Abstract
STAT proteins play critical roles in the signal transduction pathways for various cytokines. The type I interferons (IFNα/β) promote the DNA-binding activity of the transcription factors STAT1, STAT2, and STAT3. Although the requirement for STAT1 and STAT2 in IFNα/β signaling and action is well documented, the biological importance of STAT3 to IFN action has not yet been addressed. We found that STAT3 plays a critical role in signal transduction by IFNα/β. A human cell line that is resistant to the antiviral and antiproliferative activities of IFN but is still IFN-responsive by virtue of STAT1 and STAT2 activation was found to be defective in STAT3 activation and in induction of NF-κB DNA-binding activity. Expression of STAT3 in these resistant cells complemented these signaling defects and also markedly increased cellular sensitivity to the antiviral and antiproliferative effects of IFN. Because STAT3 is involved in the induction of NF-κB DNA-binding activity and in the induction of antiviral and antiproliferative activity, our results place STAT3 as an important upstream element in type I IFN signal transduction and in the induction of biological activities. Therefore, our results indicate that STAT1 and STAT2 are not the only STATs required for the expression of the key biological activities of IFNα/β.
Interferons (IFNs) are cytokines that block the viral infection of cells, inhibit cell proliferation, and modulate cell differentiation. Type I IFNs (IFN α, β, and ω) compete with each other for binding to a common cell surface receptor, whereas the receptor for type II IFN (IFNγ) is a distinct entity (1). The type I IFN receptor is composed of IFNAR1 and IFNAR2 chains (2–4), which undergo rapid, ligand-dependent tyrosine phosphorylation. Although IFNAR2 is the ligand-binding subunit, IFNAR1 acts as a species-specific transducer for the actions of type I IFN (5–7). IFNs transduce signals from the cell surface resulting in selective gene activation (8–10) through the activation of JAK tyrosine kinases and signal transducers and activators of transcription (STAT) factors (11, 12). Upon their tyrosine phosphorylation, IFN-activated STATs (STAT1, STAT2, and STAT3) dimerize and translocate to the nucleus.
The crucial role that STAT1 and STAT2 play in the transcriptional response to IFNα/β and in the induction of antiviral activity has been demonstrated in knockout mice and in mutant human cell lines deficient in these proteins. However, the importance of STAT3 in IFNα/β action has been unresolved. For example, knockout of the STAT3 gene in mice leads to early embryonic lethality and STAT3-deficient cell lines could not be isolated (13). We recently reported that STAT3 acts as a bridge (adapter) for the IFN-dependent interaction of the IFNAR1 receptor chain and the regulatory 85-kDa (p85) subunit of phosphatidylinositol-3′ (PI-3) kinase (14). PI-3 kinase is important in the regulation of many cellular events involving protein tyrosine kinases and is an upstream element in a serine kinase transduction cascade (15, 16). Serine and tyrosine phosphorylation of various target proteins (STATs, IFNAR1, and IFNAR2 chains, etc.) are key early events in IFN signaling and action (17–20).
The IFN response pathway in IFN-sensitive and IFN-resistant Daudi lymphoblastoid cell lines has been studied in detail (6, 18, 21–26). Although both cell lines have similar numbers of IFNα/β-binding sites (21), IFN-resistant Daudi cells do not respond to the antiproliferative and antiviral actions of IFN. In contrast, IFN-sensitive cells are extremely sensitive to these biological actions (22). The IFN-resistant Daudi cells stably maintain their resistance to IFNα/β in the absence of added IFN (18, 21–24). IFN-resistant Daudi cells activate the JAK-STAT signaling pathway, as shown by their ability to undergo IFN-dependent tyrosine phosphorylation of the TYK2 and JAK1 tyrosine kinases and of the STAT1 and STAT2 transcription factors and to undergo IFN-stimulated gene transcription (6, 21, 24, 25). However, although IFN-resistant Daudi cells have detectable levels of STAT3, IFNα/β does not induce STAT3 tyrosine phosphorylation (26), suggesting that these cells either express a mutant form of STAT3 or they have a defect in the coupling between the IFN receptor and STAT3. These results led us to consider the possibility that the defect in IFN-resistant Daudi cells may reflect an inability to activate a STAT3-dependent signaling pathway. By demonstrating expression of wild-type STAT3 in IFN-resistant Daudi cells, we show that STAT3 is an important upstream element in IFNα/β signal transduction and in the induction of biological activities. Our results indicate that a STAT3-dependent signaling pathway is required for the expression of the key biological activities of IFN, besides the well characterized signaling pathway involving the activation of STAT1 and STAT2.
