Abstract
Pectenotoxins (PTXs) are polyether macrolides found in certain dinoflagellates, sponges and shellfish, and have been associated with diarrhetic shellfish poisoning. In addition to their in vivo toxicity, some PTXs are potently cytotoxic in human cancer cell lines. Recent studies have demonstrated that disruption of the actin cytoskeleton may be a key function of these compounds, although no clarification their mechanism of action at a molecular level was available. We have obtained an X-ray crystal structure of PTX-2 bound to actin which, in combination with analyses of the effect of PTX-2 on purified actin filament dynamics, provides a molecular explanation for its effects on actin. PTX-2 formed a 1:1 complex with actin and engaged a novel site between subdomains 1 and 3. Based on models of the actin filament, PTX binding would disrupt key lateral contacts between the PTX-bound actin monomer and the lower lateral actin monomer within the filament, thereby capping the barbed-end. The location of this binding position within the interior of the filament indicates that it may not be accessible once polymerization has occurred, a hypothesis supported by our observation that PTX-2 caused filament capping without inducing filament severing. This mode of action is unique, as other actin filament destabilizing toxins appear to exclusively disrupt longitudinal monomer contacts allowing many of them to sever filaments in addition to capping them. Examination of the PTX-binding site on actin provides a rationalization for the structure–activity relationships observed in vivo and in vitro, and may provide a basis for predicting toxicity of PTX analogues.
Keywords: pectenotoxin, actin, crystal structure, anti-tumor, filament capping
Introduction
Pectenotoxins (PTXs) are a group of macrocyclic polyethers,1 the first members of which were isolated from the scallop Patinopecten yessoensis.2, 3 PTXs have since been identified in a wide range of shellfish and in algae of the genus Dinophysis from around the world.1, 4 Because many PTXs are toxic to mice by intraperitoneal injection in the standard mouse bioassay for diarrhetic shellfish toxins,2, 3, 5-9 the levels of some PTXs have been regulated in shellfish.10 Some PTXs also display potent cytotoxity in vitro, causing apoptosis and a range of other responses including effects on microtubules, stress fibres, and actin.11-14 This cytotoxicity has lead to investigation of the effects of PTXs on tumours and cancer cell lines.15, 16
To date, fourteen PTXs have been isolated and characterized (see Figure 1a for selected examples).1, 17 Their common structural features include a spiroketal group, three oxolanes, a bicyclic ketal, and a six-membered cyclic hemiketal. The main differences between these compounds involve the stereochemistry of the spiroketal group and the level of oxidation at C18, C32, C34 and C38. Pectenotoxin-2 (PTX-2) is suspected to be the precursor from which many PTXs are derived through biotransformation during metabolism in the gut of bivalves.
Figure 1. Chemical structures of pectenotoxins compared to that of reidispongiolide A and the X-ray crystallographic structure PTX-2 extracted from the PTX-2–actin crystal structure.
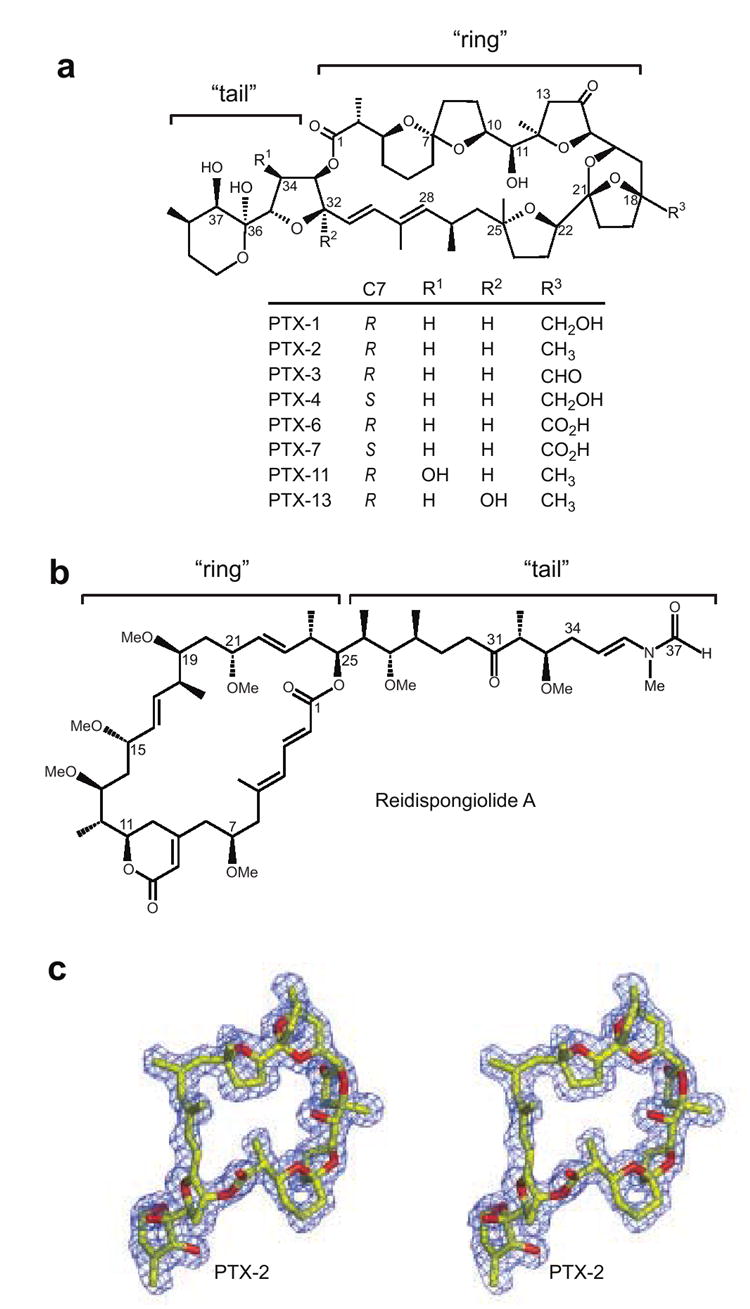
(a) The chemical scaffold of pectenotoxin is shown with eight different PTX analogues and their respective chemical modifications listed below. The overall structure has been divided into “ring” and “tail” structural components similar to other actin filament-destabilizing toxins.29, 51 (b) The chemical structure of reidispongiolide A. (c) Stereo view of the actin-bound conformation of PTX-2 is shown surrounded by experimental electron density. The Fo-Fc electron density omit map was contoured at 2.5σ. Figures 1b, 2, 3, 6, and 7 were prepared with Pymol.
Recent studies have indicated that some PTXs may exert some of their in vitro effects via the actin cytoskeleton. PTX-2 is reported to inhibit actin polymerization, to form a complex with G-actin,18-20 and PTX-6 caused depolymerization of actin in neuroblastoma cells and damaged the F-actin network in enterocytes from rabbit intestines.21, 22
Numerous natural cytotoxins target actin as a form of chemical defense for the host organism against predators or as a means of predation or communication.20, 23, 24 The broad importance of the actin cytoskeleton in eukaryotic cell function likely provides the basis for its selection as a target. The ability of these cytotoxins to interfere with actin cytoskeleton dynamics has allowed them to play important roles as probe molecules for chemical biology, and more recently their potential utility in the treatment of cancer has been recognized.20, 25, 26 For a number of these compounds, such as the reidispongiolides (Figure 1b), information about their interaction with actin and their mode of filament modification at a molecular level has been obtained. This allows predictions to be made about the effects design modifications may have on the pharmacological properties of natural and synthetic analogues in the interest of clinical utility.23 Translation of this information to PTXs is challenging because they bear little or no structural resemblance to any of the known forms of actin-binding small molecules and thus predicting the nature of their interaction with actin is difficult (Figure 1a).23
In order to understand the mechanism of action of PTXs and be able to provide a rationalization for the structure–activity relationships of different PTX isoforms we have determined the X-ray crystal structure of PTX-2 in complex with actin. The structure reveals that PTX-2 interacts with a site on actin that has not previously been observed to be the target of natural small molecules. Additionally, we have characterized the effects of PTX-2 on actin filament dynamics of purified actin filaments, which provides evidence that this toxin disrupts actin filaments by a unique mechanism. Based on these studies a molecular model for PTX-2-mediated filament destabilization has been generated that will be useful for understanding how variations in PTX structures will correlate with their bioactivity.
