a C18 SPE cartridge to remove the methylglyoxal-reduced glutathione adduct. This cartridge had been prepared by flushing with 6–8 ml of acetonitrile and 6–8 ml of 10 mM KH2PO4 (pH 2.5). o-PD was then added to a final concentration of 175 μM and the free methylglyoxal was derivatized for 2 h before analysis by HPLC.
RESULTS AND DISCUSSION
The Need for an Approach that Measures Reversibly Bound Methylglyoxal in Biological Systems.
The inability to accurately determine the level of methylglyoxal modification is a major problem in assessing the full physiological significance of this compound in biological systems. Prior studies indicate that proteins and other macromolecules are the principal targets for reversible and irreversible modification by methylglyoxal [see. Lo et al. (7)]. Techniques, such as those described by Nagaraj et al. (11), exist or are being developed for measuring irreversibly bound methylglyoxal. There is, however, a need for approaches that measure reversibly bound methylglyoxal that cannot be directly quantified in adduct form because of the inherent instability of the adducts. A previous report has raised the possibility that the standard assay measures both free and reversibly bound methylglyoxal (12). Although we believed that reversibly bound methylglyoxal represented a significant error in the standard assay (14), we also thought it likely that sample deproteinization early in the assay procedure removed important sources of reversibly bound methylglyoxal before measurement could take place. To to test this hypothesis, whole cell extracts were derivatized for extended periods of time in PCA before sample deproteinization and quantification of the methylglyoxal derivative. This was done to allow the derivatizing agent o-PD to act as a thermodynamic trap or trapping agent, by first derivatizing the available free methylglyoxal and then the reversibly bound methylglyoxal as it, in turn, spontaneously disassociated from cellular structures in response to decreased free methylglyoxal concentrations. The derivatizing agent can do this because it has a greater affinity for methyglyoxal under assay conditions than do the biological ligands with which the compound normally interacts. Our approach has the advantage that there is little danger of losing reversibly bound methylglyoxal through the various rinsing or purification steps present in many other approaches. At this point, it should be reemphasized that reversibly and irreversibly bound are functional definitions based on whether the bound methylglyoxal is in dynamic equilibrium with free methylglyoxal under assay conditions. For reversible associations, it is probable that reactions considered reversible under physiological conditions are also reversible under assay conditions. However, it is quite possible that many reactions that are essentially irreversible under physiological conditions are reversible under assay conditions. This is particularly true for the current study where the sample preparation environment is harsh due to the presence of PCA.
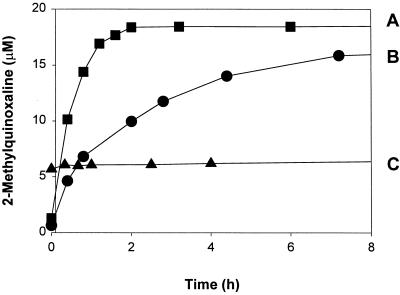
Samples from a simple model system were assayed with our approach to demonstrate that use of a trapping agent is necessary and sufficient to strip methylglyoxal from cellular structures. This system consisted of an equilibrium mixture of reduced glutathione, a tripeptide containing a single cysteine residue, and methylglyoxal at physiological temperature and pH; cysteine residues are a major modification target for methylglyoxal (7). Our approach was tested by adding PCA and the derivatizing agent to the reaction mixture. Fig. 1, curve B, shows that recovery of the majority of the methylglyoxal took 8 h under these conditions. The principal control was to incubate samples from the model system in PCA for set time intervals before removing the methylglyoxal-reduced glutathione adduct by solid-phase extraction—equivalent to sample deproteinization—and assaying the resulting adduct-free samples for free methylglyoxal. Fig. 1, curve C, indicates that the adduct is stable in PCA in the absence of PCA. This was shown by the consistent level of free methylglyoxal measured over the 8-h period. Additional controls were to separately treat methylglyoxal (20 μM) or reduced glutathione (2 mM) with PCA. No derivatized methylglyoxal was detected in the reduced glutathione control after a 2-h incubation with o-PD. Fig. 1, curve A, shows that the methylglyoxal control derivatized within the same 2-h interval. The final control was to add o-PD to the methylglyoxal/reduced glutathione mixture used to generate fig. 1, curve C, after a 47-h incubation in PCA. At that time, the kinetics of methylglyoxal recovery was similar to those seen in Fig. 1, curve B (data not shown). From these experiments, it is evident that maximal recovery of methylglyoxal from sulfhydryl groups, the probable major source of the methylglyoxal detected with our approach, requires the presence of a trapping agent, in this instance o-PD.
