Abstract
Circulating human platelets lack nuclei, cannot synthesize mRNA, and are considered incapable of regulated protein synthesis. We found that thrombin-activated, but not resting, platelets synthesize Bcl-3, a member of the IκB-α family of regulatory proteins. The time- and concentration-dependent generation of Bcl-3 in platelets signaled by thrombin was blocked by translational inhibitors, by rapamycin, and by inhibitors of phosphatidylinositol-3-kinase, indicating that it occurs via a specialized translational control pathway that involves phosphorylation of the inhibitory protein 4E-BP1. After its synthesis in activated platelets Bcl-3 binds to the SH3 domain of Fyn (p59fyn), a Src-related tyrosine kinase. This, along with its expression in anucleate cells, suggests that Bcl-3 has previously unrecognized functions aside from modulation of transcription. We also demonstrate that platelets synthesize and secrete numerous proteins besides Bcl-3 after they adhere to fibrinogen, which mediates adhesion and outside–in signaling of these cells by engagement of αIIb/β3 integrin. Taken together, these data demonstrate that regulated synthesis of proteins is a signal-dependent activation response of human platelets.
Platelets are anucleate cells that are derived from bone marrow megakaryocytes and circulate in the blood. They are best known for their ability to bind to exposed subendothelial matrix at sites of vascular injury, forming hemostatic barriers that prevent or limit bleeding (1). Platelet hemostatic responses involve activation by the coagulation protein thrombin, which is recognized by a cell surface receptor (2), or by other agonists. This results in a rapid and complex series of events that include changes in shape of the platelets, inside–out signaling of integrins, the release of preformed or internalized proteins from subcellular granules, and generation of eicosanoids by constitutive enzymes (3). Although their actions to prevent bleeding are of paramount importance, platelets have other roles as well. Platelet adhesion and aggregation in the flowing blood are critical in atherosclerosis and syndromes of pathologic thrombosis (4). They also influence vascular growth, permeability, and integrity, regulate metastasis of certain tumor types, and are intimately involved in inflammation (5). The mechanisms by which platelets contribute to these intricate processes, which are more delayed and prolonged than thrombocyte adhesion and aggregation, are not clearly defined because they have been studied less than the acute responses of these cells in hemostasis.
We recently found that human platelets induce chemokine generation by monocytes (6). This involves altered location and activity of members of the Rel family of transcription factors in the target leukocytes in response to specific adhesion and signaling events. In subsequent studies, we examined the expression of several Rel family members and their regulatory proteins in cells of this system. One of the regulatory proteins is the oncogene product B cell lymphoma-3 (Bcl-3) (7). We found that activated platelets, but not resting platelets, were responsible for the synthesized Bcl-3. The rapid and sustained accumulation was unexpected because platelets are considered incapable of regulated protein synthesis (8–10). We found that resting platelets contain mRNA for Bcl-3, and that, upon activation, they translate the message into protein via a signaling pathway that involves the phosphorylation of the eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) (11). After its synthesis Bcl-3 binds the Src-related tyrosine kinase, Fyn, in activated platelets, suggesting new functions for this protein in intracellular signaling. This, together with our findings that platelets synthesize other proteins when they are appropriately stimulated, identify new features of platelet biology that may be important in both hemostatic and pathologic conditions.
MATERIALS AND METHODS
Cell Isolation.
Washed platelets were isolated from whole blood as previously described (1). Total and differential cell counts indicated that final platelet suspensions contained 1–5 contaminating leukocytes per 10,000 platelets. Unless otherwise indicated, 2.5 × 108 platelets were used for each experimental point, a total that falls within the range of platelets found in 1 ml of whole blood (12). Platelets were stimulated with thrombin (Sigma). Lymphocytes and monocytes were isolated from whole blood by countercurrent elutriation as previously described (13). Lymphocyte and monocyte populations were greater than 90% pure. Neutrophils were isolated as previously described and were greater than 95% pure.
Western Blot Analysis.