Results and Discussion
Overview of the PTX-2-Actin Complex
The structure of the Ca-ATP form of actin in complex with PTX-2 was solved by molecular replacement to a resolution of 1.7 Å. The crystallographic statistics are presented in Table 1. The electron density for PTX-2 is unequivocal and its absolute stereochemical configuration is consistent with the previously determined assignments based on X-ray crystallographic studies of the free PTX-1 molecule,3 and NMR studies of PTX-6 derivatives (Figure 1c).27 Crystals of actin with PTX-2 and latrunculin B bound were also obtained, which diffracted to nearly 1.4 Å, however no differences in the overall structures of actin or PTX-2 were observed. The basis for inclusion of latrunculin B in our PTX-2–actin co-crystallization was that it improved the overall size and quality of the crystals relative to those with PTX-2 only.
Table 1.
Data collection and refinement statistics
| PTX-2 only | PTX-2 + LatB | |
|---|---|---|
| Data collection | ||
| Space group | C2 | C2 |
| Unit cell dimensions | ||
| a, b, c (Å) | 59.4, 56.9, 105.7 | 57.7, 54.3, 105.9 |
| α, β, γ(°) | 90,90.16,90 | 90,90.27,90 |
| Resolution (Å) a | 45-1.7 (1.76-1.7) | 40-1.41 (1.46-1.41) |
| Observed reflections | 157276 | 282080 |
| Unique reflections | 37735 | 60883 |
| Redundancy a | 4.2 (3.9) | 4.6 (4.1) |
| Average I / σI a | 33.3 (9.5) | 28.5 (5.7) |
| Completeness (%) a | 97.2 (93.4) | 96.6 (84.9) |
| Rmerge (%) a | 4.0 (10.9) | 4.0 (13.6) |
| Refinement | ||
| No. reflections b | 35865 | 53998 |
| Rwork / Rfree c | 0.168/0.204 | 0.165/0.181 |
| Number of atoms | ||
| Actin | 2780 | 2717 |
| Pectenotoxin-2 | 61 | 61 |
| Latrunculin B | - | 27 |
| CaATP | 32 | 32 |
| Solvent | 386 | 357 |
| B-factors (Å2) | ||
| Actin | 16.0 | 15.2 |
| Pectenotoxin-2 | 16.9 | 15.1 |
| Latrunculin B | - | 11.7 |
| CaATP | 9.9 | 8.6 |
| Solvent | 25.8 | 22.9 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.012 | 0.008 |
| Bond angles (°) | 1.772 | 1.678 |
| Ramachandran plot (%) | ||
| Most favored | 95.8 | 95.4 |
| Additionally allowed | 4.2 | 4.6 |
Data for each complex was collected from a single crystal.
Data in parentheses represent highest resolution shell.
The number of reflections used during refinement.
, where Rwork refers to the Rfactor for the data utilized in the refinement and Rfree refers to the Rfactor for 5% of the data that were excluded from the refinement.
Contrary to previous stoichiometric analyses indicating that PTX-2 forms a 1:4 complex with G-actin,18 the crystallographic data reveals a 1:1 complex in the asymmetric unit (Figure 2a). PTX-2-bound actin assumes the ‘closed’ conformation 28 that is typically observed for other actin complex structures (RMS deviation < 1.0 Å). Surprisingly, though the general shape of PTX-2 contains a “ring” and “tail” moiety (Figure 1a), which is common to numerous filament-destabilizing toxins (Figure 1b), PTX-2 interacts with a region of actin that has not previously been observed to be a target site for filament-destabilizing small molecules. While PTX-2 binds between subdomains 1 and 3 and is near the barbed end (Figure 2a), the PTX-binding site is on the opposite face of actin relative to the site occupied by the trisoxazole, reidispongiolide/sphinxolide, and aplyronine families of filament-destabilizing marine toxins (Figure 2b).
Figure 2. The PTX-2–actin complex.
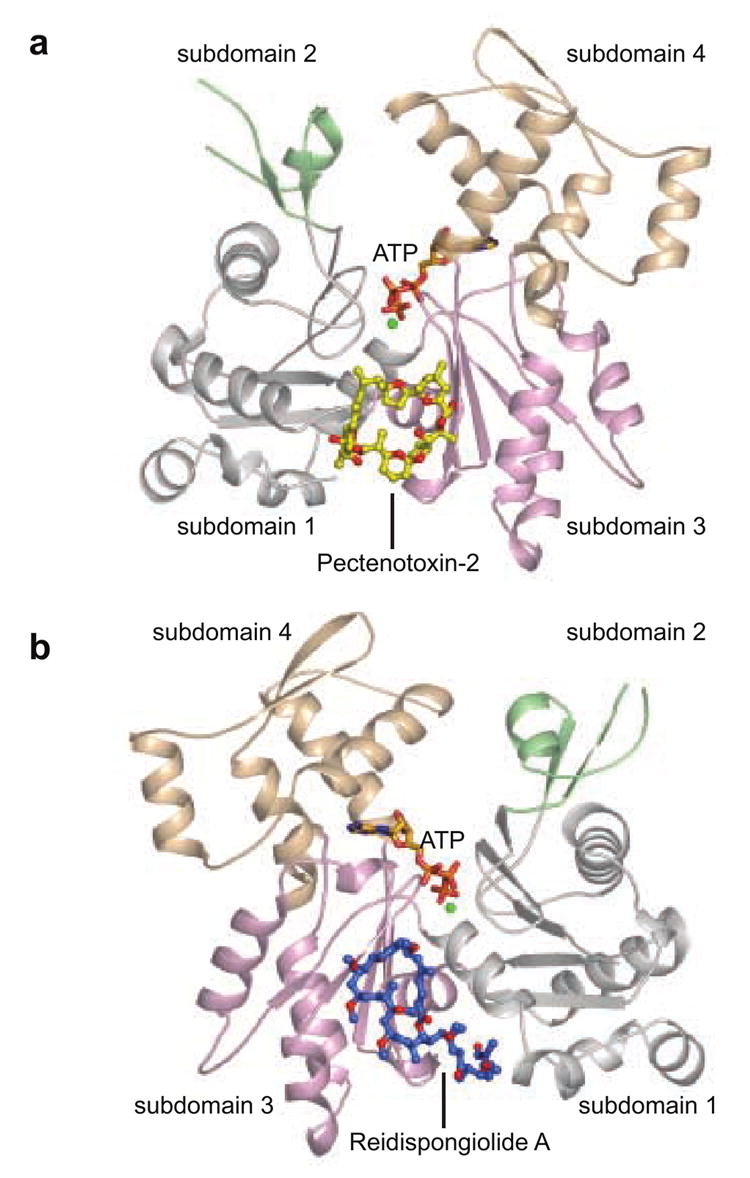
(a) PTX-2 (yellow) is shown as a ball-and-stick representation bound to actin between subdomains 1 and 3. (b) The structure of the reidispongiolide A–actin complex 29 is shown for reference and is rotated 180° along the vertical axis from the orientation of the PTX-2–actin complex in (a). Reidispongiolide A is shown as blue ball-and-sticks. In this figure subdomains 1, 2, 3 and 4 of actin are colored in light gray, green, wheat, and light pink respectively. This color scheme is used throughout the figures to depict the subdomains of actin.