Figure 1.
Reaction with and recovery of methylglyoxal from reduced glutathione. (A) Methylglyoxal (20 μM) was derivatized with o-PD (175 μM) in PCA (0.5 M). At each time, 2-MQ content was determined by HPLC analysis. (B) The methylglyoxal in an equilibrium mixture of reduced glutathione (2 mM) and methylglyoxal (20 μM) was derivatized with o-PD (175 μM) in PCA (0.5 M). At each time, 2-MQ content was determined by HPLC analysis. (C) An equilibrium mixture of reduced glutathione (2 mM) and methylglyoxal (20 μM) was incubated in PCA (0.5 M). At each time, the methylglyoxal-reduced glutathione adduct was removed by solid-phase extraction and the remaining free methylglyoxal was derivatization by o-PD (175 μM) for 2 h followed by 2-MQ determination by HPLC analysis.
These results contrast with those reported by McLellan et al. (13) and Chaplen et al. (14) in which reasonable methylglyoxal recoveries (53–72%) were demonstrated as part of the development of assays for free methylglyoxal, assays that did not use the diaminobenzene derivative as a trapping agent. An explanation for this lies with the protocols used to determine free methylglyoxal recovery in the original studies. In these, samples (whole blood or cell extracts) were treated with PCA and then spiked with free methylglyoxal; the overall percent recovery was determined by comparing the amount of spiked methylglyoxal remaining after the assay procedure versus that originally added. Exposure of samples to PCA before adding methylglyoxal would prevent the formation of reversible adducts between methylglyoxal and biological ligands because the major targets of methylglyoxal modification (sulfhydryls and primary amines) are easily oxidized under such conditions (17, 18). Furthermore, precipitation of proteins and other cellular structures by PCA would impede access to many modification sites through steric hinderance. As a result, it is likely that few sites remained for methylglyoxal modification, allowing recovery of much of the added methylglyoxal without a trapping agent. This contrasts with the current study where methylglyoxal and the reduced glutathione were reacted under physiological conditions before adding the PCA; thus, the hemithioacetal adduct could form unhindered and the trapping agent was required to maximize methylglyoxal recovery.
High Levels of Methylglyoxal Were Recovered from CHO Cells with Our Approach.
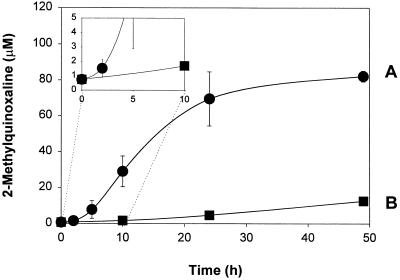
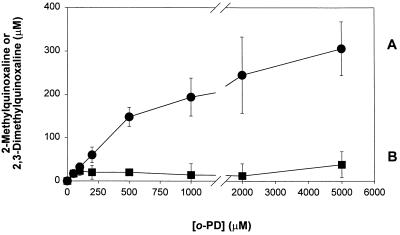
Cell samples were then assayed with our approach. In this system, the principal control was to incubate the whole cell extract with PCA for various times before deproteinizing and derivatizing the free methylglyoxal present at that time. Fig. 2 shows the time course of methylglyoxal detected (as the 2-MQ derivative) from cell extracts by our procedure (Fig. 2, curve A). After 24 h, the concentration detected was 70 ± 20 μM (mean ± σ; n = 2), as compared with 0.8 ± 0.3 μM (mean ± σ; n = 3) from samples assayed with the standard procedure (equivalent to methylglyoxal present at t = 0 on Fig. 2, curve A). The kinetics of methylglyoxal recovery from cell samples was significantly different from that observed in the model methylglyoxal/reduced glutathione system. In addition, there was an increase in free methylglyoxal detected in the control cell samples after the 24-h acid incubation period that was not present in the control samples from the model system. An explanation for these observations is that the model system represents methylglyoxal–sulfhydryl group interactions only, although there are several other interactions that could be occurring that might be the cause of observed differences between the recovery kinetics shown by the model system and the cell samples. The level of methylglyoxal detected with our procedure is 80- to 100-fold greater than that detected with the standard procedure. Incubation beyond 24 h did not cause a statistically significant increase in the level of methylglyoxal detected. By contrast, the concentration of methylglyoxal in the control samples (Fig. 2, curve B) was 5 ± 2 μM (mean ± σ; n = 3) after 24 h. This showed that o-PD must be present during the acid incubation step to recover the high levels of methylglyoxal. Another indication o-PD was required as a thermodynamic trap was that increasing the concentration of o-PD also increased the level of methylglyoxal detected (Fig. 3). At 200 μM o-PD, 60 ± 18 μM (mean ± 2σ; n = 