Abstract
The bacterial RecA protein has been implicated in the evolution of antibiotic resistance in pathogens, which is an escalating problem worldwide. The discovery of small molecules that can selectively modulate RecA’s activities can be exploited to tease apart its roles in the de novo development and transmission of antibiotic resistance genes. Toward the goal of discovering small-molecule ligands that can prevent either assembly of an active RecA-DNA filament or its subsequent ATP-dependent motor activities, we report the design and initial validation of a pair of rapid and robust screening assays suitable for the identification of inhbitors of RecA activities. One assay is based on established methods for monitoring ATPase enzyme activity and the second is a novel assay for RecA-DNA filament assembly using fluorescence polarization. Taken together, the assay results reveal complementary sets of agents that can either selectively suppress only the ATP-driven motor activities of the RecA-DNA filament or prevent assembly of active RecA-DNA filaments altogether. The screening assays can be readily configured for use in future automated HTS projects to discover potent inhibitors that may be developed into novel adjuvants for antibiotic chemotherapy that moderate the development and transmission of antibiotic resistance genes and increase the antibiotic therapeutic index.
Keywords: RecA, Antibiotic resistance, DNA repair, Recombination, High-throughput screen
Bacteria maintain a dynamic balance between the contrasting needs to preserve genomic information and to generate genetic diversity. The repair of damaged DNA is essential to the maintenance of heritable genetic information, while the variation of that information drives evolutionary adaptation [1]. In Escherichia coli, the RecA protein helps balance these needs by detecting the influence of environmental stress on DNA replication and initiating a programmed response to the resulting DNA damage [2-6]. Recently, RecA functions have been linked to various aspects of bacterial pathogenicity, including the induction of toxin biosynthesis [7], antigenic variation [8], and survival responses to antibacterial agents [9,10]. Of particular interest is the identification of RecA as a likely player in the mechanisms leading to the de novo development and transmission of antibiotic resistance genes. In these respective phenomena, RecA facilitates the development of antibiotic resistance via its roles in stress-induced DNA repair [1,11,12] and the horizontal transfer of genes between organisms [13,14]. The importance of these processes in bacterial pathogenecity continues to make RecA an attractive target for mechanistic and pharmacologic study [15-17].
Although RecA is highly conserved and may play similar roles in other bacteria [6], RecA-dependent processes have not been elucidated in many pathogens of interest. To delineate its roles in pathogenicity, including the development of antibiotic resistance, potent and selective modulators of RecA function are needed. To the best of our knowledge, however, no small-molecule natural product inhibitor of RecA activities have been reported [15,16]. The present paper describes the development of a pair of rapid, microvolume molecular screening assays to facilitate the discovery of potent RecA inhibitors from libraries of small molecules.
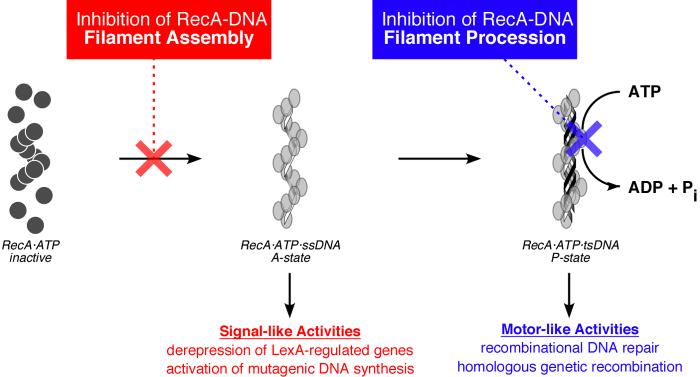
All RecA functions require formation of an active RecA-DNA filament comprising multiple RecA monomers, ATP, and DNA (i.e., states A and P in Fig. 1). This activated filament is responsible for two sets of biological functions: (1) induction of the SOS response to genomic damage by stimulation of LexA repressor autoproteolysis (state A in Fig. 1) [18]; and (2) upon further DNA binding, direct participation in recombination and DNA repair (state P in Fig. 1) [6,19]. We posit that the discovery of small molecules that interfere with the assembly or subsequent proccessive activities of RecA-DNA filaments would be an important step in the development of inhibitors for the suppression of the development and transmission of antibiotic resistance. Moreover, we expect such agents to be useful as tools for dissecting resistance gene development and transmission pathways in bacterial pathogens. To tease apart the roles of RecA in these pathways, we envisaged two complementary sets of agents: one that can selectively suppress only the processive activities of the P-state RecA-DNA filament, and a second that can prevent assembly of active RecA-DNA filaments altogether (blue and red text, respectively, in Fig. 1).
Fig. 1.
Cartoons depicting the conformational states of RecA in the absence and presence of single-stranded DNA (ssDNA), and the two classes of activities relevant to the de novo development and transmission of antibiotic resistance genes. In the absence of DNA, RecA adopts an “inactive” conformation and a quaternary state favoring monomers and low aggregates (e.g., dimers and hexamers). In the presence of DNA and ATP, RecA adopts one of two “active” conformational states in which the protein self-assembles into a homopolymeric filament that coats the DNA strands (one RecA monomer per three DNA nucleotides). The A-state RecA-DNA filament, which requires ATP binding but not its hydrolysis, activates SOS by derepression of LexA-regulated genes. An important component of SOS is the overexpression and activation of low-fidelity DNA polymerases whose activity leads to heritable genetic changes in the bacterium. The P-state RecA-DNA filament, comprising RecA, ATP, and three DNA strands (tsDNA), uses ATP hydrolysis to carry out processive activities such as DNA recombinational repair and homologous recombination. These recombinational activities promote the horizontal transfer of antibiotic resistance genes. As described in the text, inhibitors that selectively bind the inactive conformation of RecA (red) would prevent nucleoprotein filament assembly, simultaneously precluding RecA’s signaling and motor activities. Inhibitors that prevent the assembled RecA-DNA filament from hydrolyzing ATP (blue) would prevent only motor-dependent processive activities.
One strategy for developing RecA inhibitors is to exploit the structural differences between the active and inactive conformers of the protein [20,21]. To carry out its biological functions, RecA must be bound to DNA in an active conformation (states A and P in Fig. 1); however, in the absence of DNA, RecA adopts an inactive conformation. Importantly, ADP and other select nucleotides stabilize the inactive conformer and inhibit the assembly of active RecA-DNA filaments [15,17,22-26]. Inhibitors of this class would abrogate all activities of the RecA-DNA filament, including both signaling and processive recombinational activities. In contrast, because the ATP-hydrolysis-dependent motor activities associated with recombination require the P state, inhibitors that are selective for the conformational P state may allow separation of the motor-like and signaling functions of RecA [27].