The Binding Interface
Binding of PTX-2 to actin buries 1103.4 Å2 of surface area where 31% of the PTX-2 molecular surface area becomes inaccessible. The ring portion of PTX-2 lies flat within a shallow groove between subdomains 1 and 3, and its interaction with actin is primarily hydrophobic in nature. The ring is centered above the side chain of Leu110 and is cradled on either side by the side chains of Pro112 and His173 (at the conjugated diene by Pro112 and at the 5,6-spiroketal ring system by His173) (Figure 3). A key hydrophobic interaction is also formed between the methyl of C45 and Ile75. The ring is further held by a hydrogen bond between O6 and the main chain amide of Arg177 and a water molecule that is further coordinated by the carbonyl oxygen of Arg177 (Figure 3b). Undoubtedly, the conjugated diene components of the macrolide ring provide conformational constraints to the ring shape that help ensure presentation of these key atoms to the appropriate surface of actin.
Figure 3. PTX-2–actin contacts.
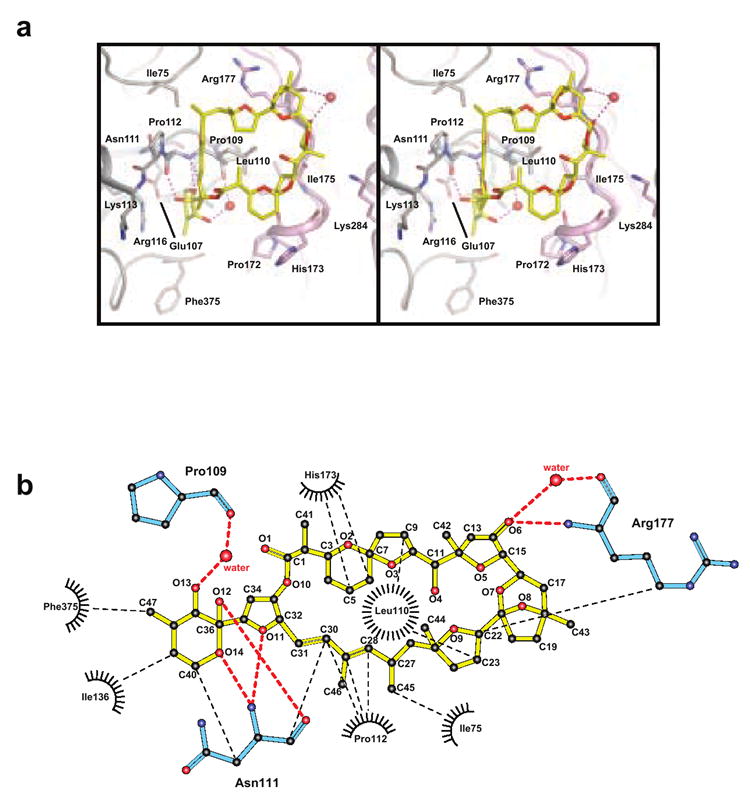
(a) Stereo view of specific PTX-2–actin interactions are shown. PTX-2 is in yellow and its contact residues on actin are labeled and colored according to the domains to which they belong. Polar interactions between PTX-2 and actin are shown in purple. Waters involved in hydrogen bonding between PTX-2 and actin are shown as red spheres. (b) Ligplot of the interactions between PTX-2 and actin. Van der Waals contacts are represented by black spokes radiating between interacting residues and are connected by thin dashed black lines. Water molecules are depicted as red spheres with hydrogen bonding interactions connected by thick dashed red lines. Actin residues involved in polar interactions are colored cyan. Figure produced with the program LIGPLOT.59
The tail of PTX-2 curves into the outer edge of the barbed-end cleft and interacts predominantly with subdomain 1. All of the polar atoms in the tail interact with actin. Altogether the tail forms four hydrogen bonds either directly with actin residues or through actin-coordinated waters (Figure 3b). These interactions occur at Pro109 and Asn111. The hydrophobic effect seems to contribute to the tail’s coordination as does the fact that the six-membered cyclic hemiketal sits in a narrow hydrophobic hole formed by Ile136, Phe375 and the β-carbon of Asn111.
PTX-2 Caps But Does Not Sever Actin Filaments
It has been reported that PTX-2 sequesters monomeric actin and inhibits polymerization.18, 20 PTX-6 has been shown to induce depolymerization of F-actin in neuroblastoma cells and to damage actin filaments in enterocytes from rabbit intestines.21, 22 Using purified actin samples, we have performed biophysical analyses to determine the effects of PTX-2 on actin filament severing and capping. Our results show that PTX-2 does not induce severing of F-actin but that it very efficiently inhibits actin polymerization by capping the fast-growing barbed-end.
At 48 μM in the presence of salts and ATP, actin forms stable filaments, which sediment during low speed centrifugation (20,000 × g). After a brief incubation (30 min) of F-actin with defined molar ratios of toxin [0.5:1, 1:1, 2:1 (toxin:actin)], analysis of the pelleted material by gel electrophoresis provides a measure of the stable F-actin content remaining in each sample, and thus the filament destabilizing effect of the toxin. However, due to the short incubation time, the concentration of F-actin involved, and the buffer conditions (50 mM KCl, 2 mM MgCl2, 1 mM ATP, 10 mM Tris-HCl, pH 7.5), the majority of filament loss cannot be attributed to depolymerization and sequestration of actin monomers from the filament ends but must correlate with F-actin severing. Previously, we demonstrated that short incubation of F-actin with the barbed-end targeting macrolides reidispongiolide A or halichondramide dramatically reduced the amount of F-actin pelleted by centrifugation, indicating that these were potent filament severing compounds.29 Here, we show that, in contrast to reidispongiolide A and halichondramide, PTX-2 does not display severing activity even when added at a two-fold molar excess over actin (Figure 4). The effect observed for PTX-2 was comparable to that of latrunculin B, which does not possess filament severing ability.30, 31
Figure 4. F-actin severing activity of PTX-2.
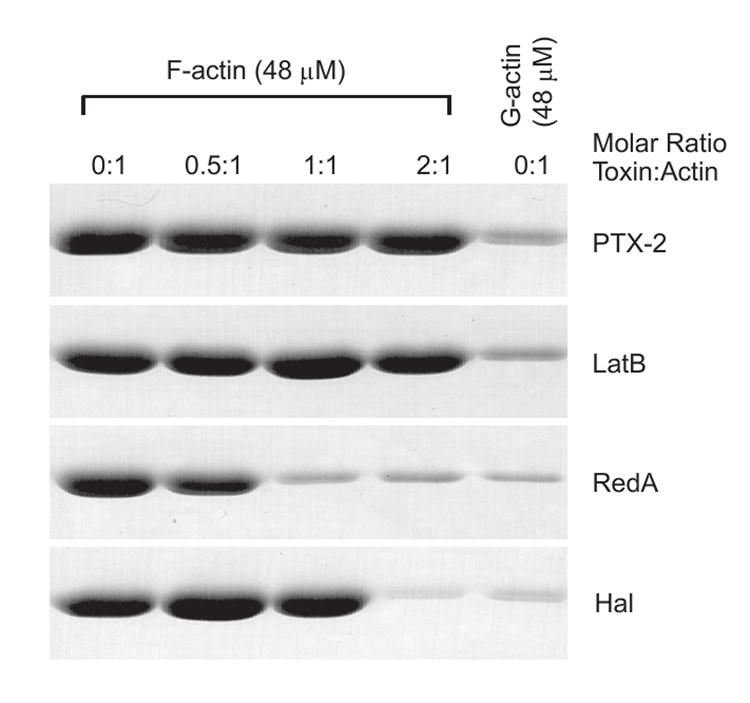
F-actin (48 μM) in F-buffer was treated with the indicated molar ratio amounts of PTX-2, latrunculin B (LatB), reidispongiolide A (RedA), or halichondramide (Hal) for 30 min and then centrifuged at 20,000 × g for 30 min. The pellet of each sample following analysis by SDS-PAGE is shown. The pellets for G-actin (48 μM) in G-buffer without the addition of toxin are shown as a control for sedimentation of non-filamentous actin within the tube.