6) methylglyoxal was detected as compared with 70 ± 20 μM in the original experiment. At the highest concentration of o-PD used (5,000 μM), 310 ± 62 μM (mean ± 2σ; n = 6) methylglyoxal was recovered. This represented 100- to 1,000-fold more methylglyoxal than previously reported for the standard assay in biological systems [for summary, see review of Thornalley (1)]. One criticism of our approach is that it requires the prolonged incubation in PCA. This is an important concern because there is a long history of artifactual methylglyoxal production in biological samples (see refs. 13 and 14). Therefore, we took several precautions to correct for known artifacts and demonstrate that other artifacts are not involved.
Figure 2.
Kinetics of methylglyoxal recovery from cell extracts in the presence of o-PD. (A) Samples were incubated with o-PD (200 μM) for the indicated times before HPLC analysis. Adjustment for nucleic acid degradation was done. After 24 h, this adjustment was equivalent to 12.5 μM methylglyoxal. (B) Control samples were incubated for the indicated times before the PCA precipitate was removed by centrifugation (12,000 × g, 10 min) and o-PD (200 μM) was added for 2 h to derivatize the free methylglyoxal that was present followed by HPLC analysis. Samples were adjusted for nucleic acid degradation. After 24 h, this adjustment was equivalent to 3.95 μM methylglyoxal. All samples were whole CHO cell extracts from a common source and were incubated at 20°C in PCA (0.45 M). Each data point represents mean ± σ (n = 2 or 3). (Inset) Initial recovery of methylglyoxal. The samples analyzed at t = 0 are equivalent to samples assayed with the standard assay.
Figure 3.
Recovery of methylglyoxal from cell extracts at various concentrations of o-PD. (A) 2-MQ recovery. (B) 2,3-Dimethylquinoxaline recovery. 2,3-Dimethylquinoxaline is the o-PD derivative of 2,3-butanediol. All samples were whole CHO cell extracts from a common source. Samples were derivatized at 20°C in PCA (0.45 M) for 24 h with o-PD at the indicated concentrations. After 24 h, the samples were analyzed for 2-MQ and 2,3-dimethylquinoxaline content by HPLC. Each data point represents the mean ± 2σ (n = 6).
Methylglyoxal Measurements Were Corrected for Nucleic Acid Degradation.
We have recently shown that one source of methylglyoxal is degradation of nucleic acid components by PCA (16). Therefore, the values reported herein were corrected for this artifact, which can readily be accounted for in the standard assay with calf thymus DNA control experiments (14). Fig. 2, curves A and B, have already been adjusted to account for DNA degradation (equivalent to 12.5 μM and 3.95 μM after 24 h for curves A and B, respectively). Certain of the assumptions made to adjust sample concentrations were examined more closely than before, however, because the absolute error introduced into our assay by nucleic acid degradation (12.5 μM after 24 h) was much greater than that introduced into the standard assay (0.2 μM after 4 h). Rather than using a literature value, the actual mass fraction of DNA present in CHO cells was measured. Only the DNA mass fraction was checked because DNA is the major contributor to the error associated with nucleic acid degradation (>92% of the total error) (16). DNA was purified from CHO cells by using phenol/chloroform extraction and ethanol precipitation (16). The DNA concentration was found to be approximately 7 mg/g (wet weight) of cells. This was close to value of 7.5 mg/g (wet weight) of cells reported for animal cells grown in culture (19), indicating that the correct numbers were being used in the adjustment calculations. Another assumption was that the yield of methylglyoxal from CHO cell DNA was equivalent to that from calf thymus DNA. Calf thymus DNA was used in the control experiments because gram quantities are readily available commercially. DNA isolated from CHO cells yielded 5 mmol of methylglyoxal per mol of DNA as compared with 5.8 mmol of methylglyoxal per mol of calf thymus DNA, indicating that calf thymus DNA control experiments could be used to adjust CHO cell sample methylglyoxal concentrations. Finally, it should be noted that the adjustments for Fig. 3, curves A and B, are significantly different from each other. Although the cause of this phenomenon is unknown, it is important to emphasize that it can readily be accounted for by treating the DNA degradation control samples in the same manner as the cell samples being adjusted.
