Summary
The termini of linear chromosomes are protected by specialized DNA structures known as telomeres that also facilitate the complete replication of DNA ends. The simplest type of telomere is a covalently closed DNA hairpin structure found in linear chromosomes of prokaryotes and viruses. Bidirectional replication of a chromosome with hairpin telomeres produces a catenated circular dimer that is subsequently resolved into unit-length chromosomes by a dedicated DNA cleavage-rejoining enzyme known as a hairpin telomere resolvase (protelomerase). Here we report a crystal structure of the protelomerase TelK from Klebseilla oxytoca phage φKO2, in complex with the palindromic target DNA. The structure shows the TelK dimer destabilizes base pairing interactions to promote the refolding of cleaved DNA ends into two hairpin ends. We propose that the hairpinning reaction is made effectively irreversible by a unique protein-induced distortion of the DNA substrate that prevents re-ligation of the cleaved DNA substrate.
Introduction
The duplication of chromosome ends during replication is essential for genomic stability, and consequently, specialized replicative processes have evolved for this purpose. The termini of linear chromosomes pose challenges during replication because incomplete synthesis of DNA 5′ ends by the lagging strand of the replication fork could lead to progressive shortening of a chromosome over generations (Olovnikov, 1973; Watson, 1972). Also, the exposed ends of double-strand DNA are substrates for aberrant recombination or repair leading to chromosomal rearrangements and fusions (McClintock, 1941; Müller, 1938).
Several effective strategies have evolved to overcome the chromosome end replication problem. Bacterial and archaeal species with circular genomes have eliminated issues of exposed DNA ends, but instead must resolve interlinked (concatenated) chromosomes following replication by the action of topoisomerases and site-specific recombinases. The linear chromosomes of eukaryotes have guanine-rich repetitive DNA sequences near the ends (Szostak and Blackburn, 1982) that bind to protective proteins and adopt a compact structure that is resistant to degradation. GT-rich terminal sequences with a 3′ overhang can fold into structures such as the T-loop (Griffith et al., 1999) and possibly the G-quadruplex (Parkinson et al., 2002; Smith and Feigon, 1992) that together with site-specific binding proteins form specialized nucleoprotein complexes constituting telomeres (Baumann and Cech, 2001; Bilaud et al., 1997; Cooper et al., 1997; Smith and de Lange, 1997). The telomere provides physical protection of the DNA ends (van Steensel et al., 1998) while permitting extension of the telomeric sequence by the activity of a specialized reverse transcriptase to prevent chromosome shortening (Blackburn, 1991; Greider and Blackburn, 1985). Other types of terminal structures have also been identified in linear chromosomes. Drosophila telomeres are maintained by the insertion of a transposon sequence at chromosomal termini (Levis et al., 1993; Pardue and DeBaryshe, 1999). In streptomyces (Bao and Cohen, 2001; Hirochika and Sakaguchi, 1982; Lin et al., 1993), adenoviruses (Rekosh et al., 1977), and bacteriophages such as φ29 (Mellado et al., 1980), DNA synthesis is primed by a protein that remains covalently attached to the 5′-ends of the genome and protects the DNA from degradation.
An elegantly simple solution to the chromosome end problem is widespread among bacteria and viruses, including the plant pathogen Agrobacterium tumefaciens (Goodner et al., 2001), eukarytic brown algae viruses (Delaroque et al., 2001), linear plasmid prophage of several bacteriophages from Vibrio, Yersinia, Klebsiella, and E. coli (Casjens et al., 2004; Hertwig et al., 2003; Lobocka et al., 1996; Oakey et al., 2002; Rybchin and Svarchevsky, 1999), and genus Borrelia, the causative agent of the Lyme disease and relapsing fever (Casjens et al., 1997; Hinnebusch and Barbour, 1991; Hinnebusch et al., 1990). These organisms have linear chromosomes or replicons with covalently closed hairpin termini (telomeres). Here, bidirectional DNA replication starts from an internal origin (Picardeau et al., 1999; Ravin et al., 2003) and produces a circular dimer with inverted telomeric repeat sequences at the junctions between two copies of the replicated chromosome. This circular replication intermediate is then resolved into two unit length linear chromosomes with covalently closed DNA hairpin ends by a specialized telomere resolvase/protelomerase (Casjens, 1999; Kobryn and Chaconas, 2001; Ravin et al., 2001) (Figure 1A). Y-shaped intermediates that have been observed in electron micrographs of these chromosomes result from telomere resolution at one end of the chromosome dimer before the replication fork reaches the other end (Ravin et al., 2003).
Figure 1. Protelomerase resolves replicated hairpin telomeres.
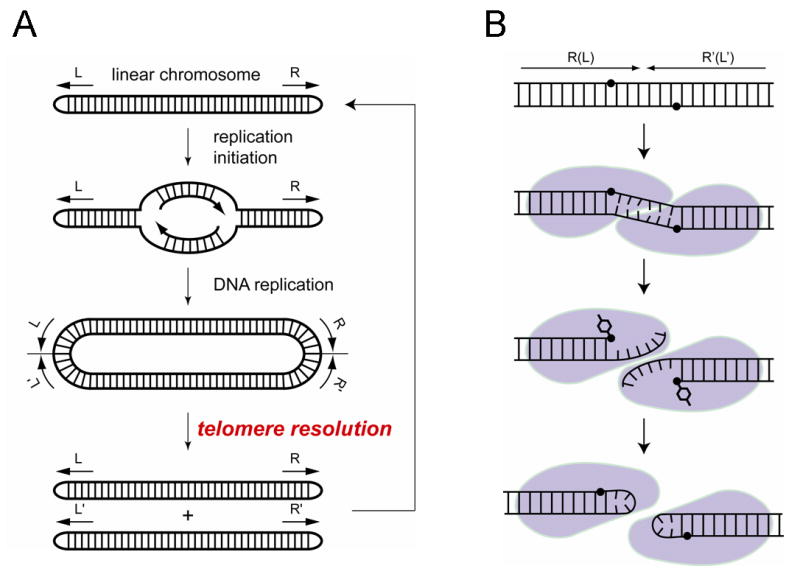
(A) Replication of a linear chromosome with hairpin telomeres produces a dimeric circular intermediate that is resolved into unit-length chromosomes by the activity of protelomerase. (B) A model for the hairpin formation reaction by the protelomerase TelK, proposed based on the crystal structure presented in this study.
Protelomerases share limited sequence homology with the λ-integrase family of site-specific recombinases (also known as tyrosine recombinases) and with type IB topoisomerases (Deneke et al., 2000; Kobryn and Chaconas, 2002; Rybchin and Svarchevsky, 1999). These enzymes have similar catalytic mechanisms for DNA cleavage and ligation, generating a 3′-phosphotyrosine DNA intermediate that enables the covalent rejoining of cleaved DNA strands without the use of a high-energy cofactor (Champoux, 1981; Craig and Nash, 1983; Huang et al., 2004; Pargellis et al., 1988). However, each family of enzymes performs a different DNA remodeling task using a monomer, dimer, or tetramer bound to one or two DNA molecules. The monomeric type-IB topoisomerases cleave and rejoin a single strand of DNA to relax supercoils (Champoux, 2001). Protelomerases bind as dimers to a double-stranded DNA and generate a staggered cut that is converted into two DNA hairpin ends (Figures 1B and 2). Site-specific recombinases catalyze the pair-wise exchange of four DNA strands in the context of a tetrameric recombinase bound to two recombining DNAs (Biswas et al., 2005; Chen and Rice, 2003; Van Duyne, 2001). Using a common reaction chemistry, these enzymes perform specialized DNA remodeling functions to achieve different biological outcomes.