Platelets were gently rocked alone or in the presence of thrombin for designated times. At the end of each time period, the suspensions were centrifuged and the supernatants were removed. The cell pellets were placed in SDS/PAGE reducing buffer, subjected to electrophoresis in a 9% SDS-polyacrylamide gel, and transferred to a polyvinylidene difluoride membrane. Western blot analysis was accomplished by using affinity-purified anti-Bcl-3 antibodies and secondary antibodies (Amersham). The anti-Bcl-3 antibody used in most studies was raised by using a synthetic peptide corresponding to amino acid residues 433–446 of human Bcl-3 (Santa Cruz Technology). Identical patterns of protein expression were found by using two other antibodies corresponding to amino acids 1–14 or residues 290–421 of human Bcl-3 (Upstate Biotechnology, Lake Placid, NY). Preincubation of the antibodies with the peptides they were raised against completely abolished Bcl-3 detection in activated platelets (not shown). In selected studies, platelets were pretreated for 2 hr with actinomycin D (1 mM), AG-490 (10 μM), cycloheximide (1 mM), LY294002 (10 μM), puromycin (1 mM), rapamycin (100 nM), staurosporine (10 μM), wortmannin (100 nM), or vehicle (dimethyl sulfoxide), and immunoreactive protein for Bcl-3 or 4E-BP1 was detected. The rabbit anti-serum against 4E-BP1 was kindly provided by John C. Lawrence, Jr., Department of Molecular Biology and Pharmacology, Washington University School.
Transient Transfections.
The cDNA for human Bcl-3 (7) was kindly provided by Timothy McKeithan (University of Chicago, Division of Biological Sciences) and cloned into the eukaryote expression vector pcDNA I/Amp (Invitrogen). One microgram of this construct or vector alone was transiently transfected into COS-7 cells using 6 μl of lipofectamine (Life Technologies, GIBCO/BRL, Gaithersburg, MD) per 35-mm well as indicated by the vendor. Cytoplasmic lysates were obtained 48 hr after transfection by using lysis buffer (50 mM Tris⋅HCl, pH 7.5/50 mM NaCl/1 mM EDTA/1 mM MgCl2/0.1% Triton X-100 supplemented with 1 mM phenylmethylsulfonyl fluoride, 20 mM NaF, 1 mM Na3VO4, 10 μg/ml of leupeptin, and 10 μg/ml of aprotinin). In selected experiments, the cellular lysates were incubated with glutathione S-transferase (GST) constructs (Santa Cruz Technology) consisting of GST alone, GST-Grb2 (amino acids 1–217 containing SH3-SH2-SH3 domains), or GST-Fyn [amino acids 145–247 containing the src homology 2 (SH2), amino acids 85–139 containing the src homology 3 (SH3), or amino acids 85–247 containing the SH2-SH3 domains of Fyn] with varying salt concentrations as previously described (14).
Immunocytochemical Procedures.
Immunocytochemical detection of Bcl-3 in platelets was performed as previously described (6,13) by using an ABC kit from Vectastain (Vector Laboratories) for alkaline phosphatase detection.
Metabolic Radiolabeling and Immunoprecipitation of Human Platelet Proteins.
Platelets (1 × 109) were placed in 1 ml of methionine-free M199 medium for 2 hr followed by the addition of 50 μCi (1 Ci = 37 GBq) of [35S]methionine (Amersham). After the addition of labeled methionine, the platelets were activated with thrombin or allowed to adhere to immobilized fibrinogen (21) in the absence of thrombin. Four hours later, the platelets were washed and lysed in buffer (described above) on ice for 30 min. The soluble platelet fraction was immunoprecipitated with anti-Bcl-3 antibody by using protein G-agarose beads. The beads were washed with lysis buffer, placed in 50 μl SDS/PAGE loading buffer, and boiled for 10 min to elute the immunoprecipitated 35S-labeled proteins. Coimmunoprecipitations of proteins from lysates of unlabeled platelets were also conducted by immunoprecipitating with anti-Fyn (Santa Cruz Technology) and blotting with anti-Bcl-3.
Reverse Transcription–PCR (RT-PCR).
Total RNA was isolated from purified human platelets (10 × 109) or leukocytes (1 × 107) by using 1 ml TRIzol reagent for 5 × 109 platelets or 5 × 106 leukocytes as indicated by the vendor before RT-PCR was performed. One microgram of total RNA served as a template for single-strand cDNA synthesis in a reaction by using Moloney murine leukemia virus reverse transcriptase (GIBCO/BRL) under conditions supplied by the manufacturer.
In Vitro Translation of Platelet RNA.