The Influence of the Derivatizing Agent Was Considered.
o-PD itself is known to cause an artifactual peak that coelutes with 2-MQ under the assay conditions used (14). To determine whether the change in assay conditions caused an increase in the size of this peak, blanks (no cell extract) were prepared and derivatized with o-PD by using our procedure. A small increase in the size of the artifact peak was noted, but this represented an absolute error of less than 1 μM—relatively minor when compared with total concentrations of 80–100 μM; in addition, it could be easily accounted for during final concentration calculations. As a further check, samples were prepared and analyzed with an HPLC system that used a different mobile phase and a multidiode array detector (16). With this system, it was possible to positively identify 2-MQ on the basis of its distinctive UV–visual spectrum. High levels of 2-MQ were still present under our conditions, corroborating our previous HPLC measurements.
Degradation of Glycolytic Pathway Intermediates Does Not Explain the Detected Methylglyoxal.
Under some conditions, glycolytic intermediates, in particular, dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G-3-P) degrade to form methylglyoxal (13). Such degradation is minimized in our assays by using low temperatures and highly acidic conditions. The expected absolute error from glycolytic pathway intermediate degradation in our assay is 0.7 μM. This error was calculated by using the observed degradations for DHAP and G-3-P, which are 0.27% and 1.12%, respectively, after a 4-h incubation in 0.5 M PCA (19), with the assumption that DHAP and G-3-P degradation is linear over the 24-h period. In addition, DHAP was assumed to be present in the cell at 38.3 μM and G-3-P at 1.7 μM—concentrations determined by using the combined intracellular concentration of DHAP and G-3-P of 40 μM (20) and an interconversion equilibrium constant of K = 22 (21). At 0.7 μM, the error from glucotriose degradation is only a small fraction of methylglyoxal detected with our approach. Furthermore, even if all the DHAP and G-3-P degraded to methylglyoxal, this would not explain the level of methylglyoxal detected, especially at the 300 μM levels present in some experiments (see Fig. 3).
Milder Assay Conditions in Our Assay Also Result in Increased Methylglyoxal Detection.
One concern is that the higher levels of methylglyoxal resulted from some unspecified degradation caused by PCA. To examine this possibility, the assay was also run with acetic acid, which is much less oxidizative than PCA (16). In addition to using acetic acid, lower levels of o-PD were used (50 versus 200 μM) in an attempt to reduce the amount of the interfering compound, described above and always present in the o-PD stock solution, that coelutes with the derivative methylglyoxal 2-MQ (16). It later became apparent that this compound can easily be accounted for during the calculation of final methylglyoxal concentrations (16). Fig. 3 shows that decreasing o-PD concentrations also decreased the absolute level of methylglyoxal recovery. This effect is evident in that the absolute amount of methylglyoxal detected with our assay was considerably less in the second series of experiments (approximately 6 μM) than detected originally (70 μM). However, Table 1 shows that samples assayed with acetic acid present during the 24-h derivatization step have comparable levels of methylglyoxal to those with PCA present. In addition, samples, prepared with the control procedure described above, had substantially lower amounts of methylglyoxal than noncontrol samples in both cases. This indicated that o-PD was required as a trapping agent to detect high levels of methylglyoxal in samples prepared with either acetic acid or PCA.
Table 1.