Figure 2. Efficient resolution of the replicated telomere sequence by TelK.
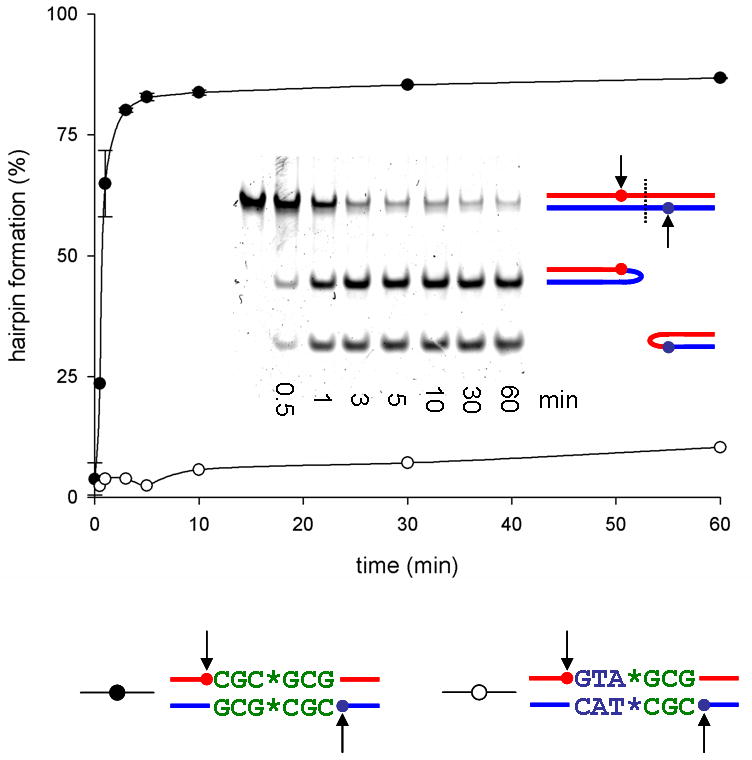
Resolution kinetics of 66bp oligonucleotide substrates into the hairpin products by TelK538. A substrate with the natural symmetric central sequence gets resolved efficiently (shown on the gel in the inset), whereas an artificially asymmetrized sequence blocks hairpin formation. Error bars denote standard deviation.
DNA hairpins are generated following an endonucleolytic strand cleavage as transient reaction intermediates during the remodeling of immunoglobulin and T-cell receptor genes, and during the transposition of some mobile genetic elements (van Gent et al., 1996). Enzymes catalyzing these reactions have specialized DNA binding elements that stabilize a hairpin fold during the direct attack of a nicked 3′-OH end on a phosphodiester bond (Davies et al., 2000; Rice and Baker, 2001). Protelomerases are unique in converting a doubly nicked DNA duplex into two stable hairpins via a transient enzyme-DNA covalent intermediate. This reaction sequence suggests a different mechanism from that of DNA transposases that form a hairpin starting from DNA with one nicked strand.
A key question concerning the mechanism of DNA hairpin formation by protelomerases is how the canonical double helical conformation of the DNA substrate is efficiently rearranged into a seemingly higher energy (enthalpically unfavorable) hairpin structure. To better understand the mechanism of this unique DNA cleavage and rejoining reaction, we have determined crystal structures of the protelomerase TelK from Klebseilla oxytoca phage φKO2 (Casjens et al., 2004; Huang et al., 2004), an essential factor for the maintenance of a linear plasmid prophage episome during lysogenic growth of φKO2. Our crystallographic and biochemical data suggest that TelK breaks the duplex structure of DNA to allow the spontaneous folding of individual DNA strands into hairpins.
Results and Discussion
Overview of the TelK Dimer Complexed with DNA
TelK was crystallized in complex with a nicked palindromic target DNA in which orthovanadate (VO43−) mimics the scissile phosphate of a pentavalent transition state for DNA cleavage (Table 1 and Figure 3) (Davies and Hol, 2004). We also crystallized covalent TelK-DNA complexes in the same overall conformation using suicide DNA substrates, representing the phosphotyrosine intermediate following DNA cleavage (Table S1 and Figure S1). The TelK dimer presents a series of DNA contacting surfaces that hold the 44bp DNA substrate in a curved conformation with a central discontinuity in the DNA helical axis (Figures 4A, 4B, and 5). The contact area between subunits is extensive, with the major interactions occurring between the N-terminal domains and between the catalytic domains (Figure 4). The DNA is completely enveloped by the protein, which gives the appearance of a relatively inflexible platform for holding a bound duplex DNA substrate in the strained conformation during the cleavage reaction.
Table 1.
Summary of the x-ray diffraction data, phasing, and model refinement statistics
| Dataset | Nicked suicide native | Selenium (two crystals merged) | ethylmercury phosphate | iodo-dU | Vanadate native | GTATAC native |
|---|---|---|---|---|---|---|
| space group | P41 | P41 | P41 | P41 | P41 | P41 |
| unit cell (Å) | a=b=157.07, c=91.56 | a=b=158.48, c=90.7 | a=b=157.32, c=90.83 | a=b=157.70, c=90.47 | a=b=157.97, c=90.84 | a=b=158.39, c=90.93 |
| wavelength (Å) | 0.97934 | 0.97934 | 0.97857 | 0.97934 | 0.97857 | 0.97857 |
| resolution (Å) | 50-3.2 (3.31-3.20) | 50-3.2 (3.31-3.20) | 50-3.2 (3.31-3.20) | 50-3.2 (3.31-3.20) | 50-3.2 (3.31-3.20) | 50-3.2 (3.31-3.20) |
| R-merge (%) | 11.3 (32.9) | 14.7 (38.8) | 10.0 (27.2) | 19.9 (46.8) | 13.0 (35.7) | 12.5 (44.6) |
| I/σ | 12.3 (5.2) | 12.3 (4.7) | 11.9 (4.4) | 6.7 (2.4) | 8.9 (2.9) | 11.6 (3.3) |
| Completeness (%) | 93.8 (93.9) | 99.9 (100) | 99.1 (99.0) | 90.4 (59.4) | 97.3 (94.3) | 100 (99.9) |
| Redundancy | 5.3 | 5.8 (anomalous) | 3.4 | 4.8 | 2.9 | 3.8 |
| Phasing (25-3.2Å) | ||||||
| Sites | 24 | 6 | 13 | |||
| Cullis_R (MLPHARE) | ||||||
| acentric / centric | 0.96 / 0.93 | 0.96 / 0.95 | 0.90 / 0.86 | |||
| anomalous | 0.92 | 0.99 | 1 | |||
| Figure of merit | 0.324 | |||||
| Refinement | ||||||
| resolution (Å) | 50-3.2 (3.23-3.20) | 50-3.2 (3.23-3.20) | 50-3.2 (3.23-3.20) | |||
| reflections - working set | 33034 (943) | 34529 (983) | 35637 (1004) | |||
| R-work (%) | 30.1 (40.9) | 25.5 (30.6) | 27.9 (38.6) | |||
| reflections - test set | 1739 (64) | 1680 (58) | 1753 (60) | |||
| R-free (%) | 33.6 (49.7) | 28.6 (35.3) | 30.4 (52.8) | |||
| rmsd bond/angle | 0.0091 / 1.60 | 0.0083 / 1.43 | 0.0092 / 1.56 |
Figure 3. Electron density for the TelK-DNA complex.