Total RNA was extracted from platelets as described above. Platelet RNA, at varying concentrations (50, 100, and 200 μg/ml), was then incubated with [35S]methionine in a wheat-germ extract translation system (Promega) for 120 min at 37°C. The samples were loaded onto a 9% SDS-polyacrylamide gel followed by autoradiography. A portion of the product from the in vitro translation reaction, where 200 μg/ml of platelet RNA was used, was immunoprecipitated with an antibody against Bcl-3 as described above. The immunoprecipitated proteins were eluted from protein-A beads by boiling in SDS-loading buffer and subsequently separated on a 9% SDS-polyacrylamide gel.
RESULTS AND DISCUSSION
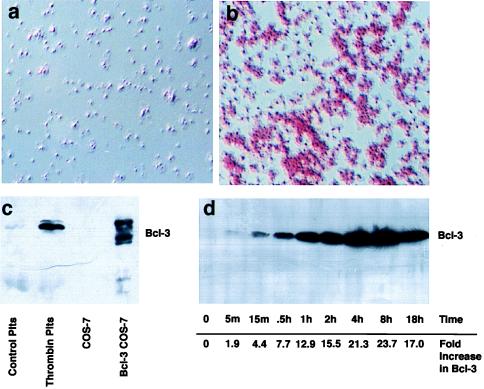
We examined Bcl-3 expression in purified platelets that were incubated in the presence or absence of thrombin. Bcl-3 was detected in platelets stimulated with thrombin, which activates them through a G protein-coupled receptor (2), but not in platelets incubated with buffer alone (Fig. 1). In contrast, thrombin did not induce Bcl-3 protein in purified monocytes or in preparations of other leukocyte types (Fig. 2a). Overexpression of Bcl-3 in COS-7 cells indicated that the protein detected in activated platelets was a hyperphosphorylated form (Fig. 1c), and kinetic studies demonstrated that activated platelets progressively accumulated Bcl-3 protein over time, suggesting it was being synthesized (Fig. 1d). By immunohistochemical analysis virtually all platelets stimulated with thrombin accumulated Bcl-3 (Fig. 1b), indicating that this is not restricted to immature cells or to another subset.
Figure 1.
Activated platelets express Bcl-3. (a and b) Immunocytochemical detection of Bcl-3. Bcl-3 was examined in resting (a) or thrombin-stimulated (b) (0.1 unit/ml for 2 hr) platelets as described in Materials and Methods. Red staining in b indicates the presence of Bcl-3 and is prominent in platelet aggregates. Rabbit IgG, or deletion of the primary antibody, did not result in red staining, indicating that the immunolocalization observed was specific for Bcl-3. (c) Detection of Bcl-3 by Western blot analysis in stimulated platelets and transfected cells. Cellular lysates from control or activated platelets (plts) stimulated with 0.1 unit/ml of thrombin for 2 hr, or from COS-7 cells transfected with vector or Bcl-3 cDNA, were obtained, and Bcl-3 expression (56-kDa protein) was determined by Western blot analysis. In transfected COS-7 cells (lane 4), the slower-migrating bands are phosphorylated forms of Bcl-3 protein that are concentrated in the cytoplasm whereas the faster-migrating band is not phosphorylated and concentrates in the nucleus (not shown). (d) Bcl-3 protein increases over time in platelets activated with thrombin. Platelets were stimulated with thrombin (0.1 unit/ml), and Bcl-3 expression was examined by Western blot analysis over 18 hr. Numbers at the bottom indicate the increase in Bcl-3 protein expression over baseline as measured by densitometry (m, minutes; h, hours). This figure is representative of three experiments.
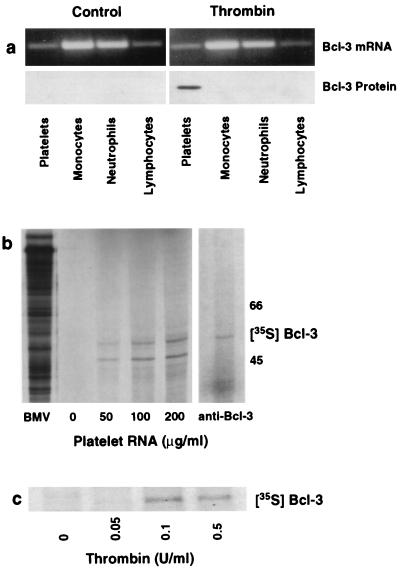
Figure 2.