Effect of different cell sample preparation techniques on the level of methylglyoxal detected in CHO cells
| Sample | Reversibly bound methylglyoxal,* μM | Free methylglyoxal,† μM |
|---|---|---|
| Acetic acid during derivatization | 6.0 ± 0.6 | 1.4 ± 0.3 |
| PCA during derivatization | 6 ± 1 | 0.7 ± 0.1 |
Mean ± σ (n = 2 to 4). All samples were whole CHO cell extracts from a common source. Samples were incubated for 24 h at 20°C in PCA (0.45 M) or acetic acid (0.1 M) during derivatization with o-PD (50 μM) before measurement of 2-MQ content.
Mean ± σ (n = 2 to 4). All samples were incubated for 24 h before deproteinization and derivatization with o-PD (50 μM), which was added for 2 h to derivatize the free methylglyoxal that was present.
Other Evidence.
It may be possible that assay conditions lead to some unaccounted for carbohydrate degradation as a result of the harsh assay conditions. However, an estimate of this error (4.0 μM) after 24 h indicates that this is significantly less than the 70 μM detected with our approach and that this is most probably not the source of the detected methylglyoxal; the error represents the difference between the methylglyoxal measured in the control (Fig. 2, curve B) after 24 h (adjusted for nucleic acid degradation) and the free methylglyoxal present when the experiment was initiated. In addition, the acetic acid results showed that higher levels of methylglyoxal are recovered in the presence of a thermodynamic trap even when the conditions are less acidic and oxidative than when PCA is present, reducing the possibility that the extra methylglyoxal detected with our assay was an artifact. Another factor that suggested that the detected methylglyoxal was not an artifact was that the amount of methylglyoxal measured increased monotonically with the concentration of o-PD (Fig. 3), whereas the level of 2,3-butanedione detected under the same conditions remained low. CHO cells grown in culture almost certainly do not contain 2,3-butanedione because the compound is produced mainly as a result of ethanol metabolism (22); no ethanol was present in the growth medium, and more importantly, CHO cells cannot metabolize ethanol, lacking the enzyme alcohol dehydrogenase (23). Therefore, one would expect to see levels of 2,3-butanedione consistent with those produced by the reported degradation of cellular components (16). If the increase in methylglyoxal measured had solely been a result of an increase in the detection of carbohydrate degradation intermediates, then it would be expected that the level of all degradation products would also increase; this clearly is not the case. Thus, the observed behavior is consistent with o-PD acting as a trapping agent and recovering reversibly bound methylglyoxal. Finally, we have recently demonstrated that CHO cells expressing recombinant glyoxalase I, the major detoxification enzyme for methylglyoxal, show lower levels of methylglyoxal as measured by both our assay and the standard assay (15). The fact that cells with an augmented detoxification ability for methylglyoxal show decreased methylglyoxal concentrations as measured by both assays strongly argues that the methylglyoxal measured by our assay was originally produced by the cell.
Possible Sources of the Detected Methylglyoxal.
Proteins are modified both reversibly and irreversibly under physiological conditions (7, 10) and represent a probable source of the detected methylglyoxal. Previous work reported from this laboratory supports this possibility (16). In that study, the cytosolic fraction of the CHO cells, consisting mainly of protein, yielded 26 μM methylglyoxal under assay conditions similar to those used with our current approach. This is equivalent to 78 μM for the cell as a whole (the cytosolic fraction contained 33% of the total cellular protein), which is the correct order of magnitude for the concentration of o-PD used (385 μM). In addition to proteins, there are probably other structures in the cell that could be reversibly modified by methylglyoxal. For example, nucleic acids react reversibly with methylglyoxal under physiological conditions (24). Lipid amines are also a possible target—although not previously shown to react with methylglyoxal, they are a known modification target for malondialdehyde, a lipid peroxidation by-product (25).
Biological Significance of High Levels of Methylglyoxal Modification in Mammalian Cells.