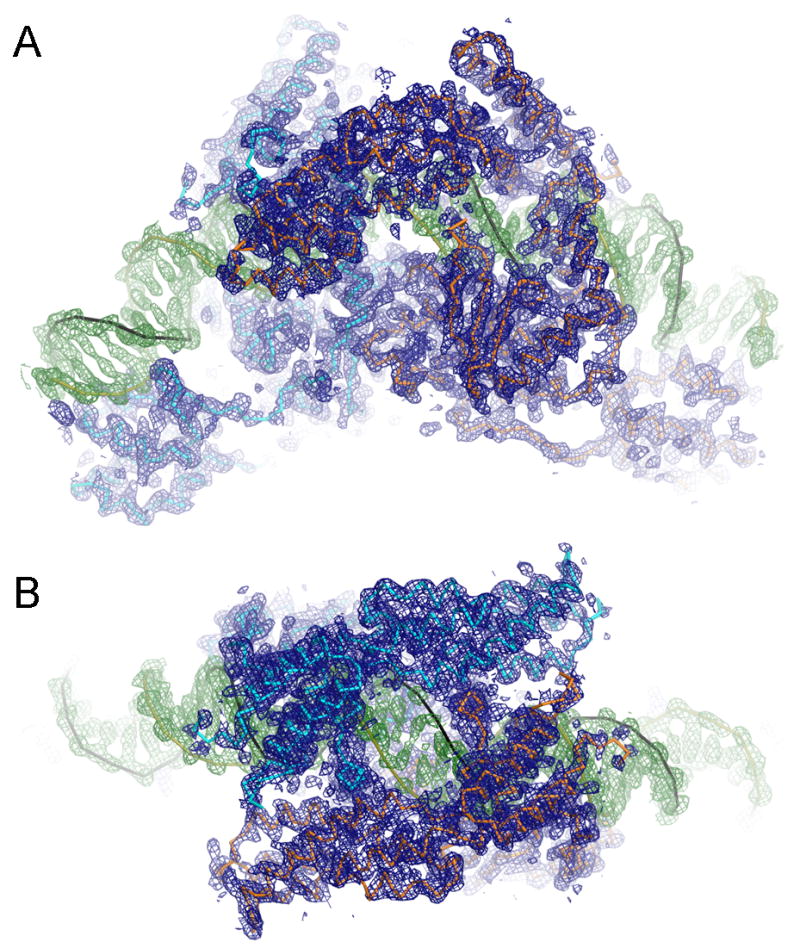
The simulated annealing composite omit map at 3.2 Å resolution for the TelK-DNA-vanadate complex that represents the intermediate of DNA cleavage reaction. The electron density within 2.5Å from atoms in the final model is shown at a contour level of 1σ above the mean. Densities for protein and DNA are colored in blue and green, respectively. (A) A view perpendicular to the 2-fold noncrystallographic symmetry axis that relates two TelK subunits. (B) Top view along the 2-fold axis.
Figure 4. Structure of the TelK dimer bound to DNA.
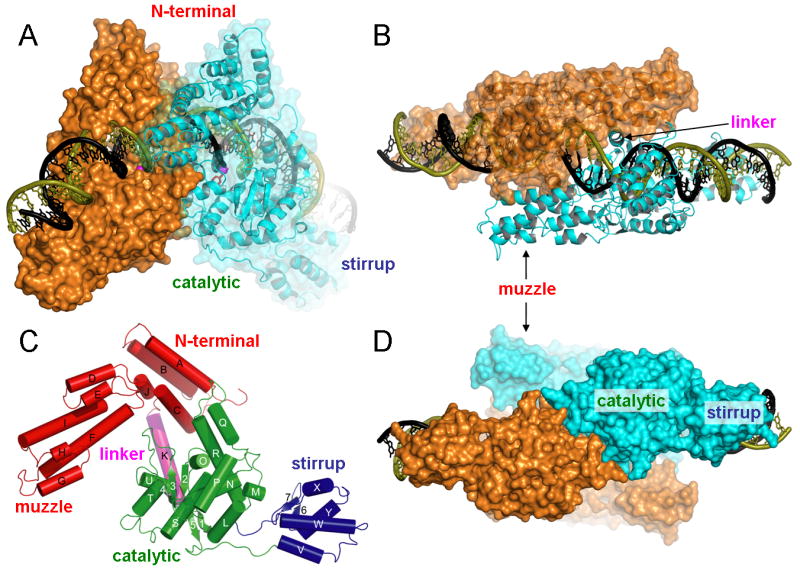
(A) A view perpendicular to the 2-fold noncrystallographic symmetry axis that relates two TelK subunits colored respectively in orange and cyan. (B) Top view along the 2-fold axis. (C) TelK monomer after a ∼45deg rotation around the vertical axis compared to the cyan monomer in (A), with individual protein domains color-coded. (D) Bottom view along the 2-fold axis showing extensive dimerization contacts between the two catalytic domains.
Figure 5. Distortion of the duplex DNA substrate induced by the binding of a TelK dimer.
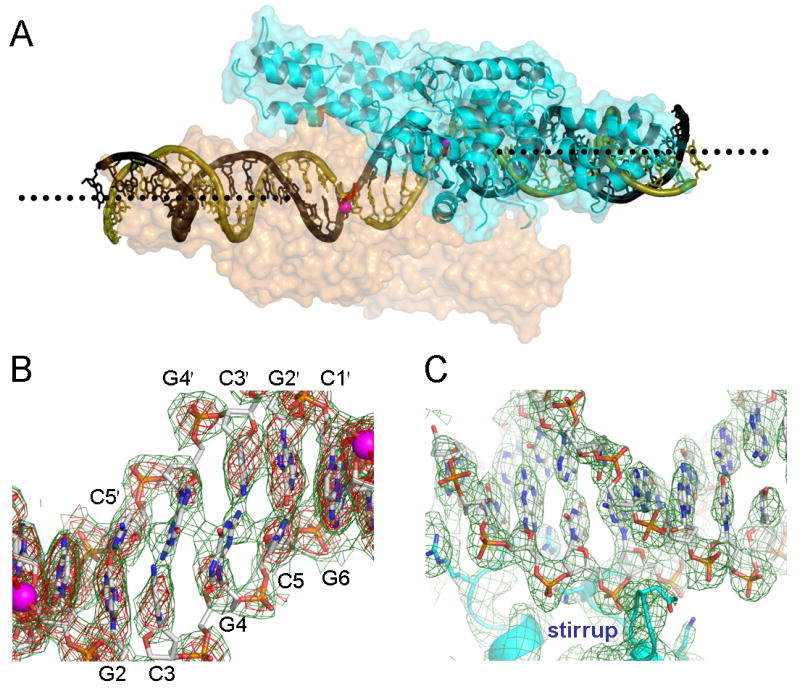
(A) A large offset in the DNA helical axis is highlighted by dotted lines showing the axes for each half site. (B) Base pairing interactions are severely compromised in the central region between the two scissile phosphates represented by magenta spheres. The unstacked Gua4 bases in turn participate in an inter-strand base stacking. A sigmaA weighted 2Fo-Fc omit map at 3.2Å resolution, contoured at 0.8σ (green) or 1.6σ (red) above the mean level, was computed with phases derived from a model that had never been refined in the presence of the central DNA residues Cyt1, Gua2, Cyt3, Gua4, and Cyt5 built for either strand. (C) The distal region of DNA bound by the stirrup domain is shown for comparison, with a simulated annealing composite omit map (1.0σ) superimposed.
