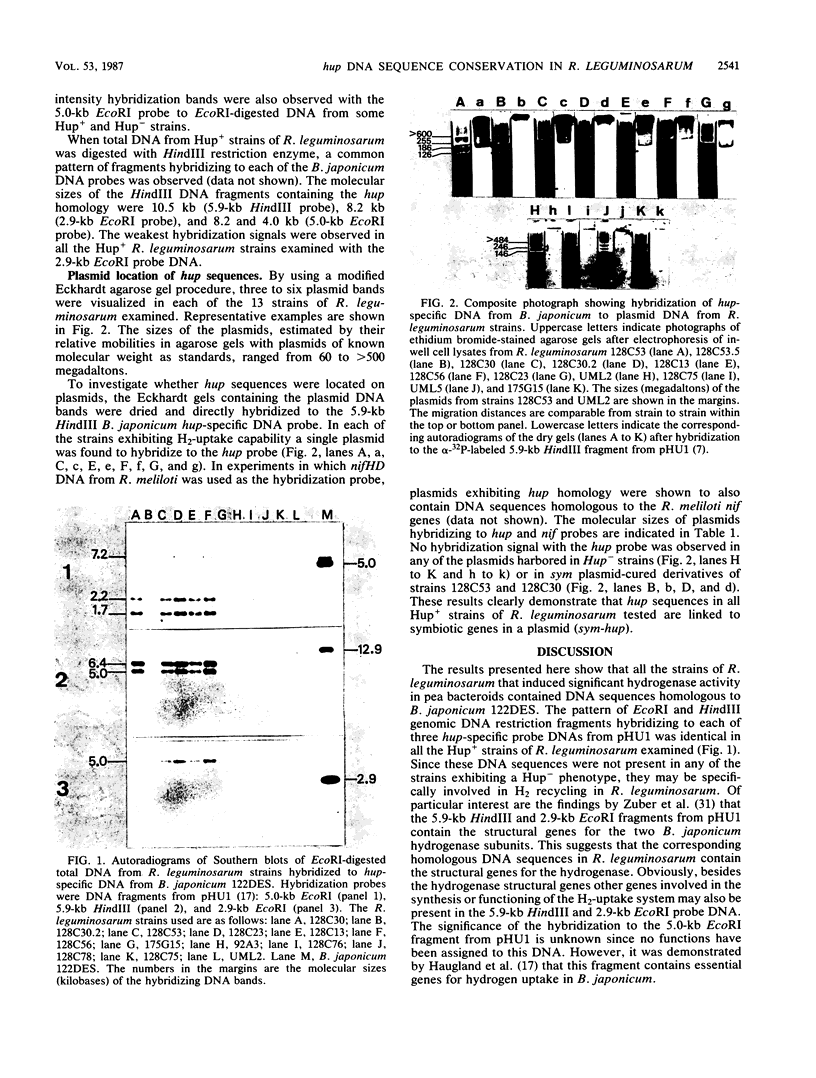
Abstract
Thirteen Rhizobium leguminosarum strains previously reported as H2-uptake hydrogenase positive (Hup+) or negative (Hup−) were analyzed for the presence and conservation of DNA sequences homologous to cloned Bradyrhizobium japonicum hup-specific DNA from cosmid pHU1 (M. A. Cantrell, R. A. Haugland, and H. J. Evans, Proc. Natl. Acad. Sci. USA 80:181-185, 1983). The Hup phenotype of these strains was reexamined by determining hydrogenase activity induced in bacteroids from pea nodules. Five strains, including H2 oxidation-ATP synthesis-coupled and -uncoupled strains, induced significant rates of H2-uptake hydrogenase activity and contained DNA sequences homologous to three probe DNA fragments (5.9-kilobase [kb] HindIII, 2.9-kb EcoRI, and 5.0-kb EcoRI) from pHU1. The pattern of genomic DNA HindIII and EcoRI fragments with significant homology to each of the three probes was identical in all five strains regardless of the H2-dependent ATP generation trait. The restriction fragments containing the homology totalled about 22 kb of DNA common to the five strains. In all instances the putative hup sequences were located on a plasmid that also contained nif genes. The molecular sizes of the identified hup-sym plasmids ranged between 184 and 212 megadaltons. No common DNA sequences homologous to B. japonicum hup DNA were found in genomic DNA from any of the eight remaining strains showing no significant hydrogenase activity in pea bacteroids. These results suggest that the identified DNA region contains genes essential for hydrogenase activity in R. leguminosarum and that its organization is highly conserved within Hup+ strains in this symbiotic species.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedmar E. J., Edie S. A., Phillips D. A. Host Plant Cultivar Effects on Hydrogen Evolution by Rhizobium leguminosarum. Plant Physiol. 1983 Aug;72(4):1011–1015. doi: 10.1104/pp.72.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánfalvi Z., Sakanyan V., Koncz C., Kiss A., Dusha I., Kondorosi A. Location of nodulation and nitrogen fixation genes on a high molecular weight plasmid of R. meliloti. Mol Gen Genet. 1981;184(2):318–325. doi: 10.1007/BF00272925. [DOI] [PubMed] [Google Scholar]