Platelets synthesize Bcl-3 protein from preexisting mRNA. (a) Thrombin stimulates Bcl-3 protein expression in platelets but not in other blood cells. Platelets and leukocyte subpopulations were isolated, each cell population was exposed to thrombin (0.1 unit/ml) for 2 hr, and total RNA and protein were isolated for reverse transcription–PCR (Upper) and Western blot analysis (Lower). We found the same result in three experiments. (b) In vitro translation of total RNA from platelets results in Bcl-3 protein synthesis. (Left) Increasing concentrations of platelet RNA (μg/ml) were translated in vitro in the presence of [35S]methionine and subsequently examined by autoradiography. mRNA from Brome mosaic virus (BMV) served as a positive control. (Right) The [35S]methionine-labeled protein from platelet RNA translated in vitro was immunoprecipitated with an anti-Bcl-3 antibody as described in Materials and Methods. A second experiment yielded the same result. (c) Radiolabeled incorporation of [35S]methionine into Bcl-3 in platelets activated with thrombin. Platelets were preincubated with [35S]methionine and stimulated with increasing concentrations of thrombin, and lysates were incubated with an anti-Bcl-3 antibody after 4 hr. This figure is representative of three experiments.
De novo synthesis of Bcl-3 protein was unexpected because platelets do not contain nuclei or DNA other than that in mitochondria and are considered incapable of regulated protein synthesis (8–10). However, platelets retain mRNA derived from nucleated megakaryocytes (15) and it remains intact throughout the lifespan of the cell (16–17). We found that platelets contain mRNA for Bcl-3 (Fig. 2a Upper). It is also present in freshly isolated monocytes, lymphocytes, and neutrophils. Thrombin did not alter Bcl-3 mRNA levels in platelets or any of the leukocyte subsets, but specifically induced the synthesis of Bcl-3 protein in platelets (Fig. 2a Lower). In separate experiments, we also found that supernatants from thrombin-stimulated platelets did not induce Bcl-3 protein in leukocyte subsets. Moreover, when additional leukocytes (2.5 × 106 total) were added to platelet preparations, Bcl-3 protein expression was not enhanced (data not shown). These studies, along with Figs. 1b and 2a, demonstrate that Bcl-3 protein expression is platelet-specific. We confirmed that mRNA for Bcl-3 in platelets is functional by in vitro translation of total platelet RNA. Several [35S]methionine-labeled proteins were translated from platelet RNA (Fig. 2b Left). Immunoprecipitation of the translated products with an antibody against Bcl-3 yielded a protein of the expected size (Fig. 2b Right). These findings were confirmed in platelets by determining whether [35S]methionine was incorporated into Bcl-3 protein after thrombin stimulation. Anti-Bcl-3 did not immunoprecipitate protein from control platelets, but radiolabeled Bcl-3 was immunoprecipitated from platelets stimulated with increasing concentrations of thrombin (Fig. 2c). Together, these findings indicate that constitutive Bcl-3 mRNA can serve as a template for synthesis of the protein in platelets.
We next demonstrated that synthesis of Bcl-3 in platelets stimulated by thrombin occurs by a regulated pathway for protein translation. We found that puromycin, which causes the premature release of nascent polypeptide chains, markedly attenuated Bcl-3 expression (Fig. 3a). Cycloheximide, which blocks the translocation reaction on ribosomes, also inhibited Bcl-3 expression. In contrast, actinomycin D, a transcriptional inhibitor, did not inhibit Bcl-3 protein (not shown). We also found that rapamycin, a bacterially derived immunosuppressant that inhibits the translation of a specific subset of mRNAs (18), blocked Bcl-3 protein in thrombin-stimulated platelets (Fig. 3a). To date, only a small number of proteins are known to be synthesized by the rapamycin-sensitive pathway (18). Each is induced in response to mitogenic stimuli, and their mRNAs contain polypyrimidine-rich tracts and other features that impose extensive secondary structure on their 5′ untranslated regions and that consequently subject them to translational control that is sensitive to rapamycin inhibition (18). Bcl-3 has similar features. The 5′ untranslated region of Bcl-3 contains five stretches of pyrimidine tracts ≥4 bases (7). Further, the G/C-rich 5′ untranslated region is predicted to form extensive secondary structure (19) with an overall free energy (ΔG) of −76.1 kcal/mol, consistent with values predicted for mRNAs for proteins translated by the rapamycin-sensitive pathway (18). In addition, we found that inhibitors of phosphatidylinositol 3-kinase (PI 3-kinase), LY294002 and wortmannin, also blocked thrombin-induced Bcl-3 expression in platelets (not shown), indicating that this response likely is PI 3-kinase-dependent. This feature is characteristic of the rapamycin-sensitive translation control mechanism in other cells (18). In contrast, staurosporine, a protein kinase C inhibitor, and AG-490, a JAK2 inhibitor, did not alter Bcl-3 expression, suggesting that other kinase pathways in platelets reported to be activated by thrombin (20–21) do not regulate Bcl-3 expression (not shown).