MATERIALS AND METHODS
Cells.
Human IFN-sensitive Daudi lymphoblastoid cells and an IFN-resistant subclone were maintained at 2.5–10 × 105 cells/ml in RPMI 1640 medium containing 10% defined bovine calf serum (HyClone). The 3′ end of the STAT3 cDNA was modified to incorporate the DEQKLISEEDL c-myc epitope sequence and was cloned into the pcDEF1 expression vector (27). The epitope-tagged STAT3 cDNA construct was transfected into the IFN-resistant cells by electroporation. As a control, IFN-resistant cells were electroporated with pcDEF1 vector alone. Transfected cultures were selected in medium containing 0.75 mg/ml geneticin (Life Technologies). After 7–10 days, individual clones were isolated by limiting dilution and tested for STAT3 expression by immunoreactivity with anti-myc (Santa Cruz Laboratories). Stable transfectants were maintained in medium containing geneticin.
IFN.
The recombinant human type I IFN, IFNCon1 (1 × 109 units/mg protein), was provided by Amgen Biologicals. IFN activities are expressed in international reference units/ml as assayed by protection against the cytopathic effect of vesicular stomatitis virus (VSV) on human fibroblasts, using the National Institutes of Health human IFNα standard for reference (21).
Nuclear Extracts and Gel-Shift Assays.
Nuclei were extracted with buffer containing 20 mM Tris⋅HCl (pH 7.85), 250 mM sucrose, 0.4 M KCl, 1.1 mM MgCl2, 5 mM 2-mercaptoethanol, and 0.4 mM phenylmethylsulfonyl fluoride (PMSF), and extracts were frozen on dry ice and stored at −80°C (28). For gel-shift analysis, the nuclear extracts were incubated with a [32P] end-labeled promoter probe for either the ISRE from ISG15 (5′-GATCCATGCCTCGGGAAGGGAAACCGAAACTGAAGCC-3′), the high-affinity sis-inducible element (SIE) from the c-fos gene (5′-AGCTTCATTTCCCGTAATCCCTAAAGCT-3′), or the κB site in the Ig light chain enhancer (5′-AGTTGAGGGGACTTTCCCAGG-3′) at 25°C for 20 min, and the free probe was separated from protein–DNA complexes on 5% polyacrylamide gels. For supershift assays, nuclear extracts were preincubated with anti-STAT (Santa Cruz Laboratories) or anti-Rel at 25°C for 20 min. Gels were quantitated by PhosphorImager autoradiography.
Immunoprecipitations and Immunoblot Analysis.
For immunoprecipitation studies, 1 × 108 cells were treated with IFN (5,000 units/ml) at 37°C for the indicated periods of time and then washed with ice-cold PBS and lysed for 20 min in lysis buffer (50 mM Tris, pH 7.4/150 mM NaCl/1 mM EDTA/0.5% Nonidet P-40/15% glycerol) containing 1 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 5 μg/ml soybean trypsin inhibitor, 5 μg/ml leupeptin, and 1.75 μg/ml benzamidine (12). Samples were centrifuged (12,000 × g, 15 min) at 4°C, and supernatants were immunoprecipitated with anti-STAT3 or anti-p85 (Transduction Laboratories) overnight at 4°C. Immune complexes were collected by using Protein A-Sepharose beads (Pharmacia) and eluted in sample buffer. Samples (equivalent to 1–2 × 107 cells) were run on SDS-7.5% polyacrylamide gels, transferred to polyvinylidene difluoride membranes (Millipore) and probed with the immunoprecipitating antibody or anti-pTyr mAb (Oncogene Science, Ab-2) followed by anti-mouse or anti-rabbit IgG coupled with horseradish peroxidase (Amersham). Blots were developed by using enhanced chemiluminescence (Amersham).