Each TelK monomer has an oblong shape (approx.110 Å by 35 Å by 60 Å) that covers an entire DNA half-site, with a noncrystallographic dyad axis relating the two halves of the complex. The core of the TelK monomer consists of an N-terminal domain (colored red in Figure 4C) and a catalytic domain connected by a long α-helical linker (helix K) forming a two-domain architecture reminiscent of the tyrosine recombinases (Chen and Rice, 2003; Van Duyne, 2001) (MacDonald et al., 2006). However, there are two functionally important extensions of this basic architecture in TelK. An additional, C-terminal DNA binding domain named the stirrup (Figure 4C) is present only in TelK and is required for the resolution of hairpin telomeres (see below). TelK's N-terminal domain (a.k.a., the core binding domain; (Tirumalai et al., 1998)) has an insertion termed the muzzle, which contacts the opposing subunit and enforces an offset in the path of the DNA across the dimer interface (Figures 4B and 5).
The catalytic domain of TelK constitutes a large dimerization surface (Figure 4D; 1100 Å2 of buried surface area in each subunit), in contrast to catalytic domains of the tyrosine recombinases that communicate with neighboring subunits through an extended peptide segment (Chen and Rice, 2003; Van Duyne, 2001). Electrostatic interactions are abundant in the dimerization surface of TelK's catalytic domain—this interface includes 12 pairs of residues forming hydrogen bonds or salt bridges (Figure S3). The hydrophilic nature of the dimer interaction surface may-reflect the fact that TelK can exist as a monomer when not bound to DNA (Huang et al., 2004). Additional, less extensive contacts are made between the muzzle subdomains (residues 79-200; Figure 4) of subunits within the TelK dimer. Two long α-helices (αF and αI) form the core of this protruding element, which cradles the linker helix K of the opposite subunit (Figure 4B).
The TelK dimer shows an interlocked domain arrangement where the muzzle is packed against the DNA-binding toroid (catalytic and N-terminal domains) of the opposite subunit. This reciprocal interaction appears to reinforce the protein scaffold that clamps the bound DNA in a distorted conformation to promote the separation of cleaved DNA strands prior to hairpin formation. The DNA appears to be sequestered within the TelK dimer so that product release is likely to require a loosening of protein subunit interactions. Opening of the catalytic domain interface, or even a complete dissociation of the TelK dimer, may in fact be a prerequisite for the hairpin products formation itself. TelK dimer offers only a limited space across the two active sites for the pair-wise strand rearrangements (Figure S4), and could only accommodate two hairpin ends with an extremely compact conformation (Figure S5). However we cannot exclude the possibility that hairpin formation occurs in the context of the TelK dimer, and requires minimal structural change in the protein. How the DNA strand rearrangement process is coordinated with the dynamics of TelK dimer remains to be investigated.
DNA Interactions by TelK
TelK makes extensive interactions with the DNA substrate, burying a total of 6000 Å2 of the accessible protein and DNA surfaces per monomer. All three domains of TelK participate in DNA binding interactions, with as many as 55 protein residues positioned in close proximity to the DNA (<3.6Å; Figure 6A). Seven bases of the binding site are recognized by sequence-specific hydrogen bonding interactions, in addition to van der Waals contacts that involve additional bases of the binding site (Figure 6A). The N-terminal and catalytic domains of TelK bind to opposite faces of the DNA by inserting α-helices into the major groove (Figure 6B), and several basic residues from the α-helical linker (helix K) contact the DNA phosphodiester backbone. The side chain of Trp 219 within helix K packs in the major groove against the edges of 4 bases adjacent to the site of strand cleavage. The N-terminal domain inserts α-helix C into the major groove at a right angle with respect to the DNA axis. Asn67 and Ser68 from helix C make base-specific hydrogen bonding interactions with the DNA (Figure 6B). A basic N-terminal segment (residues 1-10) lacks well-defined electron density, but appears to interact with the minor groove, which is significantly widened in the intervening region between the DNA sites bound by the catalytic and stirrup domains. The catalytic domain inserts α-helix R into the major groove. Helix R is curved (∼45 degrees) and tracks along the major groove where it contributes three base-specific contacts from the side chains of Asn357 and Thr362, and the main chain carboxyl of Ala358 (Figure 6B).
Figure 6. DNA substrate recognition by TelK.
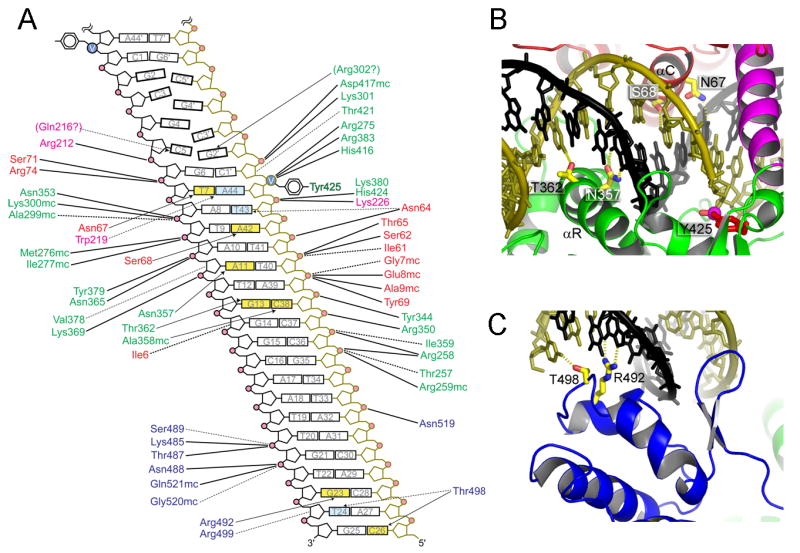
(A) A schematic diagram showing the DNA backbone and base-specific interactions. The protein residues are color-coded for each domain according to the scheme in Figure 4C and DNA backbones are colored to match the structural figures. DNA bases involved in hydrogen bonding interactions are colored yellow, and those involved in van der waals contacts are colored cyan. Solid and dashed lines denote electrostatic/hydrogen-bond and van der waals interactions, respectively. (B, C) Base-specific hydrogen bond interactions made by the core-binding and catalytic domains (B), and by the C-terminal stirrup domain (C).
Protein fold analysis by DALI (Holm and Sander, 1998) indicated that the catalytic domain of TelK is structurally most similar to that of λ-integrase (Kwon et al., 1997), except for a unique insertion (Figure 4C; helix Q and the N-terminal part of helix R, and the intervening loop; residues 335-352) that traverses the major groove opposite the linker helix (Figure S3) and completes the DNA-binding toroid formed by the N-terminal and the catalytic domains. One of the additional interactions made by this insertion involves Arg350, an essential residue for DNA nicking activity (Huang et al., 2004). An extended loop between β3 and β4 of TelK (residues 296-310), which is represented by a shorter connecting segment in the tyrosine recombinases, traverses along the minor groove and makes several interactions with the DNA backbone. Two side chains from this loop (Lys300 and Arg302) insert deep into the minor groove. Arg302 appears to be the only residue whose side chain is in a position for hydrogen bonding interactions with bases of the central 6 base pair region (Figure S4), raising the possibility of a direct role in hairpin formation. Although the N-terminal and catalytic domains of TelK completely encircle the DNA cleavage site, there are few interactions with the central six base pairs that ultimately form the hairpin products of telomere resolution.
TelK's C-terminal stirrup domain contacts the DNA flanking the cleavage site (Figures 4 and 5C), extending the DNA binding interface. This domain binds to DNA using the winged helix-turn-helix motif, similarly to the human Rap30 DNA-binding domain and linker histon H5 (Groft et al., 1998). The α-helical and β-hairpin segments of the stirrup straddle the major and minor grooves of DNA, respectively (Figures 4 and 6C). A short α-helix (helix X) the adjoining loop contribute Arg 492 and Thr 498 for base-specific hydrogen bonds and several other residues for DNA backbone interactions (Figure 6A). The stirrup is tethered to the catalytic domain by two extended linkers (Figure 4) and makes few interactions with the rest of the protein. The spatial configuration of the catalytic and the stirrup domains appears to be a major determinant for the orientation and extent of DNA bending.