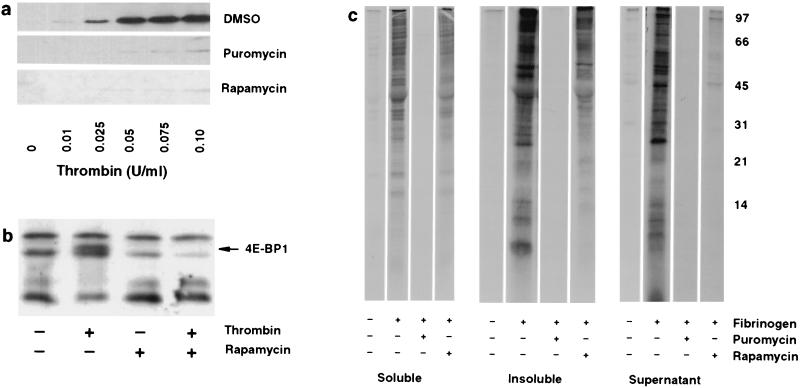
Figure 3.
Synthesis of Bcl-3 and other proteins is regulated by rapamycin in signaled platelets. (a) Inhibition of translation markedly attenuates Bcl-3 expression in platelets signaled by thrombin. Platelets were pretreated for 2 hr with dimethyl sulfoxide, puromycin, or rapamycin and activated with increasing concentrations of thrombin for 2 hr, and newly synthesized Bcl-3 was identified by Western blot analysis. (b) Rapamycin inhibits 4E-BP1 phosphorylation in activated platelets. Platelets were pretreated with rapamycin, and 4E-BP1, a protein that migrates in SDS/PAGE as multiple species (20–24 kDa), was identified by Western blot analysis 15 min after stimulation with thrombin (0.01 unit/ml). The solid arrow indicates a phosphorylated form of 4E-BP1 as described by Lin and Lawrence (11). a and b are representative of five experiments. (c) Platelets stimulated by adhesion to fibrinogen synthesize proteins. Platelets were pretreated with dimethyl sulfoxide, puromycin, or rapamycin. After pretreatment, they were incubated with [35S]methionine and allowed to adhere to immobilized fibrinogen for 8 hr. Supernatants were removed and the platelets were washed extensively with M199 buffer containing methionine, lysed, and fractionated into soluble and insoluble portions, and the labeled proteins were separated in a 15% SDS-polyacrylamide gel. This experiment is representative of three others.
Translational inhibition by rapamycin requires its association with mTOR (mammalian target of rapamycin, also called FRAP), a kinase that regulates the phosphorylation state and activity of the eukaryotic initiation factor 4E (eIF4E)-binding protein, 4E-BP1 (18, 22–24). 4E-BP1 is the mammalian homologue of rodent PHAS-I (25). Phosphorylation of 4E-BP1 induced by growth factors releases inhibition of the initiation factor complex and allows rapid translation and protein synthesis (25–27). We found that thrombin (0.01–0.1 unit/ml) induced 4E-BP1 phosphorylation in platelets as early as 5 min after stimulation (not shown). 4E-BP1 phosphorylation was maximal at 15 min and was eliminated by rapamycin pretreatment (Fig. 3b). Moreover, inhibition of PI 3-kinase blocked both 4E-BP1 phosphorylation and Bcl-3 synthesis (not shown), consistent with signaling via the rapamycin-sensitive pathway (18).
The principal mechanism for aggregation of stimulated platelets involves binding of fibrinogen to αIIb/β3 integrin (glycoprotein IIb/IIIa; GPIIb/IIIa), the most abundant integrin on the platelet surface (28). When platelets adhere to immobilized fibrinogen, outside–in signals are delivered through the αIIb/β3 heterodimer (29). We found a marked increase in [35S]methionine incorporation into proteins after adherence of platelets to fibrinogen (Fig. 3c). The labeling of proteins with [35S]methionine was completely abolished by puromycin, whereas rapamycin showed more selective inhibition consistent with its lesser impact on total protein synthesis in other cell types (18) (Fig. 3c). The importance of these data are threefold: first, they demonstrate that proteins besides Bcl-3 are synthesized by platelets when they are appropriately signaled; second, they demonstrate that platelet protein synthesis is regulated by both rapamycin-dependent and -independent pathways; third, they suggest that engagement of αIIb/β3 integrin signals translational events in platelets. In subsequent experiments, we have documented further the latter point (R.P., P. Bray, D.A.D., T.M.M., S.M.P., A.S.W., and G.A.Z., unpublished data).