Molecules that cap the ends of actin filaments are generally capable of binding to either the barbed- or pointed-end of the filament and preventing further elongation of that end.32 For PTX-2 to possess filament capping ability, its complex with an actin monomer must be capable of interacting with other actin subunits, predictably actin dimers, trimers and/or a filament end, and inhibiting further addition of actin subunits. The filament capping ability of PTX-2 was determined by measuring the effect of limited (substoichiometric) PTX-2–G-actin complexes on polymerization of a 10-fold larger pool of uncomplexed G-actin using a kinetic pyrenyl fluorescence assay.29, 33 By pre-assembling the complexes with an equimolar ratio of monomeric actin to toxin at least 12 hours prior to initiation of the experiment, and adding 1 μM of the complex to 9 μM uncomplexed monomeric actin, toxin-actin complexes that are capable of filament capping would substantially reduce the rate of polymerization and thus the intensity of the pyrene fluorescence signal relative to an actin sample with no toxin-actin complex added. Alternatively, if the functionality of the toxin is limited to actin monomer sequestration, the rate of polymerization (pyrene fluorescence signal) would not be significantly different from the sample lacking the toxin–actin complex because these complexes would not contribute to, or hinder, polymerization of the uncomplexed actin monomers due to their inability to bind other actin subunits. The plot of the pyrene fluorescence signal for the PTX-2–actin complex sample over time shows that these complexes significantly inhibit filament elongation, much more so than can be attributed to monomer sequestration alone (Figure 5a). In fact, the capping activity of the PTX-2–actin complex was more robust than that observed for the filament severing toxins reidispongiolide A and halichondramide, which also displayed capping activity in previous studies.29
Figure 5. Actin filament-capping activity of PTX-2.
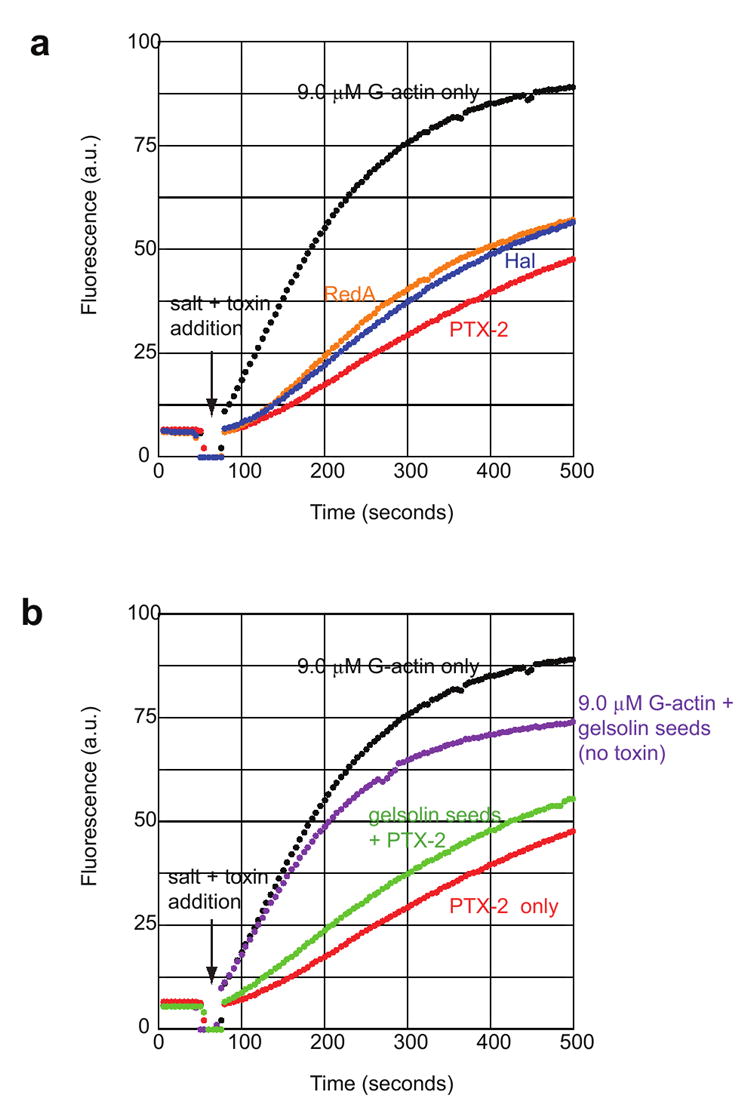
(a) The graph shows the change in pyrene fluorescence signal that occurs during polymerization of 9.0 μM G-actin (15% pyrenyl-G-actin) in the presence of 1 μM toxin–G-actin complexes as a function of time. A baseline signal was obtained for each actin sample before removal of the cuvette (at 50 seconds) from the fluorimeter and addition of 1 μM toxin–G-actin complex and KCl and MgCl2. Polymerization reaction profiles are labeled according to the toxin–actin complexes added (black, 9.0 μM G-actin without toxin–actin complex; blue, halichondramide; red, PTX-2; orange, reidispongiolide A). (b) 9.0 μM G-actin was pre-incubated with pre-assembled gelsolin–actin seeds added at a 1:50 gelsolin:actin ratio for five minutes prior to the addition of toxin–actin complexes and salts. Control samples of 9.0 μM G-actin only and G-actin plus gelsolin seeds are shown in black and purple, respectively. Polymerization of G-actin in the presence of 1 μM PTX-2–G-actin complex only is shown in red, while the same reaction contained gelsolin seeds is shown in green.
To determine whether filament capping by PTX-2–actin complexes was occurring at the pointed-end or the barbed-end, the polymerization assay was also performed in the presence of gelsolin–actin seeds, which cap the barbed-end and permit filament growth from the pointed-end only.34 Under these conditions, we observed that the PTX-2–actin seeds had less of an inhibitory effect on polymerization than observed in the absence of gelsolin–actin seeds (Figure 5b). This indicates that the PTX-2–actin complex is competing with the gelsolin seeds for the barbed-end during polymerization.
Although some filament capping molecules can inhibit depolymerization of the capped end,32 an analysis of the ability of the PTX-2–actin cap to interfere with depolymerization from the barbed-end was not conducted for the following reasons. First, reliable measurements of filament depolymerization specifically from the filament ends are difficult to obtain because the standard assays for measuring filament depolymerization do not differentiate between actin subunit disassembly from the filament ends, filament shearing that often occurs during reaction mixing, or quenching of the pyrene signal upon toxin binding.35 Second, because the PTX-2–actin complex essentially functions as a capping protein, the interactions of this complex with the barbed-end are formally equivalent to that of G-actin alone. This presents a problem in that to perform a depolymerization experiment an excess of toxin must be added over the concentration of barbed ends. Necessarily this will result in sequestration of the actin as it is released from the pointed end. In turn this will reduce the level of “free actin” in solution, which will enhance depolymerization. As a net result, the effect of capping of the barbed end will be obscured.
Model of Barbed-End Capping By Pectenotoxins
F-actin consists of a two start helix in which the long-pitch strands are axially staggered by approximately half the axial subunit spacing and exhibit a right-handed helical twist.36 Models of F-actin structure based on electron microscopy, fiber-diffraction and observed crystallographic interactions indicate that each subunit in the filament engages in both longitudinal and lateral contacts with other subunits.37-39 The surfaces involved in such interactions have been examined extensively by structural modeling, mutagenesis, cross-linking, proteolytic footprinting and deuterium-exchange studies, and the findings from these studies are generally in agreement with Holmes’ model of F-actin architecture.37, 40 A superposition of the PTX-2–actin complex onto this model creates a spatial overlap between PTX-2 and residues Lys191, Thr194, Glu195, Arg196, Gly197, and Ser199 in helix H8 of subdomain 4 from the lower lateral actin subunit, but does not generate a steric clash between longitudinal subunits (Figure 6). Interestingly, the interaction between helix H8 and the binding surface of PTX-2 on the upper lateral monomer has not been heavily investigated as a contributor to filament stability. With the exception of a proteolytic fragment binding study,41 few studies have suggested that helix H8 plays a critical role in lateral interactions. Instead, the H-loop has typically been credited with this role.