Toward the goal of discovering small-molecule ligands that can prevent either assembly of active RecA-DNA filaments or subsequent ATP-dependent motor activities (Fig. 1), we report the design and initial validation of a complementary pair of high-throughput-compatible screening assays suitable for the identification of inhibitors of RecA activities. One assay is based on established methods for monitoring ATPase enzyme activity and the second is a novel assay for RecA-DNA filament assembly using fluorescence polarization (FP). Our laboratory has recently reported the characterization of nucleotide analogs that are capable of differentiating between the active and inactive conformations of RecA as a means of modulating the protein’s activity [15,17]. Based on these results, we have used select nucleotide analogs as control compounds for assay validation. Importantly, this pair of screening assays allows the ready identification and segregation into two orthogonal classes of inhibitors of RecA’s coincident functions: (1) ATP-hydrolysis driven motor activity resulting in recombination, and (2) cell-signaling activities inducing the SOS response. Moreover, the screening assays can be readily configured for use in future automated high-throughput screening (HTS) projects to discover other classes of inhibitors.
Materials and methods
Materials
The E. coli RecA protein was purified as described previously [28] to ≥ 97% homogeneity and stored in aqueous buffer (25 mM Tris·HCl, pH 7.5, 1 mM DTT, 5% glycerol) at -80 °C. The protein concentration was determined using the monomer extinction coefficient 2.2 × 104 M-1cm-1 at 280 nm [29]. Both the 5′-biotinylated dT36 and the 32mer oligonucleotide with fluorescein linked at the 5′ end, which was identical in sequence to the oligonucleotide used by Defais and coworkers for their studies of RecA complex formation [30], were obtained from Sigma-Genosys (The Woodlands, TX). The concentrations of the oligonucleotides were determined using extinction coefficients calculated from the sum of the nearest-neighbor values at 260 nm. However, the A260 value measured for the fluorescein-conjugated oligonucleotide was first corrected for the contributions from fluorescein using the observed A495 and the ratio of extinction coeffetients for fluorescein at 260 and 495 nm.
ATP, ADP, and ATPγS were obtained at the highest purity possible from Roche (Nutley, NJ). The poly(dT) (average length = 319 nts) was purchased from Amersham Biosciences (Piscataway, NJ). Streptavidin paramagnetic particles (SA-PMP) were obtained from Promega (Madison, WI). Biomol Green colorimetric reagent was purchased from Biomol (Plymouth Meeting, PA). Unless otherwise specified, all other reagents were purchased at the highest purity possible from Sigma-Aldrich (St. Louis, MO). Clear flat- and U-bottom 96-well microplates were purchased from Evergreen Scientific (Los Angeles, CA). Black, flat-bottom, reduced volume 96-well microplates were purchased from Corning (Corning, NY).
Assay for inhibition of RecA-DNA filament assembly using biotin-dT36 bound to streptavidin paramagnetic beads
RecA (4 μM) was incubated at 37 °C for 15 min with the indicated concentration of ADP, 18 μM-nts biotin-dT36, 2 μM ATPγS and 60 mM NaCl in 1x Assay Buffer (25 mM Tris·HOAc, pH 7.5 at 25 °C, 5% (v/v) glycerol, 10 mM Mg(OAc)2, 1 mM DTT) in a total volume of 50 μL. During the incubation period, 50 μL of streptavidin paramagnetic particles (SA-PMP) were taken from the stock slurry (1 mg/mL) and washed three times with 50 μL of 1x Assay Buffer in a U-bottom, 96-well microplate by resuspension in buffer followed by pelleting with a magnet and removal of the supernatant. The entire 50 μL reaction volume was then added to the washed SA-PMP and the suspension was mixed thoroughly to ensure coating of the beads with the reaction mixture. The bead-reaction mixtures were incubated in a U-bottom 96-well microplate for 15 min at 37 °C in an Eppendorf Thermomixer R microplate incubator with shaking at 450 rpm.
Two colorimetric assay methods were used to monitor the extent of RecA-DNA filament assembly. After the incubation period, the SA-PMP were pulled down with a microplate magnet and a 10-μL sample of the supernatant was removed for gel analysis. Each sample was fractionated on a 12% SDS-PAGE denaturing gel for 1 h at 30 V and the gel was silver stained to visualize the protein. For direct analysis of the screen, a 10-μL sample of the supernatant was transferred to a clear, flat-bottom 96-well microplate. To this sample was added 200 μL of a 1:5 solution of Protein Assay Reagent (BioRad) in deionized H2O, and the absorbance of the resulting solution was measured in a Perkin Elmer HTS-7000 Plus BioAssay Reader using a 595 ± 25 nm bandpass filter. The A595 value was converted to [RecA] by comparison with a RecA standard curve to quantify the RecA remaining in the supernatant.
For purposes of comparison, the extents to which nucleotides inhibited RecA-(dT)36 filament assembly were measured as above over a fixed nucleotide concentration range (0 - 1000 μM). The apparent dissociation constants (Kdapp) were determined by nonlinear least squares analysis of the resulting titration isotherms using the following equation with Ymin, Ymax, and Kdapp as adustable parameters:
| (1) |
In equation 1, y is the observed amount of RecA, [L] is the total concentration of the ligand added, Ymin and Ymax are the minimum and maximum levels of RecA protein, respectively, and Kdapp is the apparent dissociation constant.
Fluorescence-based RecA-DNA filament assembly assay
RecA (4 μM) was incubated at 25 °C for 15 min with the indicated concentration of ADP, 18 μM-nts fluorescein-labeled 32mer oligo, 2 μM ATPγS, and 60 mM NaCl in a total volume of 30 μL of 1x Assay Buffer in the wells of a black, flat-bottom, reduced-volume 96-well microplate. After the 15 min incubation, total fluorescence emission and polarization were collected in a PHERAstar 96-well fluorescence polarization microplate reader (BMG Labtech, Offenberg, Germany) through a cut-off emission filter (520 nm; BMG Filter 520P) with a bandpass filter for excitation (485 ± 7.5 nm; BMG Filter 485P). Data were plotted and fit using the following equation with Ymin, Ymax, Kdapp, and n as adustable parameters:
| (2) |
In equation 2, y is fluorescence polarization, [L] is the total concentration of the ligand added Ymin and Ymax are the minimum and maximum fluorescence polarization respectively, Kdapp is the apparent dissociation constant, and n is a factor to account for any apparent cooperativity in the titration data.