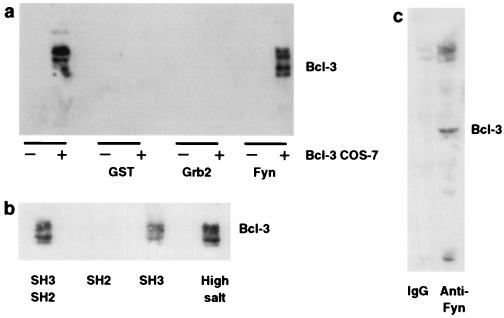
Bcl-3 was identified initially as a protooncogene that may contribute to leukemogenesis when expressed abnormally in lymphocytes (7). In vitro, it binds the Rel family members p50 and p52 and is thought to regulate transcription (30). Our finding that Bcl-3 is expressed in a regulated fashion in platelets, cells that do not transcribe mRNA, suggested that it has alternative functions. Furthermore, we did not find p50 or p52 in quiescent or activated platelets (not shown). Overexpression of Bcl-3 (31) and targeted disruption of Bcl-3 in mice (32–33) also support functions independent of regulation of p50 and p52. The Bcl-3 protein contains ankyrin repeats and proline-rich N and C termini—features indicative of protein–protein interactions. The proline-rich regions are potential targets for SH3 domains in binding partners. We examined Fyn and Grb2, two proteins with SH3 domains that are expressed abundantly in platelets (34–35). We found that a fusion protein containing the SH2-SH3 domain of Fyn, but not a Grb2 construct containing SH2-SH3, bound to phosphorylated and unphosphorylated forms of Bcl-3 that were overexpressed in transfected COS-7 cells (Fig. 4a). Bcl-3 bound the SH3, but not the SH2, domain of Fyn, and this interaction was strong enough to resist incubation in buffers containing high concentrations of salt (Fig. 4b). In addition, interaction between Bcl-3 and Fyn occurs in activated platelets because immunoprecipitation of endogenous Fyn with an anti-Fyn antibody, but not an isotype-matched antibody, yielded Bcl-3 (Fig. 4c). These results demonstrate that the magnitude of Bcl-3 accumulation in activated platelets, which appears to be quite high (see Fig. 1d), is sufficient for interaction with endogenous Fyn. Fyn is a tyrosine kinase that is thought to link intracellular signaling pathways in platelets (35) and other cell types, has been implicated in cytoskeletal organization, and is known to regulate PI 3-kinase by binding to its p85 subunit (36). It may be significant in the organization of intracellular signals that Bcl-3 and Fyn are respectively regulated by, and regulate, the PI 3-kinase pathway.
Figure 4.
Bcl-3 binds Fyn in transfected cells and activated platelets. (a) COS-7 cells, transfected with vector or Bcl-3 cDNA, were incubated alone or with agarose-conjugated fusion proteins consisting of GST, GST-Grb2, or GST-Fyn for 2 hr. After extensive washing, protein was eluted from the beads and Bcl-3 was detected by Western blot analysis. (b) Bcl-3 protein expression was detected in cellular lysates from COS-7 cells transfected with Bcl-3 cDNA and subsequently incubated with GST-Fyn fusion proteins that contained either its SH2-SH3 domains, the SH2 domain, or the SH3 domain. Binding of Bcl-3 to the GST-Fyn fusion protein containing both the SH2-SH3 domains was also examined under more stringent conditions (high salt = 400 mM, right lane). (c) Platelets were activated with thrombin (0.1 unit/ml for 2 hr), the cellular lysates were immunoprecipitated with an antibody against Fyn or mouse IgG, and Bcl-3 protein was detected by Western blot analysis as described in Materials and Methods. This figure is representative of four experiments.