Antiviral and Antiproliferative Assays.
For determining antiviral activity, cell cultures (5 × 105 cells per ml) were preincubated overnight with IFNCon1, followed by infection with VSV (Indiana strain) for 1.5 hr at 0.1 plaque-forming units per cell. At 24 hr postinfection, the virus yield in the medium was assayed by plaque formation on indicator Vero cells (28). Assay of the antiproliferative action of IFN was performed by treating Daudi cells (5 × 104 cells per well of 24-well plates) with IFNCon1. After 3 days, the cells were enumerated in a Coulter counter (29).
RESULTS AND DISCUSSION
IFN Activates the JAK-STAT Signaling Pathway in IFN-Resistant Daudi Cells but Does Not Induce STAT3 Tyrosine Phosphorylation.
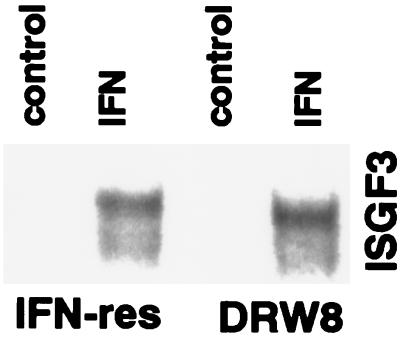
An important early event induced by IFN is the activation of the STAT1- and STAT2-containing IFN-stimulated gene factor 3 (ISGF3), which binds to the conserved IFN-stimulated response element (ISRE) present in the promoter of many IFN-responsive genes (10). Because ISGF3 induction provides direct evidence for the activation of the JAK-STAT signaling pathway, we determined the IFN-induced formation of the ISGF3 transcription factor in IFN-sensitive and IFN-resistant Daudi cells. Nuclear extracts were prepared from cells and incubated with a labeled ISRE probe, and the resultant protein–DNA complexes were analyzed by the gel-shift assay. Fig. 1 shows that IFN induced ISGF3 activity in IFN-resistant Daudi cells, demonstrating that IFN-resistant Daudi cells are capable of activating the JAK-STAT signaling pathway. In addition, the IFN-induced level of ISGF3 activity in IFN-resistant cells is similar to that observed in IFN-sensitive Daudi cells. The presence of STAT1 and STAT2 in the IFN-inducible ISGF3 DNA–protein complex was confirmed by gel supershift assays with STAT-specific antisera (Fig. 1). Further evidence for IFN activating the JAK-STAT pathway in IFN-resistant Daudi cells also was provided by our previous findings that IFN induces in IFN-resistant Daudi cells the expression of IFN-stimulated genes, such as ISG15, ISG54, and protein kinase C-ɛ (30). However, although IFN-resistant Daudi cells can activate the JAK-STAT signaling pathway and have detectable levels of STAT3, we previously established that IFN does not induce STAT3 tyrosine phosphorylation (31). In contrast, IFNα/β induces STAT3 tyrosine phosphorylation in IFN-sensitive Daudi cells. These results led us to consider the possibility that the defect in IFN-resistant Daudi cells may reflect an inability to activate a STAT3-dependent signaling pathway.
Figure 1.
The activation of ISGF3 protein–DNA complexes induced by IFN in IFN-resistant Daudi cells. Nuclear extracts were prepared from IFN-resistant (IFN-res) Daudi cells treated with IFNCon1 (5,000 units/ml) for 15 min and then subjected to EMSA with a 32P-labeled ISRE probe. Control cultures received no IFN. To validate the position of the ISGF3 complex (denoted by a ∗), nuclear extracts from IFN-treated cells were preincubated with anti-STAT1 or anti-STAT2 antibodies and then subjected to EMSA. For comparison, nuclear extracts also were prepared from IFN-sensitive (IFN-sen) Daudi cells treated with IFNCon1.
Expression of Wild-Type STAT3 in IFN-Resistant Daudi Cells Restores IFN-Dependent STAT3 and p85 Tyrosine Phosphorylation.