NADH-oxidation coupled assay for ATPase activity
The first type of enzyme-linked ATPase assay couples the formation of ADP, generated from the hydrolysis of ATP by RecA, to the oxidation of NADH to NAD+. The assay was performed as previously described [31,32] with modifications for usage in 96-well microplates. Reactions (100 μL final volume) were performed in a clear, flat-bottom 96-well microplate and contained 1 μM RecA, up to 1 mM ATP, 15 μM-nts poly(dT), 2.3 mM phosphoenolpyruvate, 5 U·mL-1 pyruvate kinase, 5 U·mL-1 lactic dehydrogenase and 2 mM NADH in 1x Assay Buffer. ATPase reactions were initiated at 37 °C by adding the pre-incubated RecA-containing cocktail to a solution of ATP using a multi-channel pipettor, and the absorbance of the reaction was recorded every 30 s through a bandpass filter (380 ± 10 nm) in a Perkin Elmer HTS-7000 Plus BioAssay Reader. The initial, steady-state reaction velocity (voobs; μM·min-1) was calculated from the change in absorbance as a function of time (δA/δt) using Δε380 = 2.57 × 10-4 μM-1 as measured in the microplate reader. The data were analyzed using a Michaelis-Menten equation modified for substrate cooperativity as described previously [33,34]:
| (3) |
where R0 is the total concentration of RecA and S0.5 is the [NTP] when the velocity is half of its maximum value.
MESG-phosphorolysis coupled assay for ATPase activity
The second type of enzyme-linked assay used involves the detection of free phosphate generated during the ATP hydrolysis reaction by RecA. This system has been used successfully for a variety of phosphate-detection based kinetic studies including ATPase [35], GTPase [35,36], phosphatase [37-39], and kinase [35,40] assays. ATP hydrolysis by RecA was measured using a Perkin-Elmer HTS-7000 Plus Bioassay Reader with a 360 ± 5 nm bandpass filter. Reactions (100 μL final volume) were performed in a clear, flat-bottom 96-well microplate and containined 0.5 μM RecA, up to 1 mM ATP, 15 μM-nts poly(dT), 0.3 mM MESG, and 1 U·mL-1 PNP in 1x Assay Buffer. To initiate the reactions at 37 °C, the pre-incubated RecA-containing cocktail was added to a solution of ATP using a multi-channel pipettor, and the A360 was monitored every 30 s in the microplate reader. The initial, steady-state reaction velocity (voobs; μM·min-1) was calculated from the change in absorbance as a function of time (δA/δt) using Δε360 = 6.0 × 10-4 μM-1 as measured in the microplate reader. The data were analyzed using equation 3 as described above.
Colorimetric phosphate-detection assay for ATPase activity
A single-point, colorimetric assay for free phosphate detection was also employed to monitor the ATPase activity of RecA for situations when an enzyme-linked system may not be suitable. This assay was conducted using Biomol Green (Biomol), an analog of the malachite green reagent that is used for detection of free phosphate in solution, most notably in protein phosphatase assays [41-43]. Reactions were performed at 37 °C in a clear, flat-bottom 96-well microplate and contained 0.5 μM RecA, up to 300 μM ATP and 15 μM-nts poly(dT) in 1x Assay Buffer (50 μL final volume). Reaction timepoints were taken by addition of 100 μL Biomol Green reagent to each well to stop the reaction, and the plate was incubated for 20 min at room temperature prior to reading the absorbance of the solutions.
To determine the amount of phosphate released during the ATPase reaction, the absorbance of the solution in the wells was measured in a Perkin Elmer HTS-7000 Plus BioAssay Reader using a bandpass filter (625 ± 15 nm), and the absorbances were compared to those of a series of phosphate standards to determine phosphate concentration at the different timepoints of the reaction. Data were analyzed as above using equation 3 to determine the relevant kinetic parameters.
Results and Discussion
We propose that the discovery of small molecules that can selectively modulate the activities of RecA could be exploited to tease apart the roles of its biological functions in the de novo development and transmission of antibiotic resistance. Because there are no known natural products that inhibit RecA, such a discovery effort would be aided by robust activity assays that could be configured for use in high-throughput screens. Toward this goal, we endeavored to develop a complementary pair of molecular screening assays for RecA-DNA filament assembly and activity. Because RecA can be inhibited by ADP as well as a variety of nucleotide analogs [17,25,44-46], we employed ADP and select NTPs characterized previously in our laboratory as inhibitors for positive control experiments to validate the assays.
Direct assay for RecA-DNA filament assembly
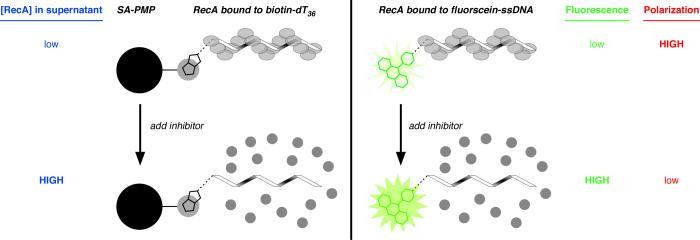
The first assay was designed to evaluate the inhibition of RecA-DNA filament assembly by direct monitoring of the protein released from a single-stranded DNA (ssDNA) substrate resulting from the addition of a putative inhibitor. Our laboratory recently reported a method to assess the effects of nucleoside di- and triphosphates on the binding of RecA to ssDNA [15,17]. The assay design takes advantage of two important observations: (1) ADP decreases the stability of NPFs [26]; and (2) the addition of ADP to a complex of RecA bound to a 30mer oligonucleotide results in disassembly of the complex and concomitant release of RecA from the DNA [23]. Combined with several other qualitative reports in the RecA field [47-50], these observations led us to consider whether ADP could prevent RecA from binding to ssDNA immobilized on streptavidin-coated paramagnetic particles (SA-PMP; see Fig. 2).
Fig. 2.