Figure 6. Superposition of PTX-2 onto the F-actin model.
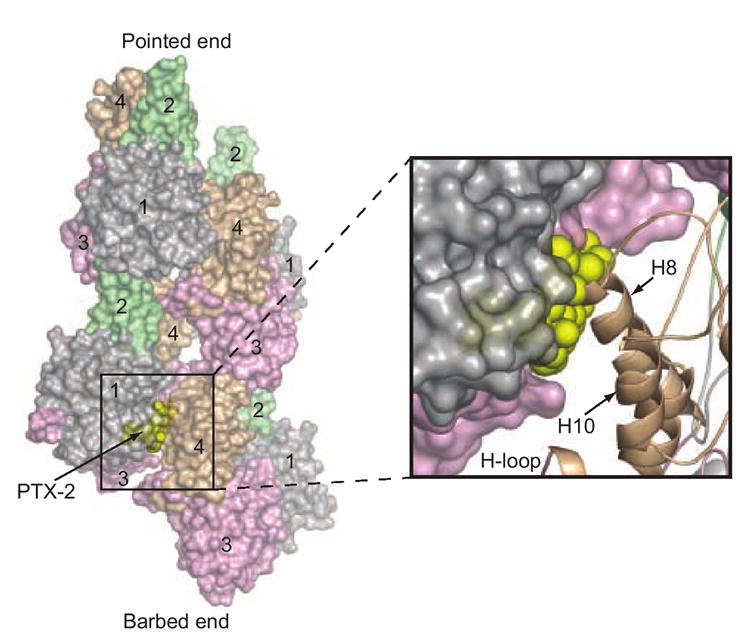
The G-actin-bound conformation of PTX-2 (yellow spheres) is superimposed on the model of F-actin to illustrate the mechanism by which it caps the barbed-end of the filament.37, 40 Four actin subunits are shown (two for each strand) in surface representation. The pointed- and barbed-ends of this “mini-filament” are labeled. Actin subdomains 1–4 are also labeled for each subunit and are colored as in Figure 2. The binding position of PTX-2 is at the interior of the filament. The boxed region shows a magnification of the steric clash between PTX-2 and helix H8 from subdomain 4 of the lower lateral actin subunit (shown in cartoon representation).
In addition to producing a steric clash with part of subdomain 4 from the lower lateral actin subunit in the F-actin model, PTX-2 interacts with residues between subdomains 1 and 3 (Ile75, Pro110, Pro112, Arg177, and Phe375) on G-actin that show protection upon actin polymerization in hydrogen/deuterium exchange mass spectrometry and radiolytic footprinting experiments.42, 43 This indicates that the PTX-2-binding surface on actin may not be accessible in F-actin, but can only be engaged in the G-actin form or at a free barbed-end. This is consistent with the observed effects of PTX-2 on purified samples of G- and F-actin where PTX-2 demonstrated robust barbed-end capping ability during G-actin polymerization but was unable to sever preassembled actin filaments. It also indicates that PTX-2 binding at the “inner” filament axis side inhibits a lateral actin subunit interaction critical for filament assembly, thereby capping the barbed-end. Such a mechanism of filament dynamics modulation is novel to barbed-end-targeting natural products.
Structure–Activity Relationship of Pectenotoxins
Metabolism of PTX-2 to other PTX derivatives by shellfish, and biosynthesis of analogues in algae, has provided a growing library of compounds 1-4, 8, 9, 12, 17 whose cytotoxicity continues to be evaluated for potential therapeutic utility.44 At a molecular level, this library of chemical modifications can help probe the versatility of PTX-binding surface on actin, providing vital information toward the design of novel analogues with improved pharmacological profiles. Though no structural information on the interaction of other PTX isoforms with actin is currently available, nor are comparisons of the effect of different PTXs on purified actin filament dynamics, the structure of the PTX-2–actin complex provides an excellent model to help rationalize previous observations of the activity of different PTX isoforms in cell and mouse model systems (Figure 7).
Figure 7. Surface representation of the interaction of PTX-2 with actin and the location of common differences among PTX variants.
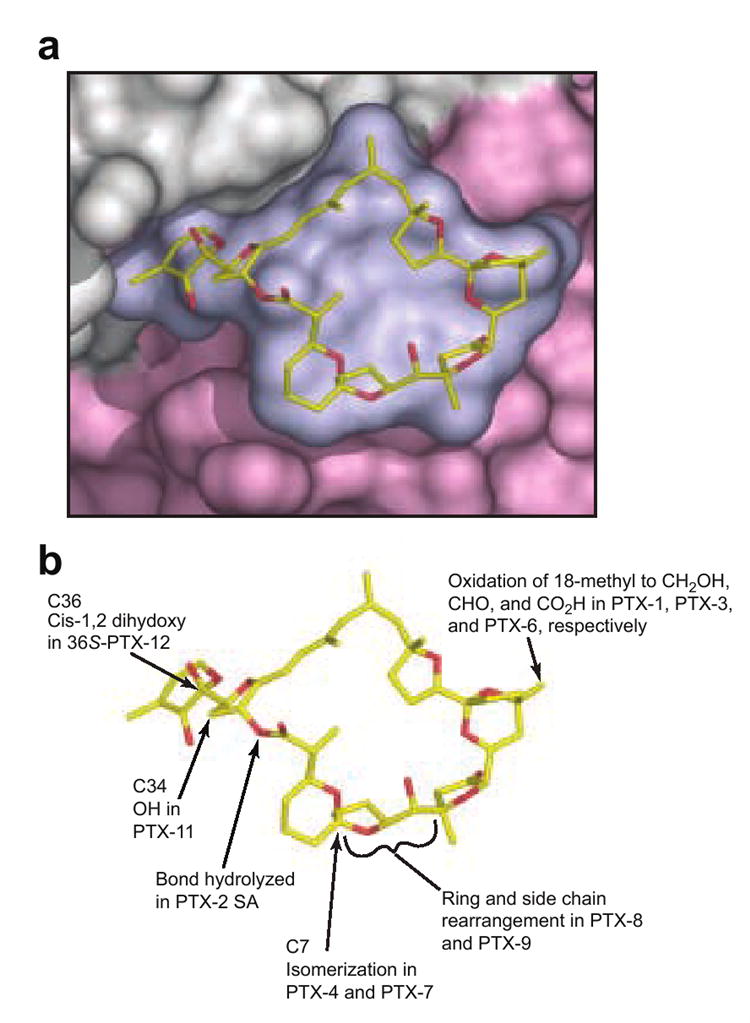
(a) Closeup view of a surface representation of PTX-2 and its binding site on actin where subdomains 1 and 3 are colored in gray and light pink respectively. (b) shows the locations of the chemical differences in common PTX variants. It is immediately clear that the change from (R) configuration at C7 in PTX-2 to the (S) configuration in PTX-4 and -7 can be expected to induce a large change in the shape of the macrolactone ring.