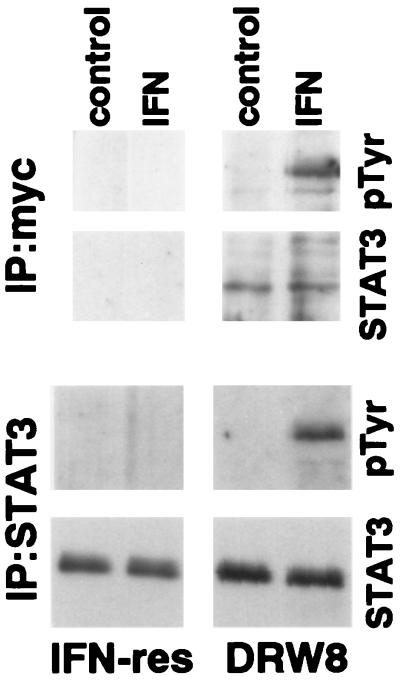
Wild-type STAT3 (with a myc epitope tag) was transfected into IFN-resistant Daudi cells, and stable clones were isolated to determine the role of STAT3 in IFNα signaling and action. Expression of the epitope-tagged STAT3 construct was confirmed by the presence of STAT3 in c-myc immunoprecipitates from the DRW8 STAT3 transfectant but not from untransfected IFN-resistant Daudi cells as shown in Fig. 2. In addition, the DRW8 STAT3 transfectant expresses more STAT3 than parental IFN-resistant Daudi cells (≈4-fold by quantitation of autoradiograms) as determined by immunoblotting. Most important, although STAT3 undergoes IFN-dependent tyrosine phosphorylation in the DRW8 transfectant as found in both c-myc and STAT3 immunoprecipitates, it does not in the parental IFN-resistant cells. Similarly, another independent STAT3 transfectant (DRW1) also undergoes IFN-dependent tyrosine phosphorylation of STAT3 (data not shown).
Figure 2.
IFN-dependent tyrosine phosphorylation of STAT3 in empty vector or wild-type STAT3 transfected IFN-resistant Daudi cells. Proteins from control or IFN-treated (5,000 units/ml for 15 min) cell lysates were precipitated with anti-myc (Upper) or anti-STAT3 (Lower). The proteins were resolved by SDS/PAGE, blotted onto polyvinylidene difluoride membranes, probed with anti-pTyr or anti-STAT3, and visualized by enhanced chemiluminescence.
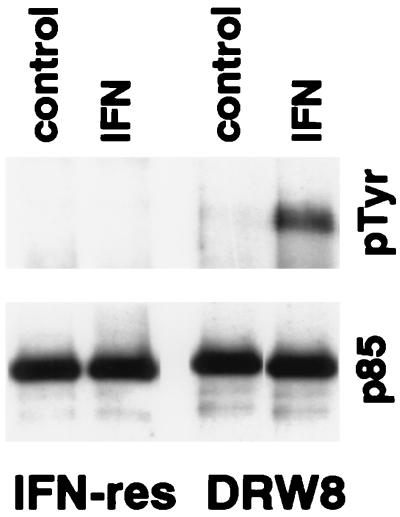
Based on our recent finding that STAT3 is involved in the IFN-dependent activation of PI-3 kinase (14), we predicted that the p85 regulatory subunit of PI-3 kinase would not undergo IFN-dependent tyrosine phosphorylation in IFN-resistant Daudi cells, which we indeed found (Fig. 3). However, although similar levels of p85 were immunoprecipitated from IFN-resistant Daudi cells and the DRW8 transfectant, p85 underwent IFN-dependent tyrosine phosphorylation in DRW8 cells expressing wild-type STAT3. This finding shows the requirement for STAT3 activation for the IFN-dependent tyrosine phosphorylation of p85 subunit of PI-3 kinase. This is particularly important because PI-3 kinase is an upstream element in a serine kinase transduction cascade (15, 16).
Figure 3.
IFN-dependent tyrosine phosphorylation of p85 in IFN-resistant Daudi cells requires wild-type STAT3 expression. Proteins from control or IFN-treated (5,000 units/ml for 15 min) cell lysates were precipitated with anti-p85. The proteins were resolved by SDS/PAGE, blotted onto polyvinylidene difluoride membranes, probed with anti-pTyr (Upper) or anti-p85 (Lower), and visualized by enhanced chemiluminescence.