Cartoons depicting three assays to assess the extent of assembly of a RecA-DNA filament. In all variations of the assay, an oligonucleotide (32 to 36 nts in length) is incubated with RecA to allow filament assembly, then a putative inhibitor of filament assembly is added. (Left) Assay for RecA assembly on biotin-dT36 associated with streptavidin-coated paramagnetic particles (SA-PMP). If filament assembly on biotin-dT36 is disrupted, any RecA in the supernatant can be detected direclty by colorometric protein quantification after the RecA bound to biotin-dT36 is pulled down using a magnet (blue text). (Right) Assay for RecA assembly on fluorescein-ssDNA using fluorescein emission (either total emission or fluorescence polarization) as the observable parameter. If filament assembly on a 32-nt ssDNA conjugated to fluorescein at the 5′ end is disrupted, the RecA-bound state emits a low total fluorescence signal (green text) and high fluorescence polarization signal (red text); conversely, the RecA-free oligonucleotide state is characterized by high total emission and low fluorescence polarization.
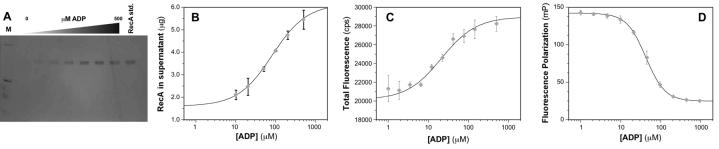
In practice, the assay is conducted by incubating the nucleotide with RecA and biotin-dT36, and then immobilizing the ssDNA and any bound RecA. The extent to which RecA-DNA filament assembly is impeded in the presence of a nucleotide analog is assessed by measuring the amount of RecA in the supernatant (Fig. 3) using either silver staining of denaturing electrophoretic gels or Bradford analysis. Optimization of the assay protocol was undertaken to maximize the signal-to-noise ratio, and inclusion of both 50 mM NaCl and a sub-stoichiometric concentration of ATPγS (2 μM ATPγS vs. 4 μM RecA) were found to yield optimal results for monitoring the concentrations of RecA remaining in the supernatant when RecA-DNA assembly was inhibited by ADP (optimization trial data not shown). Using these conditions, we demonstrated that ADP inhibits assemby of a RecA filament on biotin-dT36 (Fig. 3), recapitulating what is known about the effect of ADP on RecA-DNA interactions [25,46]. Increasing amounts of ADP led to increasing amounts of RecA in the supernatant. The silver-stained gel shows that at low concentrations of ADP, little RecA is released into the supernatant, while complete dissociation of RecA from the biotin-dT36 was achieved at 200 μM ADP (Fig. 3A).
Fig. 3.
Dependence of extent of RecA-DNA filament assembly on concentration of added ADP. (A) Silver-stained gel from SDS-PAGE fractionation of the RecA-DNA filament assembly reactions (Fig. 2, left) in the presence of increasing amounts of ADP. The lane labeled “RecA std.” contains 1.25 μg RecA protein, the maximum amount of protein that can be released in the assay. The concentration of ADP varied between 0 to 500 μM. (B) Plot of amount of unbound RecA, determined using Bradford assay for protein in the supernatant, as a function of ADP concentration for the RecA-DNA filament assembly assay. The data points represent the mean ± one standard deviation of at least three independent experiments, which were identical to those used to create (A). The smooth curve represents the best-fit binding isotherm (Kd = 80 ± 30 μM) as described in Materials and Methods. (C) Plot of total fluorescence in counts per second (cps) as a function of ADP concentration for the RecA-DNA filament assembly assay (Fig. 2, right). The data points represent the mean ± one standard deviation of at least three independent experiments. The smooth curve represents the best-fit binding isotherm (Kd = 20 ± 10 μM) as described in Materials and Methods. (D) Plot of fluorescence polarization in millipolarization units (mP) as a function of ADP concentration for the RecA-DNA filament assembly assay (Fig. 2, right). The data points represent the mean ± one standard deviation of at least three independent experiments. The smooth curve represents the best-fit binding isotherm (Kd = 42 ± 3 μM) as described in Materials and Methods.
To ensure that the screening assay’s observable would quantitatively respond to ADP concentration as established in classic biochemistry experiments, we measured the apparent equilibrium dissociation constant for RecA-ADP binding by monitoring the amount of RecA released from biotin-dT36 as a function of ADP concentration. The resulting titration isotherm was fit to a standard equation for a right hyperbola (equation 1) to give Kdapp = 80 ± 30 μM (Fig. 3B). This value compares favorably to Kdapp = 120 μM measured by Lee and Cox using a homogeneous solution-phase assay [25], verifying that the rapid nucleotide binding assay results are quantitatively related to the extent of RecA-nucleotide binding. As further validation of the assay, we cite two important control experiments. First, a correlation was observed between the abilities of six non-substrate NTPs to inhibit RecA-DNA filament assembly at a concentration of 100 μM and the Kdapp value calculated from a titration of the NTP (Table 1). Second, no differences were detected among the extents of RecA-ssDNA dissociation stimulated by nine pairs of canonical nucleoside di- and triphosphate pairs that are known to be at least moderate substrates for RecA NTPase activity [17]. This observation is consistent with the fact that the conditions of the DNA displacement screen allow RecA to hydrolyze an NTP to the corresponding NDP during the experiment.
Table 1.
Inhibition of RecA activities by nucleotide analogs as measured in microplate assays.
| Filament Assembly |
||
|---|---|---|
| Nucleotide Entry | % Inhibition (at 100 μM) | Kdapp (μM) |
| 1 | 89 ± 5 | 60 ± 30 |
| 2 | 82 ± 2 | 44 ± 6 |
| 3 | 75 ± 7 | 50 ± 30 |
| 4 | 65 ± 2 | 100 ± 40 |
| 5 | 55 ± 4 | 340 ± 190 |
| 6 | 32 ± 3 | ≥ 500 |
| ATPase Activity |
||
| Nucleotide Entry | % Inhibition (at 100 μM) | Kic (μM) |
| 6 | 98 ± 2 | 0.8 ± 0.1 |
| 2 | 34 ± 4 | 63 ± 7 |
| 1 | 11 ± 1 | 140 ± 50 |
| 3 | 7 ± 2 | 160 ± 20 |
| 5 | 6 ± 3 | ≥ 500 |
| 4 | 2 ± 2 | ≥ 500 |
Apparent dissociation constants (Kdapp) were assessed by measuring the extent of filament assembly in the FP assay over an inhibitor concentration range of 0 to 1000 μM (see Materials & Methods and Fig. 3D). ATP-competitive inhibition constants (Kic) were assessed by measuring the apparent S0.5 for ATP in the ATPase assay over an inhibitor concentration range of 0 - 1000 μM (see Materials & Methods and Fig. 5B). The identity of each nucleotide analog represented in the table is indicated by a specific numeral in the “Nucleotide Entry” column: (1) TTP (CAS# 18423-43-3); (2) N6-(1-naphthyl)ADP (see reference [15]); (3) 5-Me-UTP (CAS# 1463-10-11); (4) O6-Me-GTP (CAS# 99404-63-4); (5) 2′-O-Me-ATP (CAS# 2140-79-6); and (6) 2′-(or 3′)-O-(N-methylanthraniloyl)ATP (MANT-ATP, CAS# 85287-56-5).