PTX-2 is one of the most toxic PTXs in mice.27, 45 Examination of the binding interactions of PTX-2 with actin suggests that addition of a hydroxyl group to the methyl side chain at C34 as seen in PTX-11 probably would not have a major impact on binding to actin (Figure 7). Likewise oxidation of the 18-methyl group to either a hydroxymethyl, aldehyde, or carboxylate moiety as seen in PTX-1, PTX-3, and PTX-6, respectively, would not be expected to have a major effect on binding. This prediction is consistent with the recent observation that, like PTX-2, PTX-1 and PTX-11 influence actin assembly in cellular assays.46 Conversely, isomerisations around C7 (PTX-4, -7, -8, and -9) would be expected to change the conformation of the macrocyclic ring in ways that might be inconsistent with the interactions with His173 and Leu110, which provide critical hydrophobic interactions. Certainly, any changes in the chirality of the ring systems that make up the macrolactone ring might be expected to influence the interaction of the PTX with the shallow binding site provided by actin. In the same vein, the presence of a cis-1,2-dihydroxy system in the “tail” of 36R-PTX-12 suggests that it may bind more poorly than the trans-1,2-dihydroxy-containing 36S-PTX-12. Finally, it is likely that opening of the macrocyclic ring to form PTX seco acids would seriously interfere with binding to actin. As with all cyclic macrolides that bind to actin, the presence of a ring will reduce the conformational freedom of the unbound toxin which will decrease the entropic cost of binding a complex molecule to a comparatively shallow hydrophobic surface.29 Indeed, recent cellular studies have shown that the PTX-2 seco acid does not cause actin disassembly or changes in the cytoskeleton,46 and PTX-2 seco acids are not measurably toxic in vitro12 or in vivo.8, 47 Further studies to measure the binding affinity of different PTX isoforms to actin, as well as clarification of the nature of the interaction other PTXs make with the actin cytoskeleton, are under way.
Conclusions
The co-crystal structure of PTX-2–actin reveals the molecular details of the actin–toxin interface. This provides key information about its chemical structure determinants for actin binding, as well as sites on PTX that can be modified during analogue synthesis or used as conjugation points for addition of targeting probes. Furthermore, the structure has allowed us to perform a cursory modeling exercise to reconcile previous SAR data of PTX congeners. More in-depth structural and functional analyses of these congeners will be conducted once sufficient amounts of these toxins are available. Finally, the binding position and in vitro activity of PTX-2 suggests a new mechanism for actin filament destabilization, provides important information about F-actin subunit contacts, and underlines the abundance of sites on actin that are vulnerable to attack by natural small molecules.
Materials and Methods
Protein Preparation
Actin used in crystallizations and the low speed centrifugation assay was obtained from rabbit muscle acetone powder as previously described.29 For the actin filament capping assays pyrene-labeled rabbit skeletal G-actin (cat. no. AP05) and unlabeled rabbit skeletal G-actin (cat. no. AKL99) were purchased from Cytoskeleton Inc. (Denver, CO).
Toxin Isolation
PTX-2 was isolated from a bloom of Dinophysis acuta harvested at Sognefjord, Norway.8 Latrunculin B was purchased from Sigma. Reidispongiolide A was provided as a generous gift by Prof. Valeria D’Auria, Università degli Studi di Napoli “Federico II”, Naples, Italy. It was isolated from the sponge Reidispongia coerulea collected in the South of New Caledonia and extracted according to previously published protocols.48, 49 Halichondramide was provided as a generous gift by Prof. Junichi Tanaka, University of Ryukyus. It was collected from an unidentified black sponge collected off Iriomote Island in Okinawa and purified as previously described.50, 51 The identity of all toxins was confirmed by NMR and mass spectrometry. All toxins were solubilized in 100% methanol and stored at -20°C prior to use.
Crystallization
Pectenotoxin-2 was mixed at a 1.3-fold molar excess with G-actin and then concentrated to approximately 10 mg/ml by ultrafiltration. Crystals of actin–PTX-2 were grown by hanging drop vapor diffusion in which 5 μl of the complex was mixed with an equal volume of precipitant solution. The precipitant for the PTX-2 complex contained 100 mM Na/MES/acetate, pH 5.5, 15% methyl ether poly(ethylene glycol) 5000, 10% hexanediol, 30 mM CaCl2, and 1 mM TCEP. The actin–PTX-2–latrunculin B complex was formed by adding latrunculin B at a 1:1 molar ratio to the actin–PTX-2 complex. The precipitant for this complex contained 100 mM Na/MES/acetate, pH 5.5, 15% methyl ether poly(ethylene glycol) 5000, 10% hexanediol, 100 mM CaCl2, and 1 mM TCEP. All hanging drops were streak-seeded with crystals obtained from an initial screen 24 hours after set-up. Crystals of PTX-2–actin complex grew as rectangular boxes to maximum dimensions of ~0.4 × 0.05 × 0.05 mm in 4–5 days at 20 °C. Crystals of PTX-2–LatB–actin complex grew as skewed tetragonal prisms to maximum dimensions of ~0.3 × 0.1 × 0.05 mm in 2 weeks at 20 °C. The crystals were cryopreserved by step-wise equilibration into 25%, 75%, and finally 100% cryoprotectant solution containing 100 mM Na/MES/acetate, pH 5.5, 18% methyl ether poly(ethylene glycol) 5000, 175 mM CaCl2, 12% hexanediol, 10% ethylene glycol and flash frozen in a stream of nitrogen gas.
Data Collection and Structure Refinement
Data was collected at the SBC 19-BM beam line, Advanced Photon Source in Argonne, IL. Diffraction data were integrated and scaled with the program HKL2000.52 The structures of both complexes were solved by molecular replacement with Molrep 53 starting from the actin–sphinxolide B structure (PDB accession code 2ASO 29). The structures were refined with Refmac and Coot.54, 55 The atomic coordinates and structure factors are deposited in the Protein Data Bank, www.pdb.org [PDB ID codes XXXX PTX-2-actin, YYYY LatB-PTX-2-actin].
Structure Analysis and Figure Preparation
Structure alignments were performed with Superpose and Align.56, 57 Ligand-protein contacts were determined using the LPC suite and the program LIGPLOT v.4.4.2.58, 59 The total molecular surface area buried at the complex interface was determined with CNS version 1.1.60 Figures showing toxin and actin crystal structure models were prepared with Pymol.61
Actin Filament Capping Assays
Measurements of actin filament capping with actin–toxin complexes were performed with pyrenyl-G-actin (cat. no. AP05) from Cytoskeleton Inc. (Denver, CO, USA). Briefly, 9.0 μM of 15% pyrenyl–G-actin in G-buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP) in the presence or absence of 1 μM unlabeled G-actin–toxin complex (assembled by mixing an equal molar ratio of G-actin and toxin at least 12 hrs prior to the experiment) was mixed with 1/10th volume of Actin Polymerization Buffer (500 mM KCl, 20 mM MgCl2, and 10 mM ATP (cat. no. BSA02-010)) in a 200 μl cuvette. After five seconds of mixing by pipetting, fluorescence intensity of the pyrenyl actin was measured continuously for 500 seconds. The difference in pyrene fluorescence during polymerization was monitored at 22°C using a QuantaMaster C-60/2000 fluorimeter (Photon Technologies Inc.) with excitation and emission wavelengths of 365 and 407 nm, respectively. Slit widths were set to 1 nm. Gelsolin–actin seeds were assembled by mixing unlabeled G-actin with full-length gelsolin at a 2:1 molar ratio in G-buffer and incubating for 3 hr at 22°C followed by 12 hr at 4°C. For the capping assays measuring polymerization of pyrenyl-G-actin from the pointed-end, gelsolin–actin seeds were added to 9.0 μM of 15% pyrenyl-G-actin at a final concentration of 0.18 μM 5 min prior to the addition of 1 μM of actin–toxin complexes and 1/10th volume of Actin Polymerization Buffer.62 Graphs were refined with Kaleidagraph (version 3.6.4, Synergy Software, Reading, Pennsylvania, United States).