Distorted DNA Conformation Induced by Binding of the TelK Dimer
The DNA bound by TelK is curved by ∼73 degrees within a plane parallel to the 2-fold axis of the complex (Figure 4A). In contrast, the related λ-integrase recombinase generates a sharp kink in DNA that breaks the internal two-fold symmetry within each half of the λ-integrase tetramer, which may bias the strand exchange reaction in favor of recombined products (Biswas et al., 2005). The DNA bend introduced by TelK preserves the two-fold symmetry of the protein dimer, perhaps reflecting the symmetry of the strand exchange reaction that generates two hairpins from one DNA duplex. The distributed bending of DNA by TelK may produce tension that is relieved by the double strand-nicking reaction, serving to liberate the ends of the DNA and preventing the reversal of the initial DNA cleavage reaction. An interaction between the stirrup domain and a distal DNA site appears to stabilize the bent conformation of the DNA (Figures 4A and 4D). Deletion of the stirrup domain by a C-terminal truncation of TelK has little effect on DNA cleavage activity, but profoundly impairs hairpin formation (Figures S6 and S7). This observation suggests that DNA bending is important for the refolding of cleaved DNA strands into hairpins.
The large amount of buried surface area that is devoted to DNA interactions and subunit contacts by the TelK dimer stabilizes a marked (∼7.5Å) shift in the DNA helical axis at the center of the complex (Figure 5A). This jog in the double helix is accompanied by the shearing of base pairing interactions within the central sequence (5′-CGCGCG-3′) of the cleaved DNA substrate (Figure 5B). The central bases are partially unstacked and exhibit poor hydrogen bonding geometries across the central two G-C pairs (Figures 5B and 6A). The adjacent G-C pairs are severely buckled, but otherwise remain within base pairing distance. It is notable that two guanines from opposite strands stack together across the dyad axis, instead of stacking on the adjacent cytosines within each strand. This interstrand base stacking might support destabilization of a canonical B-form duplex as a prelude to hairpin formation. The central base pairs are less well ordered in the crystal structures of TelK covalently complexed to suicide substrates (Table 1 and Figure S1), suggesting that the unligated 5′-OH ends of the cleaved DNA are not constrained after DNA cleavage.
The Active Site of TelK
The Tyr425 nucleophile for DNA cleavage is located on a partially buried helix (U) at the dimer interface where it appears to be held in a fixed orientation by subunit packing interactions. In contrast, the catalytic tyrosine residue of the tyrosine recombinases is typically located on a flexible segment that is subject to movement within the active site in order to regulate DNA cleavage activity (Chen and Rice, 2003; Van Duyne, 2001). The pentavalent vanadium ion bound in the active site of TelK coordinates the 3′-OH group of DNA, the tyrosine nucleophile and the juxtaposed 5′-OH group of DNA (Figure 7). The electron density, within the limitations of the x-ray data at modest resolution, is suggestive of a trigonal bipyramidal reaction intermediate with the coaxial alignment of the oxygen atom of the attacking tyrosine, the vanadium atom, and the oxygen atom of the 5′ leaving group. The structure supports the predicted catalytic mechanism of TelK, in which the two consecutive SN2-type transesterification reactions first form then break the phosphotyrosine protein-DNA linkage (Huang et al., 2004; Mizuuchi and Adzuma, 1991).
Figure 7. The active site of TelK.

(A) Vanadate moiety links 5′- and 3′- ends of DNA to the Tyr425 nucleophile, mimicking the pentavalent DNA cleavage intermediate. (B) Sequences around the catalytically important residues are shown for protelomerase (top), tyrosine recombinase (middle), and type IB topoisomerase (bottom) family of proteins. The aligned proteins are; protelomerases from Klebsiella oxytoca φKO2, Escherichia coli N15, Yersinia enterocolitica PY54, Ectocarpus siliculosus virus, Vibrio harveyi bacteriophage VHML, Agrobacterium tumefaciens, and Borrelia burgdorferi, phage λ integrase, phage P1 Cre, yeast 2μ plasmid Flp, Escherichia coli XerD, phage HP1 integrase, Human topoisomeraseI, Vaccinia virus topoisomerase, Minivirus topoisomerase, and Deinococcus radiodurans topoisomerase IB.
The active site residues of the protelomerase family of proteins show mixed features of the tyrosine recombinases and type IB topoisomerases (Figure 7). The scissile phosphate is coordinated by the basic side chains Arg275, Arg383, and His416 of TelK, corresponding to Arg488, Arg590, and His632 of the human topoisomerase I (Redinbo et al., 1998). TelK residues Arg275 and Arg383 are also structurally analogous to two arginine residues of the tyrosine recombinases constituting the catalytically important and extremely well conserved Arg-His-Arg triad (e.g., Arg212-His308-Arg311 of λ-integrase) (Nunes-Duby et al., 1998). Like all type IB topoisomerases, the active site of TelK has a Lys residue (Lys380) instead of the third catalytic triad histidine residue that has been proposed as a general base catalyst for the tyrosine recombinases (Whiteson et al., 2007). Lys380 (corresponding to a non-catalytic residue Lys587 of human topo I) contacts a phosphate immediately adjacent to the scissile phosphate. His416 of TelK takes the position of a less conserved His/Trp residue of the tyrosine recombinases and the catalytically important His (Asn in some bacterial enzymes) residue of the topoisomerase IB family of proteins (Stewart et al., 1998). The side chain of Lys300 (corresponding to the catalytic residue Lys532 of human topo I (Interthal et al., 2004)) is positioned between the DNA O5′ (axial) and a vanadate non-bridging (equatorial) oxygen, although not in an ideal hydrogen-bonding geometry with either oxygen. The amino group of this lysine residue might protonate the 5′-OH leaving group during DNA cleavage, as was suggested for Lys167 of the vaccinia topoisomerase (Krogh and Shuman, 2000). The active site residues in each half of the complex are contributed by one TelK monomer, showing that the DNA cleavage is catalyzed in cis and consistent with the ability to efficiently cleave a half-site substrate (Figure S6).
Mechanism of Hairpin Telomere Formation
Palindromic DNA sequences can spontaneously fold into hairpin structures, although extended duplexes are preferred except under conditions of low salt concentration and elevated temperature (Antao and Tinoco, 1992; Bonnet et al., 1998; Pieters et al., 1989; Senior et al., 1988; Sinden and Pettijohn, 1984; Wemmer et al., 1985). The thermodynamic stability of hairpin loops is highly dependent upon the sequence of the loop and adjacent base pairs in the duplex stem region. Structures of DNA and RNA hairpins show that a reversal of the polynucleotide chain is accompanied by an interruption of base pairing interactions at the distal end of the loop (Chou et al., 2003). Based on these findings, refolding of six base pairs at the center of the TelK target sequence should be energetically unfavorable under physiological conditions, with hairpin formation stabilized by at most two base pairs prior to ligation.