Expression of Wild-Type STAT3 in IFN-Resistant Daudi Cells Restores the Formation of STAT3-Containing DNA–Protein Complexes.
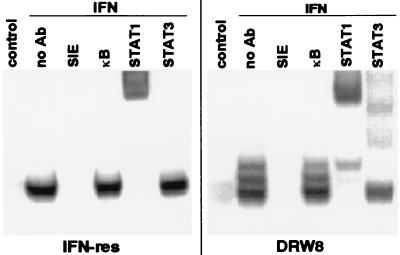
STAT3 is the transcription factor responsible for activation of the acute phase-response genes (32, 33). Its activation can be demonstrated by its presence in DNA–protein complexes by using an oligonucleotide probe for the SIE (14). The SIE is an epidermal growth factor and platelet-derived growth factor responsive element originally identified in the c-fos promoter, which contributes to c-fos transcription in vivo (34). This led us next to investigate whether DRW8 cells undergo IFN-dependent activation of STAT3-containing SIE-binding complexes. As illustrated in Fig. 4 and consistent with our previous findings (31), IFN only induces the fast-migrating STAT1/STAT1 homodimer in IFN-resistant Daudi cells, which is supershifted by anti-STAT1 but not by anti-STAT3. In contrast, IFN induces three SIE-binding complexes (a STAT1/STAT1 homodimer, a STAT1/STAT3 heterodimer, and a STAT3/STAT3 homodimer) in the IFN-treated DRW8 STAT3 transfectant, as demonstrated by supershift assays with anti-STAT1 and -STAT3. No DNA binding to the SIE was detected in the presence of excess unlabeled SIE oligonucleotide, and binding was not competed by excess of κB oligonucleotide, demonstrating that the binding to the SIE probe was specific. All three SIE-binding complexes also are formed in IFN-sensitive Daudi cells (31). Thus, transfection of wild-type STAT3 cDNA restored part of the IFN signaling pathway that is not functioning in IFN-resistant cells.
Figure 4.
The requirement for wild-type STAT3 expression in the activation of SIE-dependent protein–DNA complexes induced by IFN. Nuclear extracts were prepared from control and IFN-treated (5,000 units/ml for 15 min) cells, and then subjected to EMSA with a 32P-labeled SIE probe in the absence or presence of a 50-fold excess of unlabeled SIE or κB oligonucleotide probes. Nuclear extracts from IFN-treated Daudi cells were preincubated with anti-STAT antibodies and then subjected to EMSA to validate the positions of the STAT-containing complexes.
STAT3 Activation Is Upstream from IFN-Dependent NF-κB Activation.
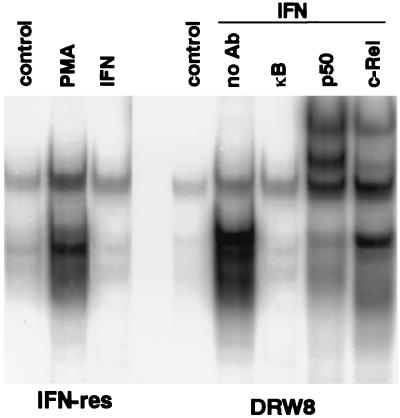
We have found recently that IFNα/β promotes DNA-binding activity of the NF-κB transcription factor in a variety of human cells (B and T cell lines, epithelioid carcinomas, fibrosarcomas, etc.) (L.M.P., S. R. Pfeffer, A.M., and C.-H.Y., unpublished data). The family of NF-κB transcription factors regulates the expression of a variety of genes involved in immune and inflammatory responses and cell growth, as well as viral genes (especially in HIV) by binding to cis-acting κB sites. Active NF-κB transcription factors are dimeric complexes, composed of various combinations of the Rel/NF-κB family of polypeptides. In addition, κB sites are found in several IFN-regulated genes, such as the IRF1 transcription factor, double-stranded, RNA-dependent protein kinase, intracellular cell adhesion molecule 1, HLA class I antigen, and IFNβ (35). As shown in Fig. 5, although IFN did not induce NF-κB gel-shift activity in IFN-resistant Daudi cells, IFN does induce NF-κB activity in the DRW8 STAT3 transfectant. Supershift assays with antisera specific for Rel proteins demonstrate the presence of c-Rel and p50 in the IFN-induced NF-κB gel-shift activity that is formed in DRW8 cells. These NF-κB complexes formed are indistinguishable from the complexes formed in treated IFN-sensitive Daudi cells (data not shown). The failure to induce NF-κB activation in IFN-resistant Daudi cells is IFN-specific and not a result of a general defect in this signaling pathway, because the phorbol ester PMA (phorbol 12-myristate 13-acetate) induces NF-κB activation in IFN-resistant Daudi cells (Fig. 5). These results indicate that STAT3 activation is upstream from induction of NF-κB and SIF DNA-binding activity. However, ISGF3 activation (STAT1/STAT2 dimers) in IFN-resistant cells is unaffected by STAT3 expression (Fig. 6), further demonstrating the selective effects of STAT3 on IFN signaling.