Fluorescence assay for RecA-DNA filament assembly using a fluorescein-oligonucleotide conjugate
To adapt the RecA-DNA filament assembly assay for high-throughput format, we decided to employ fluorescence as a detection tool for RecA dissociation. Previous research established that the emission of a fluorescein dye conjugated to an oligonucleotide was sensitive to the binding of RecA to the ssDNA [51-54]. To take advantage of this fact, we used a fluorescein-labeled 32mer as the ssDNA substrate in the RecA-DNA filament assembly assay (Fig. 2). We analyzed the effect of ADP on RecA-DNA filament assembly using fluorescein emission in a microplate assay using solution conditions identical to those optimized for the magnetic pull-down assay described above. A 40% enhancement of the fluorescein-oligonucleotide emission was observed upon addition of ADP (500 μM) to the RecA-oligonucleotide complex, providing a direct spectroscopic measurement of RecA-DNA filament assembly.
The fluorescence of the RecA-ssDNA solution was then measured in a 96-well microplate after incubation with ADP, and the resulting titration is shown in Fig. 3C. The data fit a binding isotherm similar to that for the SA-PMP direct assay with Kdapp = 20 ± 10 μM. Thus, the fluorescence assay reports similar results to those of the magnetic pull-down assay.
Optimization of the fluorescence assay: a “mix and measure” fluorescence polarization assay for RecA-DNA filament assembly
A molecular assay can only be functional for high-throughput screening if, at a minimum, it reports values that readily discriminate the activities of potential inhibitors. Although the fluorescence of the 5′-fluorescein-labeled 32mer serves as a sensitive indicator of RecA-DNA filament assembly, the mean signal for the positive control (500 μM ADP) was only 40% above signal for the background (absence of ADP). Hence, we elected to explore fluorescence anisotropy, which is known to distinguish the free and RecA-bound oligonucleotides [55,56] as an assay observable.
We performed experiments using polarized excitation and emission to observe the RecA-ssDNA complex in solution, and found that the binding of RecA protein to the oligonucleotide results in an increase in the extent of polarization of the emission from the fluorescein-oligonucleotide. Specifically, the fluorescence polarization (FP) of the oligonucleotide in the presence of RecA and 500 μM ADP was 35 mP, and this value increased to 133 mP for the RecA·oligonucleotide complexes formed in the absence of ADP. FP data were recorded for a RecA-DNA solution in a 96-well microplate after incubation with varying concentrations of ADP, and the resulting titration is shown in Fig. 3D. Fitting to the standard binding isotherm revealed an apparent Kd of 42 ± 3 μM, in good agreement with the corresponding values from the other versions of the filament assembly assay.
To summarize, we observed [ADP]-dependent dissociation of RecA, quantified both by colorimetric protein assay and by the change in FP of a fluorescein-oligonucleotide conjugate. These results confirm that the assay for RecA-DNA filament assembly is sensitive to potential inhibitors. Extension of this system to other nucleotides demonstrates that a variety of nucleotide analogs are able to prevent RecA-DNA filament assembly (Table 1). It is noteworthy that both purine and pyrimidine nucleotide analogs, as well as those bearing substitutions on either the DNA base or the 2′ and 3′ positions of the ribose ring, can inhibit this important RecA activity. A more detailed discussion of the structure-activity relationships for nucleotide analogs binding to RecA - and the implications of these relationships on inhibitor design - will be presented elsewhere (Wigle & Singleton, manuscript in preparation).
Statistical evaluation of the assays for RecA-DNA filament assembly
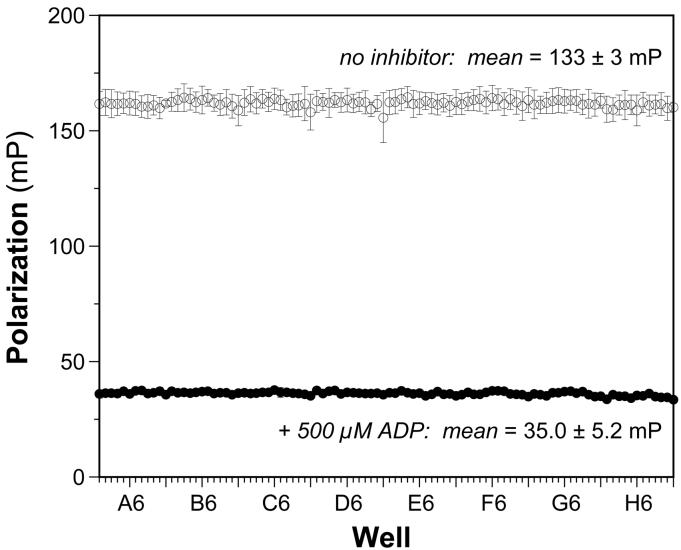
One useful measure of the suitability of an assay for high-throughput screening is the Z’ factor, which is a screening window coefficient that reflects both the assay signal dynamic range and data variation associated with signal measurements for control experiments [6]. To evaluate the two different fluorescence-based activity screens above for robust and reproducible behavior in a 96-well format, we compared the signal:background and signal:noise ratios as well as the Z’ factors calculated across a 96-well microplate for each assay. We performed a single-point assay with 500 μM ADP in each well of a 96-well microplate and compared it to the same assay done in the absence of ADP. Over a span of seven days, three replicates of each plate assay was performed to test for interday and intercolumn variability. The mean positive control and background signals, as well as the coefficients of variation in each, were compiled for each of three assays. Although the fluorescence-based filament assembly assay resulted in a more favorable Z’ factor than the direct, magnetic pull-down assay, the absolute value of the former (Z’ = 0.55) indicated a less-than-ideal screening assay. Importantly, a significant difference between the positive and negative controls for the FP assay were observed (Fig. 4), leading to a favorable Z’ factor of 0.87.