How does TelK effectively overcome the energetic barrier of converting a DNA duplex to a hairpin structure (Figure 2)? The enzyme bends the DNA (Figure 4A) and enforces an offset between half-sites (Figure 5A) that disrupts base pairing interactions between the DNA strands at the center of the binding site (Figure 5B). We propose that in this configuration the DNA is poised for spontaneous folding of the cleaved DNA strands into hairpin ends, which are subsequently captured by reaction with the 3′ phosphotyrosine to generate the covalently closed hairpin products. A modeling exercise suggests that within the physical constraints of the binding site, the apposing DNA hairpins of the product may experience electrostatic repulsion or steric clashes of extrahelical bases (Figure S5). This could provide a driving force for the physical separation of the nascent DNA hairpins, perhaps concomitant with dissociation of the TelK dimer, serving as a kinetic trap for the accumulation of product. Whether the dimer dissociates prior to the hairpin formation or the hairpin formation triggers the dimer dissociation, a favorable entropy of DNA cleavage may also contribute favorably to hairpin formation. It has been reported that tyrosine recombinases such as λ integrase and Flp form DNA hairpins as an aberrant reaction, on DNA substrates carrying mismatches between the top and bottom cleavage sites or on half-site substrates with a 5′-overhang (Nash and Robertson, 1989; Zhu et al., 1995). These observations suggest that following the staggered double-strand nicking, the DNA strands can spontaneously fold into hairpins given a driving force for the strand separation.
The crystal structure of the TelK-DNA complex does not support the reaction mechanism proposed for the Borrelia burgdorferi telomere resolvase (protelomerase) ResT (Bankhead and Chaconas, 2004), in which the DNA is extruded into a cruciform structure that is stabilized by the hairpin-binding motif of the enzyme prior to DNA cleavage. ResT binds specifically to DNA sequences flanking replicated telomere junctions, and cleaves DNA at different sites that appear to be determined by the distance from the center of the inverted repeat rather than the specific sequence of the junction region (Tourand et al., 2003). This observation implied that ResT may resolve a preformed DNA structure such as a cruciform or bulged hairpin loop located at the junction site. The amino acid sequence of ResT includes a DNA hairpin-binding motif with homology to the transposase Tn5 and related enzymes (Bankhead and Chaconas, 2004; Davies et al., 2000), supporting the hypothesis that a hairpin structure may be the target of DNA cleavage by ResT. This putative hairpin-binding motif, however, is not well conserved in TelK (Figure S8), raising the possibility of different reaction mechanisms for TelK and ResT.
The native target sites of all protelomerases are palindomic DNA sequences in which a perfect inverted repeat extends to the center of the dyad axis, which defines the apical ends of the hairpin products. The first and the sixth nucleotides from the scissile phosphates additionally must be complementary to one another, as they likely form the first base pair in the stem of the hairpin. We therefore examined how the symmetry of the central six base pairs of the target site affects DNA binding and cleavage by TelK. A substrate containing the nonpalindromic sequence 5′-GTAGCG-3′ in place of the native 5′-CGCGCG-3′ sequence was used to discourage hairpin formation. The DNA substrate with an asymmetric 5′-GTAGCG-3′ sequence is rapidly cleaved by TelK (Figure S6) even though hairpin products do not accumulate (Figure 2). Furthermore, half-site DNAs lacking a self-complementary sequence are also cleaved by TelK (Figure S6). These results show that an inverted repeat sequence in the hairpin forming region is not required for efficient DNA cleavage activity, lending strong support to a mechanism in which the enzyme interacts and cleaves a symmetrical duplex DNA before the ends of the cleaved DNA fold into hairpins.
The resolution of duplicated telomere sequences by TelK is remarkably efficient (Figure 2), and correspondingly, telomere fusion activity is undetectable in vitro even when the DNA hairpin product is present at high concentrations (not shown). We suggest that TelK dimer bound to an uncleaved duplex DNA is “spring loaded” to promote separation of cleaved DNA strands, and thereby enable spontaneous hairpin folding and stable capture by covalent ligation of the DNA (Figure 1B). In this context, one might expect TelK monomer to bind the hairpin product in preference to the duplex substrate, consistent with the observed low turnover of enzymatic activity (Huang et al., 2004). We previously proposed a model for phage λ recombination in which protein-DNA and protein-protein interactions shape the complex in a conformation that is competent for the final enzymatic step of recombination to propel the reaction forward (Biswas et al., 2005). A recent single-molecule study has indeed demonstrated that the λ-integrase forms a stable product complex that does not readily disassemble (Mumm et al., 2006). Protelomerases may similarly remain bound to the resolved chromosome termini until the next round of DNA replication, perhaps serving to protect the ends of chromosomes.
Experimental Procedures
Crystallization and structure determination
The TelK538 protein used in the present study includes 20-residue N-terminal His-tag followed by the TelK residues 1 to 538. The TelK538 construct that lacks ∼100 C-terminal residues of the full-length TelK (640 amino acids) shows a robust in vitro resolution activity (Figure 2). The last structured residue (Pro534) in the crystal structure of TelK538 is located in the linker joining the stirrup domain to the catalytic domain, and matches well with the native C-terminus of Borrelia burgdorferi telomere resolvase ResT (Figure S8). TelK538 was overexpressed in E. coli strain BL21 under the control of the arabinose PBAD promoter (Guzman et al., 1995), and was purified using the nickel-chelating and the Heparin column chromatography.
We employed three different strategies to obtain crystals of TelK in complex with a DNA substrate containing the palindromic target sequence. First, a nicked suicide DNA substrate (Huang et al., 2004; Nunes-Duby et al., 1987; Pargellis et al., 1988) was used to trap the covalent phosphotyrosine intermediate. The nick was introduced one base 3′ from the scissisle phosphate. The 5′-ends of DNA in the resulting TelK-DNA complex are one nucleotide shorter than the natural reaction intermediate because the nicked nucleotide was unable to remain with the cleavaged TelK-DNA complex and therefore the 5′-ends cannot attack the phosphotyrosine bond for subsequent ligation reaction. The second phosphotyrosine complex was trapped using a modified DNA substrate in which the nucleotide sequence between the two scissile positions is altered from 5′-CGCGCG-3′ to 5′-GTATAC-3′. The mutated DNA substrate is cleaved efficiently by TelK but, for unexplained reasons, is a very poor substrate for either hairpin product formation or religation to regenerate the substrate. This leads to accumulation of a stable phosphotyrosine intermediate that can be crystallized. A third TelK-DNA complex was assembled using orthovanadate and a DNA substrate lacking the scissile phosphates. All oligonucleotides were synthesized on the ABI394 synthesizer and were gel purified. The sequences of the oligonucleotides used in crystallization are available in supplementary table S1.
To prepare the phosphotyrosine complexes with cleaved DNA, the purified protein was mixed with 1.4x molar excess of the nicked or GTATAC suicide DNA substrates at an approximate protein concentration of 0.2mM, and was dialyzed against 10mM Hepes (pH7.2), 100mM NaCl, 150mM (NH4)2SO4, 0.5mM EDTA, 5mM DTT, and 10% glycerol. The covalent TelK-DNA complexes tended to precipitate under lower salt conditions, but could readily be resolublized by increasing the ionic strength. For the vanadate complex mimicking a DNA cleavage intermediate, the protein was mixed with a DNA substrate nicked at the scissile positions in the presence of 10mM sodium orthovanadate, and was dialyzed against the same buffer as above except that DTT was omitted to prevent reduction of vanadium(V).
Crystals of the TelK-DNA complexes were grown using the hanging drop vapor diffusion method at 22°C. The well solution consisted of 30% PEG4000, 50mM Tris (pH8.5), 10% glycerol, and 750mM (NH4)2SO4. The PEG phase of the phase-separated well solution was mixed in 1:1 volume ratio with the TelK-DNA complex to form the drops. Addition of β-NAD (nicotinamide adenine dinucleotide) into the drops at a concentration of 10mM improved the morphology of otherwise extremely thin needle crystals. The crystals were cryoprotected by increasing the glycerol concentration to 15% then flash frozen in a cold nitrogen steam for data collection. Selenomethionyl protein was expressed in minimal medium containing glycerol as the carbon source, using the metabolic inhibition method (Doublie, 2007). The Hg derivative was prepared by treating the crystal with 1mM ethyl mercury phosphate. For the iodine derivative, 5-iodo-dU was incorporated at multiple positions during the DNA synthesis.