Our findings that stimulated platelets produce Bcl-3 in a regulated fashion and that Bcl-3 associates with Fyn suggest new functions for Bcl-3 in intracellular signaling and that protein synthetic responses of platelets contribute to their physiologic and pathophysiologic actions. Because Bcl-3 synthesis is regulated at the translational level and is more delayed than initial platelet responses, it is unlikely that Bcl-3 regulates acute thrombogenic events. In this regard, it is more likely that Bcl-3 is involved in other prolonged inflammatory responses regulated by platelets, including angiogenesis, clot retraction, phagocytosis, and tissue repair and remodeling (37). It is also possible that regulated synthesis of Bcl-3 is a residual feature from megakaryocytes. If so, our experiments indicate that production of Bcl-3 by the rapamycin-sensitive pathway and its association with Fyn are important in this parent cell type. Furthermore, these studies establish that platelets are a unique model system for study of signal-dependent translation in a primary human cell type where the confounding variable of transcription does not exist.
Acknowledgments
We thank Kurt Albertine (University of Utah Health Sciences Center Research Microscopy Facility) for aid in microscopic studies, Diana Lim for preparing figures, and Donnie Benson, Ted Burke, Jeanne Falk, Ruth Ann Green, and Wenhua Li for excellent technical assistance. We also thank Neal Alto and Aaron Pontsler for their technical advice and insightful discussions, Spencer Galt, Larry Kraiss, Diane Lorant, and Vijay Modur for insightful discussions, Dr. Timothy McKeithan (University of Chicago, Division of Biological Sciences) for generously supplying a cDNA for Bcl-3, and Dr. John C. Lawrence, Jr. (Department of Molecular Biology and Pharmacology, Washington University School of Medicine) for providing the rabbit anti-serum against 4E-BP1. We also thank Dr. Raymond F. Gesteland for critical review of the manuscript. This work was supported by the Nora Eccles Treadwell Foundation and by individual grants from the National Institutes of Health (HL44525 to G.A.Z. and HL56713 to A.S.W.), the American Heart Association (Grant-in-Aid 96013280 to A.S.W.), and the office of Research and Development, Medical Research Service, Department of Veterans Affairs (to M.R.E.).
ABBREVIATIONS
- Bcl-3
B cell lymphoma-3
- eIF4E
eukaryotic initiation factor 4E
- SH2 and SH3
src homology 2 and 3, respectively
- 4E-BP1
eIF4E-binding protein 1
- PI 3-kinase
phosphatidylinositol 3-kinase
References
- 1.Elstad M R, McIntyre T M, Prescott S M, Zimmerman G A. Curr Opin Hematol. 1995;2:47–54. doi: 10.1097/00062752-199502010-00007. [DOI] [PubMed] [Google Scholar]
- 2.Coughlin S R, Vu T-K H, Hung D T, Wheaton V I. J Clin Invest. 1992;89:351–355. doi: 10.1172/JCI115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Packham M A. Can J Physiol Pharmacol. 1994;72:278–284. doi: 10.1139/y94-043. [DOI] [PubMed] [Google Scholar]
- 4.Savage B, Sladivar E, Ruggeri Z M. Cell. 1996;84:289–297. doi: 10.1016/s0092-8674(00)80983-6. [DOI] [PubMed] [Google Scholar]
- 5.Weksler B B. In: Inflammation: Basic Principles and Clinical Correlates. 2nd Ed. Gallin J I, Goldstein I M, Snyderman R, editors. New York: Raven; 1992. pp. 727–746. [Google Scholar]
- 6.Weyrich A S, Elstad M R, McEver R P, McIntyre T M, Moore K L, Morrissey J H, Prescott S M, Zimmerman G A. J Clin Invest. 1996;97:1525–1534. doi: 10.1172/JCI118575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohno H, Takimoto G, McKeithan T W. Cell. 1990;60:991–997. doi: 10.1016/0092-8674(90)90347-h. [DOI] [PubMed] [Google Scholar]
- 8.Soslau G. Arch Biochem Biophys. 1983;226:252–256. doi: 10.1016/0003-9861(83)90291-6. [DOI] [PubMed] [Google Scholar]
- 9.Siess W. Physiol Rev. 1989;69:58–178. doi: 10.1152/physrev.1989.69.1.58. [DOI] [PubMed] [Google Scholar]