Figure 5.
The requirement for wild-type STAT3 expression in the activation of NF-κB protein–DNA complexes induced by IFN. Nuclear extracts were prepared from control, PMA-treated (100 nM for 4 hr), and IFN-treated (5,000 units/ml for 15 min) cells, and then subjected to EMSA with a 32P-labeled κB probe in the absence or presence of a 50-fold excess of unlabeled κB oligonucleotide probe. Nuclear extracts from IFN-treated Daudi cells were preincubated with anti-Rel antibodies and then subjected to EMSA to validate the positions of the NF-κB complexes.
Figure 6.
Wild-type STAT3 expression has no effect on the activation of ISRE protein–DNA complexes induced by IFN. Nuclear extracts were prepared from control and IFN-treated (5,000 units/ml for 15 min) cells, and then subjected to EMSA with a 32P-labeled ISRE probe. Nuclear extracts from IFN-treated Daudi cells were preincubated with anti-STAT antibodies and then subjected to EMSA to validate the position of the ISGF3 complex.
The Involvement of STAT3 in the Induction of Antiviral and Antiproliferative Actions of IFN.
We next examined the functional consequences of expression of STAT3 in IFN-resistant Daudi cells by assaying for sensitivity to IFN′s biological activities. Although IFNs inhibit the replication of a wide variety of RNA and DNA viruses, IFN-resistant Daudi cells are highly resistant to the antiviral action of IFN (5). Cells were treated overnight with IFNCon1 and infected with VSV, and the virus released into the culture medium at 1 day postinfection was titered by plaque assays (Fig. 7A). IFN (1,000 units/ml) induces a 250-fold reduction in viral titer in STAT3 transfectants (DRW1 and DRW8), whereas IFN induces only a 5-fold reduction in viral titer in IFN-resistant Daudi cells or an empty-vector transfectant (DRV). Furthermore, the dose-response curve for antiviral activity is shifted by >100-fold in STAT3 transfectants, showing that STAT3 expression leads to markedly increased antiviral sensitivity to IFN.
Figure 7.
The sensitivity of transfectants to the antiviral and antiproliferative action of IFN. (A) Transfectants were treated overnight with the indicated concentrations of IFNCon1. The cells were then infected with VSV, and the virus yield in the medium was determined by plaque formation. The results of three separate experiments were averaged and expressed as the fold reduction in VSV titer, which was set as 1 in untreated cells. The SEM was <20%. (B) Transfectants were treated with the indicated concentrations of IFNCon1. At day 3, cultures were counted in a Coulter counter. The results of three separate experiments performed in duplicate were averaged. The SEM was <15%.
Besides their broad antiviral action, IFNs also inhibit cell proliferation. Therefore, we extended our studies to determine whether wild-type STAT3 expression also increased sensitivity of IFN-resistant Daudi cells to IFN′s antiproliferative action. Cultures were treated with IFNCon1, and cell numbers were quantitated after 3 days. As shown in Fig. 7B, IFN treatment had little, if any, inhibitory effect on the proliferation of IFN-resistant Daudi cells or the empty vector transfectant (<3% inhibition at 1,000 units/ml). In contrast, two independent wild-type STAT3 transfectants (DRW1 and DRW8) showed a marked increase in the sensitivity to the antiproliferative action of IFN with an 8% inhibition at 10 units/ml and a >40% inhibition at 1,000 units/ml. Furthermore, the dose-response curve for antiproliferative activity also is shifted by >100-fold in STAT3 transfectants.