Actin Filament Severing Assay
Measurements of F-actin severing were performed with rabbit skeletal muscle G-actin polymerized in 50 mM KCl, 2 mM MgCl2, 1 mM ATP, 10 mM Tris-HCl, pH 7.5, overnight. Toxins were added at 24 μM, 48 μM, and 96 μM (final concentrations) to 100 μL of 48 μM F-actin giving molar ratios of toxin:F-actin of 0.5:1, 1:1, and 2:1, respectively. The samples were incubated for 30 min at 4°C and then centrifuged at 20,000 × g for 30 min. The supernatant fraction was removed and the pellets were resuspended in 100 μL of G-buffer and analyzed by SDS-PAGE. Pellets of 48 μM G-actin without toxin added were included as controls to measure the amount of non-filamentous actin retained on the tube wall after centrifugation.
Acknowledgments
We thank Laura Walker for assistance with PTX-2–actin crystal growth, and V. D’Auria and J. Tanaka for graciously providing reidispongiolide A and halichondramide, respectively. Parts of this study were supported by Norwegian Research Council grant 139593/140, by the BIOTOX project (partly funded by the European Commission, through 6th Framework Programme contract no. 514074, topic Food Quality and Safety), and by the NZ Foundation for Research, Science and Technology International Investment Opportunities Fund (IIOF contract number C10X0406). We thank P. Hovgaard for collection of algae from which PTX-2 was isolated, and M. Sandvik, I. A. Samdal, and T. Rundberget for assistance with isolation of PTX-2. This work was also supported by a fellowship to JSA from the Canadian Institutes of Health Research (64606) and a research grant to IR from the National Institutes of Health (AR35186). Use of the SBC beamline Argonne National Laboratory Advanced Photon Source was supported by the U. S. Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Draisci R, Lucentini L, Mascioni A. Pectenotoxins and yessotoxins: chemistry, toxicology, pharmacology, and analysis. In: Botana LM, editor. Seafood and Freshwater Toxins: Pharmacology, Physiology, and Detection. Marcel Dekker, Inc.; New York: 2000. pp. 289–324. [Google Scholar]
- 2.Yasumoto T, Murata M, Oshima Y, Matsumoto GK, Clardy J. Diarrhetic shellfish poisoning; ACS Symposium Series, 262; New York: ACS; 1984. [Google Scholar]
- 3.Yasumoto T, Murata M, Oshima Y, Sano M, Matsumoto GK, Clardy J. Diarrhetic shellfish toxins. Tetrahedron. 1985;41:1019–1025. [Google Scholar]
- 4.Miles CO, Wilkins AL, Samdal IA, Sandvik M, Petersen D, Quilliam MA, Naustvoll LJ, Rundberget T, Torgersen T, Hovgaard P, Jensen DJ, Cooney JM. A novel pectenotoxin, PTX-12, in Dinophysis spp. and shellfish from Norway. Chem Res Toxicol. 2004;17:1423–1433. doi: 10.1021/tx049870a. [DOI] [PubMed] [Google Scholar]
- 5.Yasumoto T, Murata M, Lee J-S, Torigoe K. Polyether toxins produced by dinoflagellates. In: Natori S, Hashimoto K, Ueno Y, editors. Mycotoxins and Phycotoxins ’88. Elsevier; Amsterdam: 1989. pp. 375–382. [Google Scholar]
- 6.Yoon MY, Kim YC. Acute toxicity of pectenotoxin-2 and its effects on hepatic metabolising enzyme system in mice. Kor J Toxicol. 1997;13:183–186. [Google Scholar]
- 7.Yoon MY, Kim YC. Toxicity and changes in hepatic metabolizing enzyme system induced by repeated administration of pectenotoxin 2 isolated from marine sponges. Kor J Pharmacog. 1997;28:280–285. [Google Scholar]
- 8.Miles CO, Wilkins AL, Munday R, Dines MH, Hawkes AD, Briggs LR, Sandvik M, Jensen DJ, Cooney JM, Holland PT, Quilliam MA, MacKenzie AL, Beuzenberg V, Towers NR. Isolation of pectenotoxin-2 from Dinophysis acuta and its conversion to pectenotoxin-2 seco acid, and preliminary assessment of their acute toxicities. Toxicon. 2004;43:1–9. doi: 10.1016/j.toxicon.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki T, Walter JA, LeBlanc P, MacKinnon S, Miles CO, Wilkins AL, Munday R, Beuzenberg V, MacKenzie AL, Jensen DJ, Cooney JM, Quilliam MA. Identification of pectenotoxin-11 as 34S-hydroxypectenotoxin-2, a new pectenotoxin analogue in the toxic dinoflagellate Dinophysis acuta from New Zealand. Chem Res Toxicol. 2006;19:310–318. doi: 10.1021/tx050240y. [DOI] [PubMed] [Google Scholar]
- 10.European Union. Commission decision of 15 March 2002 laying down detailed rules for the implementation of Council Directive 91/492/EEC as regards the maximum permitted levels and the methods for analysis of certain marine biotoxins in bivalve molluscs, echinoderms, tunicates and marine gastropods (2002/225/EC) Off J the Eur Commun. 2002;L:75–62. [Google Scholar]
- 11.Aune T, Yasumoto T, Engeland E. Light and scanning electron microscopic studies on effects of marine algal toxins toward freshly prepared hepatocytes. J Toxicol Environ Health. 1991;34:1–9. doi: 10.1080/15287399109531544. [DOI] [PubMed] [Google Scholar]
- 12.Daiguji M, Satake M, James KJ, Bishop A, MacKenzie L, Naoki H, Yasumoto T. Structures of new pectenotoxin analogs, pectenotoxin-2 seco acid and 7-epi-pectenotoxin-2 seco acid, isolated from a dinoflagellate and greenshell mussels. Chem Lett. 1998:653–654. [Google Scholar]
- 13.Fladmark KE, Serres MH, Larsen NL, Yasumoto T, Aune T, Døskeland SO. Sensitive detection of apoptogenic toxins in suspension cultures of rat and salmon hepatocytes. Toxicon. 1998;36:1101–1114. doi: 10.1016/s0041-0101(98)00083-x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou ZH, Komiyama M, Terao K, Shimada Y. Effects of pectenotoxin-1 on liver cells in vitro. Nat Toxins. 1994;2:132–135. doi: 10.1002/nt.2620020308. [DOI] [PubMed] [Google Scholar]
- 15.Chae HD, Choi T-S, Kim B-M, Jung JH, Bang Y-J, Shin DY. Oocyte-based screening of cytokinesis inhibitors and identification of pectenotoxin-2 that induces Bim/Bax-mediated apoptosis in p53-deficient tumors. Oncogene. 2005;24:4813–4819. doi: 10.1038/sj.onc.1208640. [DOI] [PubMed] [Google Scholar]
- 16.Jung JH, Sim CJ, Lee C-O. Cytotoxic compounds from a two-sponge association. J Nat Prod. 1995;58:1722–1726. doi: 10.1021/np50125a012. [DOI] [PubMed] [Google Scholar]
- 17.Halim R, Brimble MA. Synthetic studies towards the pectenotoxins: a review. Org Biomol Chem. 2006;4:4048–4058. doi: 10.1039/b611531b. [DOI] [PubMed] [Google Scholar]
- 18.Hori M, Matsuura Y, Yoshimoto R, Ozaki H, Yasumoto T, Karaki H. Actin depolymerizing action by marine toxin, pectenotoxin-2. Nippon Yakurigaku Zasshi. 1999;114(Suppl 1):225P–229P. doi: 10.1254/fpj.114.supplement_225. [DOI] [PubMed] [Google Scholar]
- 19.Karaki H, Matsuura Y, Hori M, Yoshimoto R, Ozaki H, Yasumoto T. Pectenotoxin-2, a new actin depolymerizing compound isolated from scallop Patinopecten yessoensis. Jap J Pharmacol. 1999;79:268P. doi: 10.1254/fpj.114.supplement_225. [DOI] [PubMed] [Google Scholar]
- 20.Spector I, Braet F, Shochet NR, Bubb MR. New anti-actin drugs in the study of the organization and function of the actin cytoskeleton. Microsc Res Tech. 1999;47:18–37. doi: 10.1002/(SICI)1097-0029(19991001)47:1<18::AID-JEMT3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 21.Leira F, Cabado AG, Vieytes MR, Roman Y, Alfonso A, Botana LM, Yasumoto T, Malaguti C, Rossini GP. Characterization of F-actin depolymerization as a major toxic event induced by pectenotoxin-6 in neuroblastoma cells. Biochem Pharmacol. 2002;63:1979–1988. doi: 10.1016/s0006-2952(02)00993-0. [DOI] [PubMed] [Google Scholar]
