Abstract
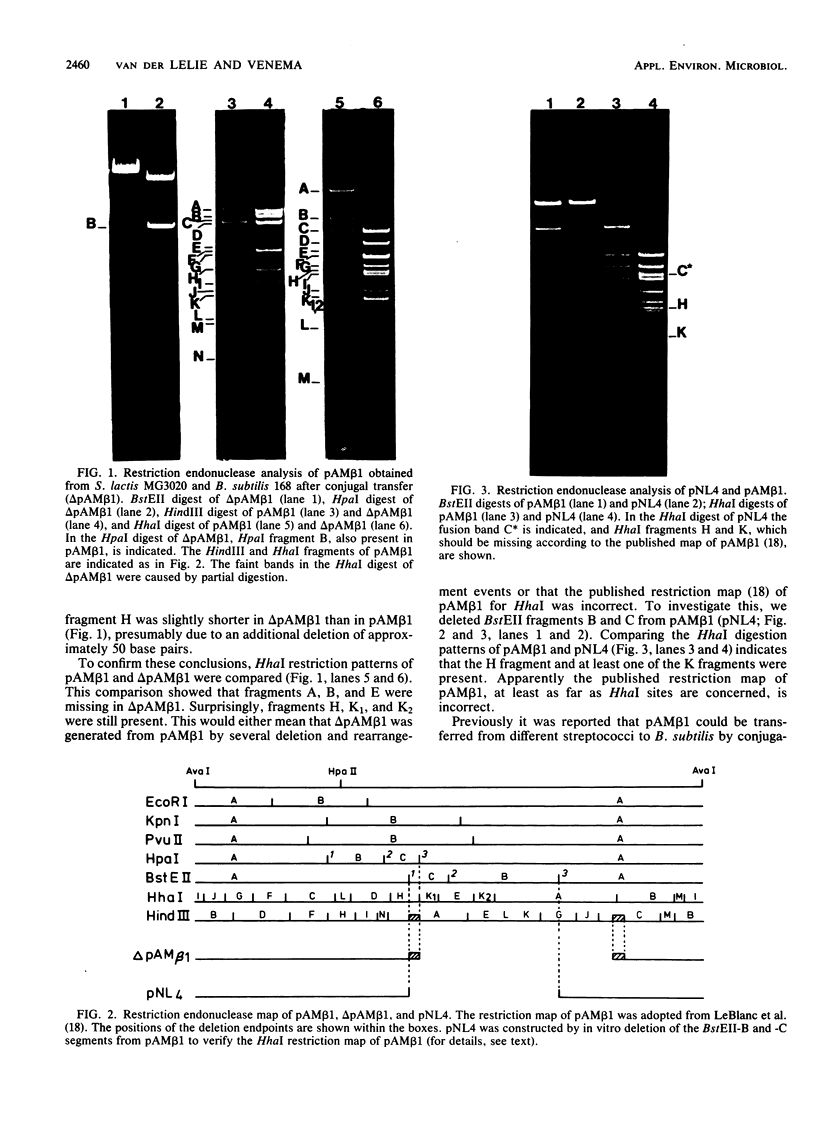
pAM beta 1, a 26.5-kilobase plasmid originally isolated from Streptococcus faecalis, was conjugally transferred from Streptococcus lactis to Bacillus subtilis. No conjugal transfer of pAM beta 1 from B. subtilis to S. lactis was observed. In addition, pAM beta 1 which had been reintroduced in S. lactis after cycling through B. subtilis had lost its conjugal transferability to Streptococcus cremoris, although under the same conditions noncycled pAM beta 1 was transferred at high efficiency. Restriction and Southern blot analyses showed that pAM beta 1 had suffered one major, specific 10.6-kilobase deletion and several minor but also specific deletions in B. subtilis. Comparing the major deletion derivative, delta pAM beta 1, with B. subtilis strains which have been reported to contain pAM beta 1 showed that these strains also contained delta pAM beta 1. Hybridization experiments showed that the deleted fragment was not transposed to the B. subtilis chromosome. Based on the size of the minor deletion derivatives from pAM beta 1, it is suggested that these use a different origin of replication in B. subtilis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron S., Venema G. Ultraviolet inactivation and excision-repair in Bacillus subtilis. I. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat Res. 1972 May;15(1):1–10. doi: 10.1016/0027-5107(72)90086-3. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Qasba P. K. Alkaline transfer of DNA to plastic membrane. Biochem Biophys Res Commun. 1984 Jul 18;122(1):340–344. doi: 10.1016/0006-291x(84)90480-7. [DOI] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Bauer B. Plasmid-determined tetracycline resistance in Streptococcus faecalis: evidence for gene amplification during growth in presence of tetracycline. Proc Natl Acad Sci U S A. 1975 May;72(5):1720–1724. doi: 10.1073/pnas.72.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Engel H. W., Soedirman N., Rost J. A., van Leeuwen W. J., van Embden J. D. Transferability of macrolide, lincomycin, and streptogramin resistances between group A, B, and D streptococci, Streptococcus pneumoniae, and Staphylococcus aureus. J Bacteriol. 1980 May;142(2):407–413. doi: 10.1128/jb.142.2.407-413.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson E. M., Chace N. M., London S. B., London J. Transfer of plasmid-mediated antibiotic resistance from streptococci to lactobacilli. J Bacteriol. 1979 Jan;137(1):614–619. doi: 10.1128/jb.137.1.614-619.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J., van der Vossen J. M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984 Oct;48(4):726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo J. K., McKay L. L. Plasmid transformation of Streptococcus lactis protoplasts: optimization and use in molecular cloning. Appl Environ Microbiol. 1984 Aug;48(2):252–259. doi: 10.1128/aem.48.2.252-259.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Hawley R. J., Lee L. N., St Martin E. J. "Conjugal" transfer of plasmid DNA among oral streptococci. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3484–3487. doi: 10.1073/pnas.75.7.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc D. J., Lee L. N. Physical and genetic analyses of streptococcal plasmid pAM beta 1 and cloning of its replication region. J Bacteriol. 1984 Feb;157(2):445–453. doi: 10.1128/jb.157.2.445-453.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lereclus D., Menou G., Lecadet M. M. Isolation of a DNA sequence related to several plasmids from Bacillus thuringiensis after a mating involving the Streptococcus faecalis plasmid pAM beta 1. Mol Gen Genet. 1983;191(2):307–313. doi: 10.1007/BF00334831. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Keeler C. L., Jr, Jones K. R., Wood P. H. Molecular characterization of unique deletion mutants of the streptococcal plasmid, pAM beta 1. Plasmid. 1980 Jul;4(1):8–16. doi: 10.1016/0147-619x(80)90079-7. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ostroff G. R., Pène J. J. Molecular cloning with bifunctional plasmid vectors in Bacillus subtilis: isolation of a spontaneous mutant of Bacillus subtilis with enhanced transformability for Escherichia coli-propagated chimeric plasmid DNA. J Bacteriol. 1983 Nov;156(2):934–936. doi: 10.1128/jb.156.2.934-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J. B., Youngman P. Streptococcus plasmid pAM alpha 1 is a composite of two separable replicons, one of which is closely related to Bacillus plasmid pBC16. J Bacteriol. 1983 Aug;155(2):607–615. doi: 10.1128/jb.155.2.607-615.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rottländer E., Trautner T. A. Genetic and transfection studies with B, subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol Gen Genet. 1970;108(1):47–60. doi: 10.1007/BF00343184. [DOI] [PubMed] [Google Scholar]
- Saito H., Shibata T., Ando T. Mapping of genes determining nonpermissiveness and host-specific restriction to bacteriophages in Bacillus subtilis Marburg. Mol Gen Genet. 1979 Feb 26;170(2):117–122. doi: 10.1007/BF00337785. [DOI] [PubMed] [Google Scholar]
- Shrago A. W., Chassy B. M., Dobrogosz W. J. Conjugal plasmid transfer (pAM beta 1) in Lactobacillus plantarum. Appl Environ Microbiol. 1986 Sep;52(3):574–576. doi: 10.1128/aem.52.3.574-576.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovo M., Morelli L., Bottazzi V., Gasson M. J. Conjugal Transfer of Broad-Host-Range Plasmid pAMbeta1 into Enteric Species of Lactic Acid Bacteria. Appl Environ Microbiol. 1983 Sep;46(3):753–755. doi: 10.1128/aem.46.3.753-755.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosman B., Kooistra J., Olijve J., Venema G. Integration of vector-containing Bacillus subtilis chromosomal DNA by a Campbell-like mechanism. Mol Gen Genet. 1986 Sep;204(3):524–531. doi: 10.1007/BF00331035. [DOI] [PubMed] [Google Scholar]
- van der Vossen J. M., Kok J., Venema G. Construction of cloning, promoter-screening, and terminator-screening shuttle vectors for Bacillus subtilis and Streptococcus lactis. Appl Environ Microbiol. 1985 Aug;50(2):540–542. doi: 10.1128/aem.50.2.540-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]