By using the assay reported herein, up to 310 μM methylglyoxal was detected in CHO cells. This represented 100- to 1,000-fold more methylglyoxal than previously reported in biological systems. A variety of methods were used to avoid artifacts. An obvious question is whether this level of methylglyoxal modification is reasonable. Much of the reversibly bound methylglyoxal present in biological systems is probably associated with sulfhydryl groups (7). To obtain an order of magnitude estimate of the level of reversibly bound methylglyoxal, the equilibrium constants for the reversible modification of free cysteine under physiological conditions can be used (7) along with measurements of free methylglyoxal in CHO cells. With an equilibrium constant, KEQ, of 7.3 × 104 M−1 from the reaction [methylglyoxal] (all solution forms) + [N-acetylcysteine] ⇌ [hemithioacetal], a cellular protein cysteine concentration of 39.74 mM—obtained from the average amino acid composition of proteins from cells grown in culture (19), the average ratio of cysteine to cystine residues in mammalian systems (26), and a free methylglyoxal concentration of 0.07 μM (14). The equilibrium concentration of [hemithioacetal] is 200 μM, which is in the same range of concentrations measured with our procedure. This estimate uses the levels of free methylglyoxal previously reported for the extracellular growth medium. Recent reports indicate that current measurements of intracellular free methylglyoxal may be too high by one or more orders of magnitude (14, 27), possibly due to interferences from assay artifacts or unavoidable recovery of reversibly bound methylglyoxal during the free methylglyoxal assay procedure. Methylglyoxal is freely membrane permeable (28); thus, the level of free methylglyoxal in the extracellular growth medium can be expected to accurately reflect the levels found in the cell.
Order of magnitude estimates indicate that the levels of reversibly bound methylglyoxal measured by our assay are reasonable. The protein concentration in animal cells is 2.9 mM, with an average protein molecular weight of 70 kDa (29). If proteins were the principal source of the methylglyoxal measured with our approach, then approximately 0.02% of amino acid residues could be modified at the upper limit. Furthermore, only low levels of methylglyoxal modification (one or two AGE per protein molecule) are necessary to cause AGE-receptor-mediated protein endocytosis and lysosomal degradation in monocytes and macrophages (9, 30). At one or two “hits” per protein molecule, therefore, 5–10% of cellular proteins could be modified to physiologically significant levels.
Despite the growing body of evidence supporting a possible role for endogenously produced methylglyoxal in AGE formation (7–9, 30), mutagenesis (5, 10), apoptosis (12), and growth inhibition (15), the full physiological significance of this compound remains unknown. Thus, our results represent direct evidence that, as suggested on the basis of in vitro experiments with BSA and methylglyoxal (7), the majority of biogenic methylglyoxal (>99%) is involved in reversible or irreversible interactions in vivo.
Acknowledgments
We thank Wyeth W. Wasserman, Carsten-Peter Carstens, T. Herbert Manoharan, Andrea Mast, and Jean Gallo for their assistance. Plasmid pSV2-dhfr was a gift from Peggy Farnham (McArdle Laboratory for Cancer Research, Madison WI). Partial support was provided by National Institutes of Health Grant P01-CA-22484. F.W.R.C. was supported by a National Institutes of Health Biotechnology Training Grant fellowship (National Research Award T32 GM08349–03), the University of Wisconsin-Madison Graduate School, and a W. R. Grace Fellowship.
ABBREVIATIONS
- CHO
Chinese hamster ovary cells
- DHAP
dihydroxyacetone phosphate
- G-3-P
glyceraldehyde 3-phosphate
- PCA
perchloric acid
- MQ
methylquinoxaline
- o-PD
o-phenylenediamine
References
- 1.Thornalley P J. Mol Aspects Med. 1993;14:287–371. doi: 10.1016/0098-2997(93)90002-u. [DOI] [PubMed] [Google Scholar]