Fig. 4.
Reproducibility of FP-based assay for RecA-DNA assembly under optimal conditions. The FP was measured for each well of a 96-well plate in the absence (open circles) or presence (open circles) of 500 μM ADP. Each data point indicates the mean FP (in millipolarization units, mP) for each well from three different experiments on three different days; the error bars indicate the standard deviation of each mean value. The overall means for the positive signal control (500 μM ADP) and negative background control (no inhibitor) were 35.0 ± 5.2 mP (CV = 15%) and 133 ± 3 (CV = 2%), respectively. The signal-to-background ratio was 3.8 and the signal-to-noise ratio was 18. The overall Z’ factor was 0.87.
Continuous enzyme-linked assay for RecA’s ATPase activity
The first step in both RecA-mediated SOS induction and recombinational DNA repair is the binding of RecA to ATP and ssDNA to form an active RecA-DNA filament. Active filament assembly normally results in ATP hydrolysis, which is necessary for controlling SOS induction as well as for the subsequent stages of recombinational DNA repair. ATP hydrolysis serves as a useful indicator of filament activity, and the abrogation of ATPase activity would be an important aspect of RecA inhibition. Hence, our initial approach to developing an assay for the ATPase activity of the RecA-DNA filament was to adapt a continuous, spectrophotometric method for monitoring production of ADP.
Many studies of RecA’s ATPase activity have employed a multi-enzyme assay that couples the production of one molecule of ADP to the oxidation of one molecule of NADH [31,32]. We have previously reported the adaptation of this method to evaluating RecA ligands using a microplate format [17]. Unfortunately, a statistical evaluation of an optimized version of this RecA ATPase assay yielded a Z’ factor of only 0.52 (see below), which indicated an assay that would be marginally useful for high-throughput screening [6].
To improve the reproducibility and robustness of the filament activation assay, we evaluated an alternate coupled enzyme assay for detection of free phosphate by the conversion of the substrate MESG to 2-amino-6-mercapto-7-methylguanine. We have previously reported the use of this method to determine the reaction velocities and kinetic parameters for a wide variety of NTP substrates for RecA’s ATPase activity [17]. One potential advantage of the MESG-coupled assay is that it also allows for the rapid screening of inhibition of RecA ATPase activity, as the phosphate detection system is not dependent on the coupled activity of an ADP-dependent enzyme (e.g., pyruvate kinase). We anticipate that the PNP enzyme will not be as sensitive to inhibition by nucleotide-analog inhibitors of RecA. Therefore, we undertook the adaptation of this protocol to rapid screening using a microplate format.
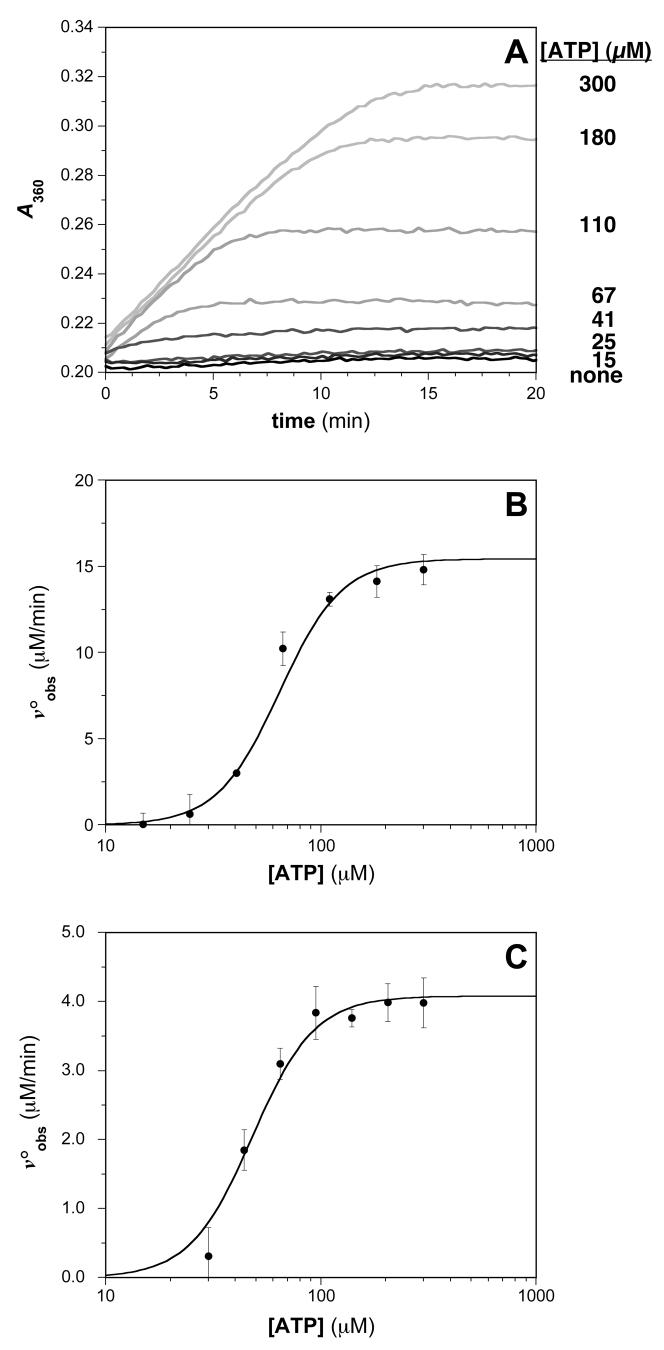
Optimization of the assay protocol was undertaken to maximize the signal-to-noise ratio, which represents a challenge when using RecA because the equilibrium constant for its self-association during filament assembly and activation is modest. Thus, in spite of a relatively low turnover number for ATP hydrolysis (kcat ≤ 0.5 s-1), the absolute rate of ATP hydrolysis is high under conditions where RecA-DNA filament formation is favorable. In balancing these competing factors, optimal conditions in 96-well plates were identified when RecA and poly(dT) were present at 0.5 μM and 1.5 μM-nts, respectively (data not shown). Fig. 5A shows an absorbance-versus-time plot of the data obtained from the MESG-based assay under these conditions in the presence of various ATP concentrations. From this data, the slope of the initial, linear portion of each data set was measured and converted to voobs as described in Materials and Methods. The data from these assays was further analyzed using equation 3 as shown in Fig. 5B, where the voobs is plotted vs. concentration of substrate to extract the relevant kinetic parameters (kcat and S0.5). For the assay shown, Vmax = 16 ± 1 μM·min-1, which corresponds to kcat = 32 min-1, and the S0.5 for ATP is 64 ± 2 μM. These kcat and S0.5 parameters are in good agreement with those reported previously for E. coli RecA [31,32,57,58]. Given that the assay recapitulates what is known about RecA’s ATPase activity, future comparisons of corresponding results obtained in the presence of varying concentrations of potential RecA ATPase inhibitors will allow for an accurate determination of the Ki for each inhibitor as well as the nature of the inhibition (competitive vs. noncompetitive vs. uncompetitive).