CONCLUSIONS
In summary, we provide evidence that STAT3 plays a critical role in signal transduction by IFNα/β as evidenced by its involvement in the induction of DNA-binding activity of the NF-κB transcription factor, as well as by STAT3’s ability to bind to genes that contain an SIE. Furthermore, STAT3 expression in an IFN-resistant cell line markedly enhances the sensitivity of the cells to antiviral and antiproliferative actions of IFNα/β. These data add an important new facet to the multiple roles of STAT3 in cell physiology and place STAT3 upstream of a number of IFN-activated signaling events. Furthermore, because STAT3 is activated by a variety of cytokines and growth factors (36–39), the STAT3-complemented cells will provide an important new tool to help define the role of this transcription factor in the signaling by these important cellular effectors. For example, in a recent preliminary study we found that, although epidermal growth factor failed to induce STAT3 tyrosine phosphorylation in IFN-resistant Daudi cells, epidermal growth factor induced STAT3 tyrosine phosphorylation in a STAT3 transfectant. This result shows that the STAT3 defect in these cells also is observed for epidermal growth factor signaling and thus is not IFN-specific, suggesting that IFN-resistant Daudi cells express a mutant form of STAT3.
Acknowledgments
We thank J. E. Darnell, Jr. (Rockefeller University), J. Ihle (St. Jude Children’s Research Hospital), and N. Rice (National Cancer Institute) for providing antibodies, and J. A. Langer (University of Medicine and Dentistry of New Jersey) for the pcDEF1 expression plasmid. This research was supported by National Institutes of Health Grant CA73753 (L.M.P.) and by funds from the Department of Pathology, University of Tennessee.
ABBREVIATIONS
- IFN
interferon
- STAT
signal transducers and activators of transcription
- PI-3 kinase
phosphatidylinositol-3′ kinase
- p85
85-kDa subunit of PI-3 kinase
- VSV
vesicular stomatitis virus
- SIE
sis-inducible element
- PMA
phorbol 12-myristate 13-acetate
References
- 1.Branca A A, Faltynek C R, D’Alessandro S B, Baglioni C. J Biol Chem. 1982;257:13291–13296. [PubMed] [Google Scholar]
- 2.Uze G, Lutfalla G, Gresser I. Cell. 1990;60:225–234. doi: 10.1016/0092-8674(90)90738-z. [DOI] [PubMed] [Google Scholar]
- 3.Novick D, Cohen B, Rubinstein M. Cell. 1994;77:391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 4.Domanski P, Witte M, Kellum M, Rubinstein M, Hackett R, Pitha P, Colamonici O R. J Biol Chem. 1995;270:21606–21611. doi: 10.1074/jbc.270.37.21606. [DOI] [PubMed] [Google Scholar]
- 5.Colamonici O R, Porterfield B, Domanski P, Constantinescu S N, Pfeffer L M. J Biol Chem. 1994;269:9598–9602. [PubMed] [Google Scholar]
- 6.Constantinescu S N, Croze E, Wang C, Murti A, Basu L, Mullersman J E, Pfeffer L M. Proc Natl Acad Sci USA. 1994;91:9602–9606. doi: 10.1073/pnas.91.20.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colamonici O, Yan H, Domanski P, Handa R, Smalley D, Mullersman J, Witte M, Krishnan K, Krolewski J J. Mol Cell Biol. 1994;14:8133–8142. doi: 10.1128/mcb.14.12.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman R L, Stark G R. Nature (London) 1985;314:637–639. doi: 10.1038/314637a0. [DOI] [PubMed] [Google Scholar]
- 9.Larner A C, Chaudhuri A, Darnell J E. J Biol Chem. 1986;261:453–459. [PubMed] [Google Scholar]
- 10.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 11.Schindler C, Shuai K, Prezioso V R, Darnell J E. Science. 1992;257:809–813. [PubMed] [Google Scholar]
- 12.Fu X-Y. Cell. 1992;70:323–335. doi: 10.1016/0092-8674(92)90106-m. [DOI] [PubMed] [Google Scholar]