All three types of TelK-DNA complexes were crystallized in the same crystal form under similar conditions. The structure of the first phosphotyrosine complex was determined by experimental phasing using selenium, mercury and iodine derivatives (Figure S1), and the models for the other two complexes were built according to difference fourier maps (summary of crystallographic data in Table 1). All three structures are essentially identical except for the base pairing interactions within the central region between cleavage sites.
All x-ray diffraction data were collected at the APS beamline 19ID (Argonne, IL). The data indexing, integration, and scaling were done using HKL2000 (Otwinowski and Minor, 1997). For the selenomethionine (SeMet) derivative, diffraction frames from two different crystals were merged and scaled together to obtain a single anomalous dataset. Further data processing including the structure factor calculation and scaling between different datasets was achieved by the CCP4 suite of programs (Collaborative_Computational_Project_Number_4, 1994). The automated Patterson search by SOLVE (Terwilliger, 2002) identified 24 selenium, 13 iodine, and 6 mercury sites. The programs SHARP (De LaFortelle and Bricogne, 1997) and MLPHARE were used for the heavy atom refinement and phase calculation, generating electron density maps of comparable quality after density modification with DM or RESOLVE (Terwilliger, 2002). The atomic model was built using O (Jones et al., 1991). Model refinement was performed using CNS (Brunger et al., 1998), imposing strict 2-fold noncrystallographic symmetry restraints throughout the process. The structure of the vanadate complex gave the best quality electron density (Figure 3) as well as model refinement statistics (R-work/R-free = 25.5/28.6% at 3.2Å resolution), thus was used in all figures to represent the structure of the TelK-DNA complex. Ramachandran analysis shows 78.6 % of the protein mainchain dihedral angles are in the most favored regions, 19.7% in additional allowed regions, 1.2% in generously allowed regions, and 0.4% in disallowed regions. Figures were produced using PYMOL (DeLano, 2002). Curves (Lavery and Sklenar, 1989) and 3DNA (Lu and Olson, 2003) were used for DNA geometry analyses.
Resolution and DNA cleavage assays
A 5′-fluorescein labeled 66bp oligonucleotide (5′-TCTTTAGCCCTATCAGCACACAATTGCCCATTATAXXXXXXTATAATGGACTATTGTGTGCTGGCG-3′) was annealed with an unlabeled complementary strand to form the duplex substrate in which the central 6bp XXXXXX is either 5′-CGCGCG-3′ or 5′-GTAGCG-3′, and italic nucleotides denote non-target sequences added to facilitate the size separation of the reaction products. The protelomerase reaction was performed at 22 °C with 50nM substrate DNA, 540nM TelK538, 83.3ng/μL λ-DNA or poly (dI-dC), 10mM Tris (pH7.5), 100mM NaCl, 0.5mM EDTA, 2mM DTT, and 10% glycerol. The presence of non-specific DNA, either phage λ-DNA or poly (dI-dC), was found to dramatically stimulate the resolution reaction although TelK538 is stable and remains fully active under the same buffer condition devoid of any DNA for longer than 5 hours. The reaction was quenched at each time point by phenol/chloroform extraction and the samples were analyzed on a 12% native acrylamide-TBE gel. The bands were quantified by detection of 5′-fluorescein using the Typhoon imager (GE healthcare). The gel was subsequently stained by SYBR green I nucleic acid stain (Invitrogen) to visualize both labeled and unlabeled hairpin products (shown in the inset of Figure 2).
The phosphotyrosine complex formation was monitored on 44bp nicked suicide substrates assembled from 24mer and 20mer oligonucleotides. 7.2μM TelK538 was incubated with 2x molar excess of substrate DNA in the same buffer condition used for the resolution reaction without non-specific DNA. The reaction was quenched at each time point by addition of SDS and the samples were analyzed by SDS-PAGE. Following the staining with coomassie blue, the band intensities were quantified by densitometry. All biochemical experiments were done in the protein and substrate DNA concentrations well above the apparent KD of TelK538 to the recognition sequence (∼17nM at 100mM NaCl), which was estimated using the fluorescence anisotropy experiment.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM59902 to TE) and a grant from the National Science Foundation (MCB-021324 to WMH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Coordinates for the TelK-DNA structure have been deposited in the Protein Data Bank under accession code 2V6E.
References
- Antao VP, Tinoco I., Jr Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead T, Chaconas G. Mixing active-site components: a recipe for the unique enzymatic activity of a telomere resolvase. Proc Natl Acad Sci U S A. 2004;101:13768–13773. doi: 10.1073/pnas.0405762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao K, Cohen SN. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 2001;15:1518–1527. doi: 10.1101/gad.896201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Bonnet G, Krichevsky O, Libchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc Natl Acad Sci U S A. 1998;95:8602–8606. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Casjens S. Evolution of the linear DNA replicons of the Borrelia spirochetes. Curr Opin Microbiol. 1999;2:529–534. doi: 10.1016/s1369-5274(99)00012-0. [DOI] [PubMed] [Google Scholar]
- Casjens S, Murphy M, DeLange M, Sampson L, van Vugt R, Huang WM. Telomeres of the linear chromosomes of Lyme disease spirochaetes: nucleotide sequence and possible exchange with linear plasmid telomeres. Mol Microbiol. 1997;26:581–596. doi: 10.1046/j.1365-2958.1997.6051963.x. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Gilcrease EB, Huang WM, Bunny KL, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J Bacteriol. 2004;186:1818–1832. doi: 10.1128/JB.186.6.1818-1832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. DNA is linked to the rat liver DNA nicking-closing enzyme by a phosphodiester bond to tyrosine. J Biol Chem. 1981;256:4805–4809. [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Chen Y, Rice PA. New insight into site-specific recombination from Flp recombinase-DNA structures. Annu Rev Biophys Biomol Struct. 2003;32:135–159. doi: 10.1146/annurev.biophys.32.110601.141732. [DOI] [PubMed] [Google Scholar]
- Chou SH, Chin KH, Wang AH. Unusual DNA duplex and hairpin motifs. Nucleic Acids Res. 2003;31:2461–2474. doi: 10.1093/nar/gkg367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative_Computational_Project_Number_4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Craig NL, Nash HA. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983;35:795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- Davies DR, Hol WG. The power of vanadate in crystallographic investigations of phosphoryl transfer enzymes. FEBS Lett. 2004;577:315–321. doi: 10.1016/j.febslet.2004.10.022. [DOI] [PubMed] [Google Scholar]
- De LaFortelle E, Bricogne G. Methods Enzymol. 1997;276:472–499. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002 http://wwwpymolorg.