Fig. 5.
Typical results for assays of RecA-DNA ATPase activities. (A) Time-dependent generation of MESG using an enzyme-linked phosphate detection system in the presence of various ATP concentrations. The absorbance at 360 nm for reaction solutions containing 0.5 μM RecA in the presence of the indicated concentration of ATP is monitored. (B) ATP concentration dependence of the steady-state ATP hydrolysis rate. The plot of voobs vs. [ATP] was constructed, where the initial velocities were determined from the slopes of the plots in (A). The steady-state kinetic parameters S0.5 and kcat were obtained using equation 3 as described in Materials and Methods: kcat = 32 ± 2 min-1; S0.5 = 64 ± 2 μM. (C) ATP concentration dependence of the steady-state ATP hydrolysis rate measured using the Biomol Green phosphate detection assay. The S0.5 and Vmax parameters for the reaction were determined using equation 3 as described in Materials and Methods: kcat = 8.2 ± 0.4 min-1; S0.5 = 48 ± 3 μM.
To optimize a single-point screen (one [ATP] and one [inhibitor]) for identifying non-substrate nucleotides that bind the active RecA-DNA filament, we measured the inhibition constants for the ATP-competitive inhibition of RecA-DNA activation, and compared the resulting Kic values to a single point of relative ATPase inhibition. The analysis included data for six non-substrate NTPs [17] and ADP, which is a well-established inhibitor [44,46,59,60], and the optimal conditions were found to include 100 μM nucleotide inhibitor and 500 μM ATP. The use of the single-point screening assay was validated by a correlation (Table 1) demonstrating that a single ATPase measurement at 100 μM nucleotide is sufficient to characterize rapidly its potential to inhibit ATP hydrolysis by RecA-DNA filaments.
Non-enzymatic assay for RecA’s ATPase activity
One possible drawback to the system described above is that the inhibitors we are testing have the potential to interfere with the additional enzymes present in the system, and could create false positives. With this drawback in mind, we decided to also employ a direct assay for monitoring production of free phosphate using a modified version of the colorimetric malachite green reagent, Biomol Green.
As with the MESG-based detection method described above, optimization of the Biomol Green assay protocol was challenging because the absolute rate of ATP hydrolysis is high under conditions where RecA-DNA filament formation is favorable. Nevertheless, optimal conditions in 96-well plates included RecA and poly(dT) concentrations of 0.5 μM and 15 μM-nts, respectively. Importantly, to keep the free phosphate concentration in the linear dynamic range of the assay reagent, the time of the reactions had to be limited to no more than 5 min. Using the Biomol Green discontinous assay system, single timepoints were taken in a 96-well plate by the addition of the Biomol Green reagent to stop the ATPase reaction, and free phosphate was measured by comparison to a phosphate standard curve. Absorbance-versus-time plots were constructed for the hydrolysis reaction in the presence of various initial ATP concentrations. From this data, the slope of the initial, linear portion of each data set was measured and converted to voobs as described in Materials and Methods. Fig. 5C shows the data obtained using the Biomol Green assay system and their analysis using equation 3. The S0.5 value for ATP was determined to be 48 ± 3 μM, a value that agrees well with those values reported previously [57,58].
Statistical evaluation of the assays for RecA’s ATPase activity
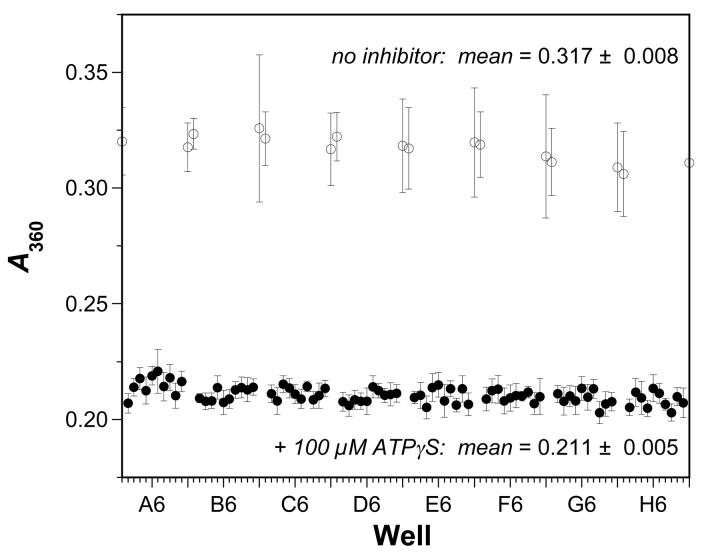
To evaluate the two different ATPase activity screens above for robust and reproducible behavior in a 96-well format, we compared the signal:background and signal:noise ratios as well as the Z’ factors calculated across a 96-well microplate for each assay. For the MESG-coupled assay, Z’ was calculated in a similar fashion. The assays were performed as above with 500 μM ATP, and the data were collected as usual. The absorbance at 15 min was taken as the positive control value, as the reactions have proceeded to completion at that point, and this value was compared to the value at 15 min when 100 μM ATPγS was added to calculate Z’ for the assay. Fig. 6 shows the results for the MESG-coupled assay. As with the fluorescence polarization assay, there is a significant difference between the positive and negative controls for the assay, and the Z’ factor of 0.63 indicates that the MESG-coupled system is useful for high-throughput screening.
Fig. 6.