- 10.de Groot P G, Sixma J J. Br J Haematol. 1990;75:308–312. doi: 10.1111/j.1365-2141.1990.tb04341.x. [DOI] [PubMed] [Google Scholar]
- 11.Lin T-A, Lawrence J C., Jr J Biol Chem. 1996;271:30199–30204. doi: 10.1074/jbc.271.47.30199. [DOI] [PubMed] [Google Scholar]
- 12.Isselbacher K J, Braunwald E, Wilson J D, Martin J B, Fauci A S, Kasper D L. Harrison’s Principle of Internal Medicine. 13th Ed. New York: McGraw–Hill; 1994. pp. 2493–2494. [Google Scholar]
- 13.Weyrich A S, McIntyre T M, McEver R P, Prescott S M, Zimmerman G A. J Clin Invest. 1995;95:2297–2303. doi: 10.1172/JCI117921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banin S, Truong O, Katz D R, Waterfield M D, Brickell P M, Gout I. Curr Biol. 1996;8:981–988. doi: 10.1016/s0960-9822(02)00642-5. [DOI] [PubMed] [Google Scholar]
- 15.Power C A, Clemetson J M, Clemetson K J, Wells T N C. Cyokine. 1995;7:479–482. doi: 10.1006/cyto.1995.0065. [DOI] [PubMed] [Google Scholar]
- 16.Booyse F M, Rafelson M E. Biochim Biophys Acta. 1967;145:188–190. doi: 10.1016/0005-2787(67)90673-9. [DOI] [PubMed] [Google Scholar]
- 17.Sottile J, Mosher D F, Fullenweider J, George J N. Thromb Haemostasis. 1989;62:1100–1102. [PubMed] [Google Scholar]
- 18.Brown E J, Schreiber S L. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 19.Kozak M. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ezumi Y, Takayami H, Okuma M. FEBS Lett. 1995;374:48–52. doi: 10.1016/0014-5793(95)01072-m. [DOI] [PubMed] [Google Scholar]
- 21.Iorio P, Gresele P, Stasi M, Nucciarelli F, Vezza R, Nenci G G, Goracci G. FEBS Lett. 1996;381:244–248. doi: 10.1016/0014-5793(96)00117-2. [DOI] [PubMed] [Google Scholar]
- 22.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Nature (London) 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 23.Chou M M, Blenis J. Curr Opin Cell Biol. 1995;7:806–814. doi: 10.1016/0955-0674(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 24.Keith C T, Schreiber S L. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 25.Proud C G. Nature (London) 1994;371:747–748. doi: 10.1038/371747a0. [DOI] [PubMed] [Google Scholar]
- 26.Lin T-A, Kong X, Haystead T A J, Apause A, Belsham G, Sonenberg N, Lawrence J C., Jr Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 27.Sonenberg N. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 245–269. [Google Scholar]
- 28.Clark E A, Shattil S J, Brugge J S. Trends Biochem Sci. 1994;19:464–469. doi: 10.1016/0968-0004(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 29.Haimovich B, Lipfert L, Brugge J S, Shattil S J. J Biol Chem. 1993;268:15868–15877. [PubMed] [Google Scholar]
- 30.Lenardo M, Siebenlist U. Immunol Today. 1994;15:145–147. doi: 10.1016/0167-5699(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 31.Caamano J H, Perez P, Lira S A, Bravo R. Mol Cell Biol. 1996;16:1342–1348. doi: 10.1128/mcb.16.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz E M, Krimpenfort P, Berns A, Verma I M. Genes Dev. 1997;11:187–197. doi: 10.1101/gad.11.2.187. [DOI] [PubMed] [Google Scholar]
- 33.Franzoso G, Carlson L, Scharton-Kersten T, Shores E W, Epstein S, Grinberg A, Tran T, Shacter E, Leonardi A, Anver M, et al. Immunity. 1997;6:479–490. doi: 10.1016/s1074-7613(00)80291-5. [DOI] [PubMed] [Google Scholar]
- 34.Horak I D, Corcoran M L, Thompson P A, Wahl L M, Bolen J B. Oncogene. 1990;5:597–602. [PubMed] [Google Scholar]
- 35.Law D A, Nannizzi-Alaimo L, Phillips D R. J Biol Chem. 1996;271:10811–10815. doi: 10.1074/jbc.271.18.10811. [DOI] [PubMed] [Google Scholar]
- 36.Prasad K V S, Janssen O, Kapeller R, Cantley L C, Rudd C E. Proc Natl Acad Sci USA. 1993;90:7366–7370. doi: 10.1073/pnas.90.15.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herd C M, Page C. Immunopharmacology of Platelets. San Diego: Academic; 1995. pp. 1–20. [Google Scholar]