- 22.Ares IR, Louzao MC, Vieytes MR, Yasumoto T, Botana LM. Actin cytoskeleton of rabbit intestinal cells is a target for potent marine phycotoxins. J Exp Biol. 2005;208:4345–4354. doi: 10.1242/jeb.01897. [DOI] [PubMed] [Google Scholar]
- 23.Allingham JS, Klenchin VA, Rayment I. Actin-targeting natural products: structures, properties and mechanisms of action. Cell Mol Life Sci. 2006;63:2119–2134. doi: 10.1007/s00018-006-6157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenteany G, Zhu S. Small-molecule inhibitors of actin dynamics and cell motility. Curr Top Med Chem. 2003;3:593–616. doi: 10.2174/1568026033452348. [DOI] [PubMed] [Google Scholar]
- 25.Jordan MA, Wilson L. Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr Opin Cell Biol. 1998;10:123–130. doi: 10.1016/s0955-0674(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 26.Giganti A, Friederich E. The actin cytoskeleton as a therapeutic target: state of the art and future directions. Prog Cell Cycle Res. 2003;5:511–525. [PubMed] [Google Scholar]
- 27.Yasumoto T, Murata M, Lee J-S. In: Bioactive Molecules: Mycotoxins and Phycotoxins ’88. Natori S, Hashimoto K, Ueno Y, editors. Vol. 10. Elsevier; Amsterdam: 1989. pp. 375–382. [Google Scholar]
- 28.Chik JK, Lindberg U, Schutt CE. The structure of an open state of beta-actin at 2.65 Å resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- 29.Allingham JS, Zampella A, D’Auria MV, Rayment I. Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity. Proc Natl Acad Sci USA. 2005;102:14527–14532. doi: 10.1073/pnas.0502089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spector I, Shochet NR, Blasberger D, Kashman Y. Latrunculins—novel marine macrolides that disrupt microfilament organization and affect cell growth: I. Comparison with cytochalasin D. Cell Motil Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- 31.Coue M, Brenner SL, Spector I, Korn ED. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987;213:316–8. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- 32.Pollard TD, Cooper JA. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- 33.Korn ED. Actin polymerization and its regulation by proteins from nonmuscle cells. Physiol Rev. 1982;62:672–737. doi: 10.1152/physrev.1982.62.2.672. [DOI] [PubMed] [Google Scholar]
- 34.Tellam R, Frieden C. Cytochalasin D and platelet gelsolin accelerate actin polymer formation. A model for regulation of the extent of actin polymer formation in vivo. Biochemistry. 1982;21:3207–3214. doi: 10.1021/bi00256a027. [DOI] [PubMed] [Google Scholar]
- 35.Saito S, Watabe S, Ozaki H, Fusetani N, Karaki H. Mycalolide B, a novel actin depolymerizing agent. J Biol Chem. 1994;269:29710–4. [PubMed] [Google Scholar]
- 36.Sheterline P, Clayton J, Sparrow JC. Actin. 4. Protein Profile, Oxford University Press; New York: 1998. [PubMed] [Google Scholar]
- 37.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 38.Belmont LD, Orlova A, Drubin DG, Egelman EH. A change in actin conformation associated with filament instability after Pi release. Proc Natl Acad Sci USA. 1999;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 40.Tirion MM, ben-Avraham D, Lorenz M, Holmes KC. Normal modes as refinement parameters for the F-actin model. Biophys J. 1995;68:5–12. doi: 10.1016/S0006-3495(95)80156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hori K, Morita F. Actin–actin contact: inhibition of actin-polymerization by subdomain 4 peptide fragments. J Biochem (Tokyo) 1992;112:401–408. doi: 10.1093/oxfordjournals.jbchem.a123912. [DOI] [PubMed] [Google Scholar]
- 42.Chik JK, Schriemer DC. Hydrogen/deuterium exchange mass spectrometry of actin in various biochemical contexts. J Mol Biol. 2003;334:373–385. doi: 10.1016/j.jmb.2003.09.044. [DOI] [PubMed] [Google Scholar]
- 43.Guan JQ, Takamoto K, Almo SC, Reisler E, Chance MR. Structure and dynamics of the actin filament. Biochemistry. 2005;44:3166–3175. doi: 10.1021/bi048021j. [DOI] [PubMed] [Google Scholar]
- 44.Chae HD, Choi TS, Kim BM, Jung JH, Bang YJ, Shin DY. Oocyte-based screening of cytokinesis inhibitors and identification of pectenotoxin-2 that induces Bim/Bax-mediated apoptosis in p53-deficient tumors. Oncogene. 2005;24:4813–4819. doi: 10.1038/sj.onc.1208640. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki K, Wright JL, Yasumoto T. Identification and characterization of pectenotoxin (PTX) 4 and PTX7 as spiroketal stereoisomers of two previously reported pectenotoxins. J Org Chem. 1998;63:2475–2480. doi: 10.1021/jo971310b. [DOI] [PubMed] [Google Scholar]
- 46.Ares IR, Louzao MC, Espiña B, Vieytes MR, Miles CO, Yasumoto T, Botana LM. Lactone ring of pectenotoxins: a key factor for their activity on cytoskeleton dynamics. Cell Physiol Biochem. 2007;19:283–292. doi: 10.1159/000100647. [DOI] [PubMed] [Google Scholar]
- 47.Miles CO, Wilkins AL, Munday JS, Munday R, Hawkes AD, Jensen DJ, Cooney JM, Beuzenberg V. Production of 7-epi-pectenotoxin-2 seco acid and assessment of its acute toxicity to mice. J Agric Food Chem. 2006;54:1530–4. doi: 10.1021/jf0523871. [DOI] [PubMed] [Google Scholar]
- 48.Dauria MV, Paloma LG, Minale L, Zampella A, Verbist JF, Roussakis C, Debitus C, Patissou J. Reidispongiolide A and B, two new potent cytotoxic macrolides from the New Caledonian sponge Reidispongia coerulea. Tetrahedron. 1994;50:4829–4834. [Google Scholar]
- 49.Carbonelli S, Zampella A, Randazzo A, Debitus C, Gomez-Paloma L. Sphinxolides E–G and reidispongiolide C: four new cytotoxic macrolides from the New Caledonian Lithistida sponges N. superstes and R. coerulea. Tetrahedron. 1999;55:14665–14674. [Google Scholar]
- 50.Kernan MR, Faulkner JD. Halichondramide, an antifungal macrolide from the sponge Halichondria sp. Tetrahedron Letters. 1987;28:2809–2812. [Google Scholar]
- 51.Klenchin VA, Allingham JS, King R, Tanaka J, Marriott G, Rayment I. Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin. Nat Struct Biol. 2003;10:1058–1063. doi: 10.1038/nsb1006. [DOI] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 53.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–68. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 57.Cohen GH. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J Appl Cryst. 1997;30:1160–1161. [Google Scholar]
- 58.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 59.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 60.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 61.DeLano WL. The PyMol molecular graphics system. DeLano Scientific; San Carlos, CA, USA: 2002. [accessed 17 July 2006]. http://pymol.sourceforge.net/ [Google Scholar]
- 62.Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000;10:1273–1282. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]