- Cantrell M. A., Haugland R. A., Evans H. J. Construction of a Rhizobium japonicum gene bank and use in isolation of a hydrogen uptake gene. Proc Natl Acad Sci U S A. 1983 Jan;80(1):181–185. doi: 10.1073/pnas.80.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin D., Ditta G., Helinski D. R. Clustering of nitrogen fixation (nif) genes in Rhizobium meliloti. J Bacteriol. 1982 Jan;149(1):221–228. doi: 10.1128/jb.149.1.221-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham S. D., Kapulnik Y., Brewin N. J., Phillips D. A. Uptake Hydrogenase Activity Determined by Plasmid pRL6JI in Rhizobium leguminosarum Does Not Increase Symbiotic Nitrogen Fixation. Appl Environ Microbiol. 1985 Oct;50(4):791–794. doi: 10.1128/aem.50.4.791-794.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. O. Hydrogenase in legume root nodule bacteroids: occurrence and properties. Arch Mikrobiol. 1972;85(3):193–201. doi: 10.1007/BF00408844. [DOI] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Emerich D. W., Ruiz-Argüeso T., Ching T. M., Evans H. J. Hydrogen-dependent nitrogenase activity and ATP formation in Rhizobium japonicum bacteroids. J Bacteriol. 1979 Jan;137(1):153–160. doi: 10.1128/jb.137.1.153-160.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland R. A., Cantrell M. A., Beaty J. S., Hanus F. J., Russell S. A., Evans H. J. Characterization of Rhizobium japonicum hydrogen uptake genes. J Bacteriol. 1984 Sep;159(3):1006–1012. doi: 10.1128/jb.159.3.1006-1012.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom S. S., Graham L. A., Maier R. J. Isolation of genes (nif/hup cosmids) involved in hydrogenase and nitrogenase activities in Rhizobium japonicum. J Bacteriol. 1985 Mar;161(3):882–887. doi: 10.1128/jb.161.3.882-887.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge J., Langridge P., Bergquist P. L. Extraction of nucleic acids from agarose gels. Anal Biochem. 1980 Apr;103(2):264–271. doi: 10.1016/0003-2697(80)90266-3. [DOI] [PubMed] [Google Scholar]
- Nelson L. M., Salminen S. O. Uptake hydrogenase activity and ATP formation in Rhizobium leguminosarum bacteroids. J Bacteriol. 1982 Aug;151(2):989–995. doi: 10.1128/jb.151.2.989-995.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson R. L., Chesshyre J. A., Wheeler C., Jones R., Woodley P. R., Postgate J. R. Genome size and complexity in Azotobacter chroococcum. J Gen Microbiol. 1984 Jul;130(7):1603–1612. doi: 10.1099/00221287-130-7-1603. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Zuber M., Harker A. R., Sultana M. A., Evans H. J. Cloning and expression of Bradyrhizobium japonicum uptake hydrogenase structural genes in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7668–7672. doi: 10.1073/pnas.83.20.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]