- 2.Schauenstein E, Esterbauer H, Zollner H. Aldehydes in Biological Systems: Their Natural Occurrence and Biological Activities. London, U.K.: Pion; 1977. pp. 112–157. [Google Scholar]
- 3.Wells-Knecht K J, Zyzak D V, Litchfield J E, Thorpe S R, Baynes J W. Biochemistry. 1995;34:3702–3709. doi: 10.1021/bi00011a027. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhary A K, Nokubo M, Reddy G R, Yeola S N, Morrow J D, Blair I A, Marnett L J. Science. 1994;265:1580–1582. doi: 10.1126/science.8079172. [DOI] [PubMed] [Google Scholar]
- 5.Papsoulis A, Al-Abed Y, Bucala R. Biochemistry. 1995;34:648–655. doi: 10.1021/bi00002a032. [DOI] [PubMed] [Google Scholar]
- 6.McLellan A C, Thornalley P J, Benn J, Sonksen P H. Clin Sci. 1994;87:21–29. doi: 10.1042/cs0870021. [DOI] [PubMed] [Google Scholar]
- 7.Lo T W C, Westwood M E, McLellan A C, Selwood T, Thornalley P J. J Biol Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- 8.Westwood M E, McLellan A C, Thornalley P J. J Biol Chem. 1994;269:32293–32298. [PubMed] [Google Scholar]
- 9.Westwood M E, Thornalley P J. Immunol Lett. 1996;50:17–21. doi: 10.1016/0165-2478(96)02496-0. [DOI] [PubMed] [Google Scholar]
- 10.Thornalley P J. Crit Rev Oncol Hematol. 1995;20:99–128. doi: 10.1016/1040-8428(94)00149-n. [DOI] [PubMed] [Google Scholar]
- 11.Nagaraj R H, Shipanova I N, Faust F M. J Biol Chem. 1996;271:19338–19345. doi: 10.1074/jbc.271.32.19338. [DOI] [PubMed] [Google Scholar]
- 12.Thornalley P J, Edwards L G, Kang Y, Wyatt C, Davies N, Ladan M J, Double J. Biochem Pharmacol. 1996;51:1365–1372. doi: 10.1016/0006-2952(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 13.McLellan A C, Phillips S A, Thornalley P J. Anal Biochem. 1992;206:17–23. doi: 10.1016/s0003-2697(05)80005-3. [DOI] [PubMed] [Google Scholar]
- 14.Chaplen F W R, Cameron D C, Fahl W E. Anal Biochem. 1996;238:171–178. doi: 10.1006/abio.1996.0271. [DOI] [PubMed] [Google Scholar]
- 15.Chaplen F W R, Cameron D C, Fahl W E. Cytotechnology. 1996;22:33–42. doi: 10.1007/BF00353922. [DOI] [PubMed] [Google Scholar]
- 16.Chaplen F W R, Cameron D C, Fahl W E. Anal Biochem. 1996;236:262–269. doi: 10.1006/abio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 17.Friedman M. The Chemistry and Biochemistry of the Sulfhydryl Group in Amino Acids, Peptides and Proteins. New York: Pergamon; 1973. [Google Scholar]
- 18.Plesnicar B. In: Oxidation in Organic Chemistry. Trahanovsky W S, editor. New York: Academic; 1978. pp. 211–295. [Google Scholar]
- 19.Xie L, Wang D I C. Biotechnol Bioeng. 1994;43:1175–1189. doi: 10.1002/bit.260431123. [DOI] [PubMed] [Google Scholar]
- 20.Shonk C E, Boxer G E. Cancer Res. 1964;24:709–721. [PubMed] [Google Scholar]
- 21.Veech R I, Raijman L, Dalziel K, Krebs H A. Biochem J. 1969;115:837–842. doi: 10.1042/bj1150837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otsuka M, Mine T, Ohuchi K, Ohmori S. J Biochem. 1996;119:246–251. doi: 10.1093/oxfordjournals.jbchem.a021230. [DOI] [PubMed] [Google Scholar]
- 23.Meskar A, Holownia A, Bardou L G, Menez J F. Alcohol. 1996;13(6):611–616. doi: 10.1016/s0741-8329(96)00076-6. [DOI] [PubMed] [Google Scholar]
- 24.Krymkiewicz N. FEBS Lett. 1973;29:51–54. doi: 10.1016/0014-5793(73)80013-4. [DOI] [PubMed] [Google Scholar]
- 25.Bidlack W R, Tappel Lipids. 1973;8:203–207. doi: 10.1007/BF02544636. [DOI] [PubMed] [Google Scholar]
- 26.Jocelyn P C. Biochemistry of the SH Group. New York: Academic; 1972. pp. 1–46. [Google Scholar]
- 27.Shih M J, Edinger J W, Creighton D J. Eur J Biochem. 1997;244:852–857. doi: 10.1111/j.1432-1033.1997.00852.x. [DOI] [PubMed] [Google Scholar]
- 28.Thornalley P J. Biochem J. 1988;254:751–755. doi: 10.1042/bj2540751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alberts G, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular Biology of the Cell. 3rd Ed. New York: Garland; 1994. [Google Scholar]
- 30.Westwood M E, Thornalley P J. J Protein Chem. 1995;14(5):359–372. doi: 10.1007/BF01886793. [DOI] [PubMed] [Google Scholar]