- Delaroque N, Muller DG, Bothe G, Pohl T, Knippers R, Boland W. The complete DNA sequence of the Ectocarpus siliculosus Virus EsV-1 genome. Virology. 2001;287:112–132. doi: 10.1006/viro.2001.1028. [DOI] [PubMed] [Google Scholar]
- Deneke J, Ziegelin G, Lurz R, Lanka E. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc Natl Acad Sci U S A. 2000;97:7721–7726. doi: 10.1073/pnas.97.14.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2007;363:91–108. doi: 10.1007/978-1-59745-209-0_5. [DOI] [PubMed] [Google Scholar]
- Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, et al. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Groft CM, Uljon SN, Wang R, Werner MH. Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly. Proc Natl Acad Sci U S A. 1998;95:9117–9122. doi: 10.1073/pnas.95.16.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig S, Klein I, Lurz R, Lanka E, Appel B. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol Microbiol. 2003;48:989–1003. doi: 10.1046/j.1365-2958.2003.03458.x. [DOI] [PubMed] [Google Scholar]
- Hinnebusch J, Barbour AG. Linear plasmids of Borrelia burgdorferi have a telomeric structure and sequence similar to those of a eukaryotic virus. J Bacteriol. 1991;173:7233–7239. doi: 10.1128/jb.173.22.7233-7239.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch J, Bergstrom S, Barbour AG. Cloning and sequence analysis of linear plasmid telomeres of the bacterium Borrelia burgdorferi. Mol Microbiol. 1990;4:811–820. doi: 10.1111/j.1365-2958.1990.tb00651.x. [DOI] [PubMed] [Google Scholar]
- Hirochika H, Sakaguchi K. Analysis of linear plasmids isolated from Streptomyces: association of protein with the ends of the plasmid DNA. Plasmid. 1982;7:59–65. doi: 10.1016/0147-619x(82)90027-0. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WM, Joss L, Hsieh T, Casjens S. Protelomerase uses a topoisomerase IB/Y-recombinase type mechanism to generate DNA hairpin ends. J Mol Biol. 2004;337:77–92. doi: 10.1016/j.jmb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Interthal H, Quigley PM, Hol WG, Champoux JJ. The role of lysine 532 in the catalytic mechanism of human topoisomerase I. J Biol Chem. 2004;279:2984–2992. doi: 10.1074/jbc.M309959200. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kobryn K, Chaconas G. The circle is broken: telomere resolution in linear replicons. Curr Opin Microbiol. 2001;4:558–564. doi: 10.1016/s1369-5274(00)00251-4. [DOI] [PubMed] [Google Scholar]
- Kobryn K, Chaconas G. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol Cell. 2002;9:195–201. doi: 10.1016/s1097-2765(01)00433-6. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Shuman S. Catalytic mechanism of DNA topoisomerase IB. Mol Cell. 2000;5:1035–1041. doi: 10.1016/s1097-2765(00)80268-3. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Tirumalai R, Landy A, Ellenberger T. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- Lin YS, Kieser HM, Hopwood DA, Chen CW. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol Microbiol. 1993;10:923–933. doi: 10.1111/j.1365-2958.1993.tb00964.x. [DOI] [PubMed] [Google Scholar]
- Lobocka MB, Svarchevsky AN, Rybchin VN, Yarmolinsky MB. Characterization of the primary immunity region of the Escherichia coli linear plasmid prophage N15. J Bacteriol. 1996;178:2902–2910. doi: 10.1128/jb.178.10.2902-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. Structural basis for broad DNA-specificity in integron recombination. Nature. 2006;440:1157–1162. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado RP, Penalva MA, Inciarte MR, Salas M. The protein covalently linked to the 5′ termini of the DNA of Bacillus subtilis phage phi 29 is involved in the initiation of DNA replication. Virology. 1980;104:84–96. doi: 10.1016/0042-6822(80)90367-0. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K, Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991;66:129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- Müller HJ. The remaking of chromosomes. Collect Net Woods Hole. 1938;13:181–198. [Google Scholar]
- Mumm JP, Landy A, Gelles J. Viewing single lambda site-specific recombination events from start to finish. Embo J. 2006;25:4586–4595. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HA, Robertson CA. Heteroduplex substrates for bacteriophage lambda site-specific recombination: cleavage and strand transfer products. Embo J. 1989;8:3523–3533. doi: 10.1002/j.1460-2075.1989.tb08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Duby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Duby SE, Matsumoto L, Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987;50:779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Oakey HJ, Cullen BR, Owens L. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J Appl Microbiol. 2002;93:1089–1098. doi: 10.1046/j.1365-2672.2002.01776.x. [DOI] [PubMed] [Google Scholar]
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG. Drosophila telomeres: two transposable elements with important roles in chromosomes. Genetica. 1999;107:189–196. [PubMed] [Google Scholar]
- Pargellis CA, Nunes-Duby SE, de Vargas LM, Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988;263:7678–7685. [PubMed] [Google Scholar]
- Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- Picardeau M, Lobry JR, Hinnebusch BJ. Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol Microbiol. 1999;32:437–445. doi: 10.1046/j.1365-2958.1999.01368.x. [DOI] [PubMed] [Google Scholar]
- Pieters JM, Mellema JR, van den Elst H, van der Marel GA, van Boom JH, Altona C. Thermodynamics of the various forms of the dodecamer d(ATTACCGGTAAT) and of its constituent hexamers from proton nmr chemical shifts and UV melting curves: three-state and four-state thermodynamic models. Biopolymers. 1989;28:717–740. doi: 10.1002/bip.360280304. [DOI] [PubMed] [Google Scholar]
- Ravin NV, Kuprianov VV, Gilcrease EB, Casjens SR. Bidirectional replication from an internal ori site of the linear N15 plasmid prophage. Nucleic Acids Res. 2003:6552–6560. doi: 10.1093/nar/gkg856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin NV, Strakhova TS, Kuprianov VV. The protelomerase of the phage-plasmid N15 is responsible for its maintenance in linear form. J Mol Biol. 2001;312:899–906. doi: 10.1006/jmbi.2001.5019. [DOI] [PubMed] [Google Scholar]
- Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- Rekosh DM, Russell WC, Bellet AJ, Robinson AJ. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977;11:283–295. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- Rybchin VN, Svarchevsky AN. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol Microbiol. 1999;33:895–903. doi: 10.1046/j.1365-2958.1999.01533.x. [DOI] [PubMed] [Google Scholar]
- Senior MM, Jones RA, Breslauer KJ. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc Natl Acad Sci U S A. 1988;85:6242–6246. doi: 10.1073/pnas.85.17.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR, Pettijohn DE. Cruciform transitions in DNA. J Biol Chem. 1984;259:6593–6600. [PubMed] [Google Scholar]
- Smith FW, Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T. TRF1, a mammalian telomeric protein. Trends Genet. 1997;13:21–26. doi: 10.1016/s0168-9525(96)10052-4. [DOI] [PubMed] [Google Scholar]
- Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Blackburn EH. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982;29:245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallogr D Biol Crystallogr. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- Tirumalai RS, Kwon HJ, Cardente EH, Ellenberger T, Landy A. Recognition of core-type DNA sites by lambda integrase. J Mol Biol. 1998;279:513–527. doi: 10.1006/jmbi.1998.1786. [DOI] [PubMed] [Google Scholar]
- Tourand Y, Kobryn K, Chaconas G. Sequence-specific recognition but position-dependent cleavage of two distinct telomeres by the Borrelia burgdorferi telomere resolvase, ResT. Mol Microbiol. 2003;48:901–911. doi: 10.1046/j.1365-2958.2003.03485.x. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD. A structural view of cre-loxp site-specific recombination. Annu Rev Biophys Biomol Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Wemmer DE, Chou SH, Hare DR, Reid BR. Duplex-hairpin transitions in DNA: NMR studies on CGCGTATACGCG. Nucleic Acids Res. 1985;13:3755–3772. doi: 10.1093/nar/13.10.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteson KL, Chen Y, Chopra N, Raymond AC, Rice PA. Identification of a potential general acid/base in the reversible phosphoryl transfer reactions catalyzed by tyrosine recombinases: Flp H305. Chem Biol. 2007;14:121–129. doi: 10.1016/j.chembiol.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XD, Pan G, Luetke K, Sadowski PD. Homology requirements for ligation and strand exchange by the FLP recombinase. J Biol Chem. 1995;270:11646–11653. doi: 10.1074/jbc.270.19.11646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