- 13.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeffer L M, Mullersman J E, Pfeffer S R, Murti A, Shi W, Yang C H. Science. 1997;276:1418–1420. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- 15.Burgering B M T, Coffer P J. Nature (London) 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 16.Franke T F, Yang S-I, Chan T O, Datta K, Kazlaukas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer L M, Strulovici B, Saltiel A R. Proc Natl Acad Sci USA. 1990;87:6537–6541. doi: 10.1073/pnas.87.17.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeffer L M, Eisenkraft B L, Reich N C, Improta T, Baxter G, Daniel-Issakani S, Strulovici B. Proc Natl Acad Sci USA. 1991;88:7988–7992. doi: 10.1073/pnas.88.18.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reich N C, Pfeffer L M. Proc Natl Acad Sci USA. 1990;87:8761–8765. doi: 10.1073/pnas.87.22.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen Z, Zhong Z, Darnell J E., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 21.Pfeffer L M, Stebbing N, Donner D B. Proc Natl Acad Sci USA. 1987;84:3249–3253. doi: 10.1073/pnas.84.10.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeffer L M, Donner D B, Tamm I. J Biol Chem. 1987;262:3665–3670. [PubMed] [Google Scholar]
- 23.Pfeffer L M, Donner D B. Cancer Res. 1990;50:2654–2657. [PubMed] [Google Scholar]
- 24.Kessler D S, Pine R, Pfeffer L M, Levy D E, Darnell J E. EMBO J. 1988;7:3779–3783. doi: 10.1002/j.1460-2075.1988.tb03262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Constantinescu S N, Croze E, Murti A, Wang C, Basu L, Hollander D, Russell-Harde D, Garcia-Martinez V, Mullersman J E, Pfeffer L M. Proc Natl Acad Sci USA. 1995;92:10487–10491. doi: 10.1073/pnas.92.23.10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croze E, Russell-Harde D, Wagner T C, Pu H, Pfeffer L M, Perez H D. J Biol Chem. 1996;271:33165–33168. doi: 10.1074/jbc.271.52.33165. [DOI] [PubMed] [Google Scholar]
- 27.Goldman L A, Cutrone E C, Kotenko S V, Krause C D, Langer J A. BioTechniques. 1996;21:1013–1015. doi: 10.2144/96216bm10. [DOI] [PubMed] [Google Scholar]
- 28.Improta T, Pine R, Pfeffer L M. J Interferon Res. 1992;12:87–94. doi: 10.1089/jir.1992.12.87. [DOI] [PubMed] [Google Scholar]
- 29.Eisenkraft B L, Nanus D M, Albino A P, Pfeffer L M. Cancer Res. 1991;51:5881–5887. [PubMed] [Google Scholar]
- 30.Wang C, Constantinescu S N, MacEwan D J, Strulovici B, Dekker L V, Parker P J, Pfeffer L M. Proc Natl Acad Sci USA. 1993;90:6944–6948. doi: 10.1073/pnas.90.15.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang C H, Shi W, Basu L, Murti A, Constantinescu S N, Blatt L, Croze E, Mullersman J E, Pfeffer L M. J Biol Chem. 1996;271:8057–8061. doi: 10.1074/jbc.271.14.8057. [DOI] [PubMed] [Google Scholar]
- 32.Zhong Z, Wen Z, Darnell J E., Jr Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 33.Akira S, Nishio Y, Inoue M, Wang X-J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 34.Wagner B J, Hayes T E, Hoban C J, Cochran B H. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baeuerle P A, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 36.Campbell G S, Meyer D J, Raz R, Levy D E, Schwartz J, Carter-Su C. J Biol Chem. 1995;270:3974–3979. doi: 10.1074/jbc.270.8.3974. [DOI] [PubMed] [Google Scholar]
- 37.Boulton T, Zhong Z, Wen Z, Darnell J E, Jr, Stahl N, Yancopoulos G D. Proc Natl Acad Sci USA. 1995;92:6915–6919. doi: 10.1073/pnas.92.15.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong Z, Wen Z, Darnell J E., Jr Proc Natl Acad Sci USA. 1994;91:4806–4810. doi: 10.1073/pnas.91.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamanka Y, Nakajima K, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]