Reproducibility of MESG-based assay for RecA-DNA ATPase activity under optimal conditions. The MESG absorbance was measured at 360 nm after 15 min in each well of a 96-well plate in the absence (open circles) or presence (open circles) of 100 μM ATPγS. The concentration of ATP was 500 μM in the experiments. Each data point indicates the A360 for each well from three different experiments on three different days; the error bars indicate the standard deviation of each mean value. The overall means for the positive signal control (100 μM ATPγS) and negative background control (no inhibitor) were 0.211 ± 0.005 (CV = 2%) and 0.317 ± 0.008 (CV = 3%), respectively. The signal-to-background ratio was 1.5 and the signal-to-noise ratio was 13. The overall Z’ factor was 0.63.
In similar fashion the Z’ factor for the Biomol Green phosphate detection system (data not shown) was calculated to be Z’ = 0.68, with a high signal:noise ratio and a signal:background ratio greater than 1. Overall, the combination of a continuous system (MESG) and an alternative, single-point system (Biomol Green) that can be used for inhibitor screening will allow for rapid determination of both the inhibition of RecA’s ATPase activity by compounds and also the reaction velocity and inhibition constants associated with a particular inhibitor of RecA’s ATPase activity.
Segregation of inhibitors of RecA activities
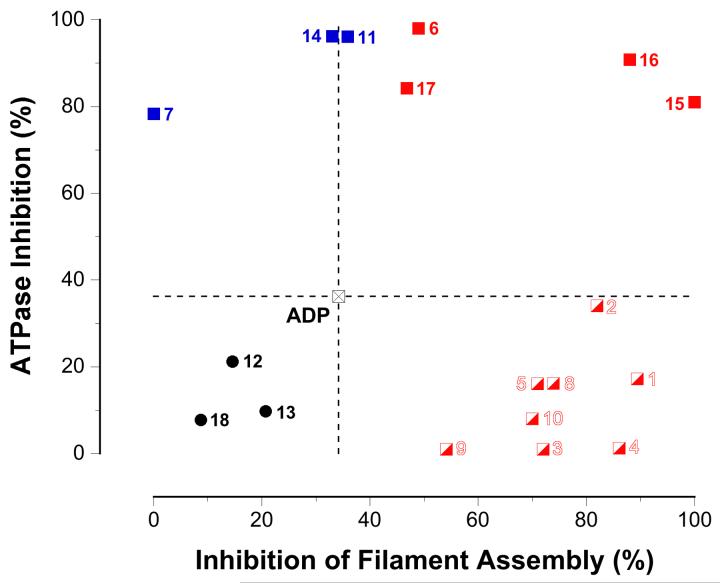
We envisaged the two screening assays as a coupled pair that would serve to provide complementary information on two of RecA’s coincident functions: (1) signaling activities inducing the cellular SOS response, and (2) ATP-hydrolysis driven motor activity resulting in recombination. This feature is critical to the strategic development of conformationally selective ligands that would allow separation of the motor-like and signaling functions of RecA and thereby permit dissection of resistance gene development and transmission pathways in bacterial pathogens. To assess the extent to which the nucleotide analog inhibitors are conformationally selective, a plot comparing each compound’s abilities to inhibit RecA’s activity in the two screening assays was constructed (Fig. 7).
Fig. 7.
Segregation of inhibitor activity in two dimensions. The graph shows a plot of the ATPase inhibition of each compound vs. the dissociation of the RecA-DNA interaction induced by each compound. The activities of ADP, plotted as the crossed black square at the origin and the dotted lines, are used as a reference for comparing the activities for each of the other inhibitors. Nucleotide analogs that are more inhibitory than ADP in both assays are plotted quadrant I (red squares). Nucleotide analogs that are less inhibitory than ADP in both assays are found in quadrant III (black circles). Nucleotide analogs that are more inhibitory than ADP in the ATPase assay but not the filament assembly assay are plotted in quadrant II (blue squares). Nucleotide analogs that are more inhibitory than ADP in the filament assembly assay but not the ATPase assay are plotted in quadrant IV (red-and-white squares). See the text for more details. The identity of each nucleotide analog represented in the plot is indicated by a specific numeral as follows: (1) TTP (CAS# 18423-43-3); (2) N6-(1-naphthyl)ADP (see reference [15]); (3) 5-Me-UTP (CAS# 1463-10-11); (4) O6-Me-GTP (CAS# 99404-63-4); (5) 2′-O-Me-ATP (CAS# 2140-79-6); (6) 2′-(or 3′)-O-(N-methylanthraniloyl)ATP (MANT-ATP, CAS# 85287-56-5); (7) adenosine 5′-O-(γ-thio)triphosphate (ATPγS, CAS# 93839-89-5); (8) 5-propynyl-dUTP (CAS# 111289-88-4); (9) Puromycin 5′-O-triphosphate (CAS# 101043-49-6); (10) N6-phenylADP (CAS# 105701-92-6); (11) 2′-(or 3′)-O-(4-benzoylbenzoyl)ATP (BzBz-ATP, CAS# 112898-15-4); (12) 2′-(or 3′)-O-(BODIPY® FL)-adenosine 5′-O-(β:γ-imido)triphosphate (BODIPY FL AMPPNP, Invitrogen # B-22356); (13) adenosine 5′-O-triphosphate, P3-(5-sulfo-1-naphthylamide) (ATPγS AmNS, Invitrogen # A-12412); (14) 2′-(or 3′)-O-(N-methylanthraniloyl)ADP (MANT-ADP, CAS# 125902-32-1); (15) 2′-(or 3′)-O-(N-methylanthraniloyl)GDP (MANT-GDP; CAS# 128451-34-3); (16) 2′-(or 3′)-O-(trinitrophenyl)ADP (TNP-ADP, CAS# 807261-76-30); (17) 2′-(or-3′)-O-(N-(2-aminoethylcarbamoyloxy)BODIPY® TR)ADP (BODIPY TR ADP, Invitrogen # A-22359); and (18) Tenofovir (CAS# 147127-20-6).
Figure 7 shows the extent to which each nucleotide analog inhibited RecA’s ATPase activity as a function of the extent to which the compound inhibited RecA-DNA filament assembly. The inhibitory properties of ADP were used as a reference for comparison of the activities of each nucleotide analog inhibitor. With respect to the coordinate system origin corresponding to ADP, four quadrants can be defined: quadrant I contains compounds with inhibitory activities greater than ADP in both screening assays; quadrant II contains compounds that were more inhibitory than ADP in the ATPase assay but not the filament assembly assay; quadrant III contains compounds that were less inhibitory than ADP in both screening assays; and quadrant IV contains compounds that were more inhibitory than ADP in the filament assembly assay but not the ATPase assay.
Nucleotide analogs that were less inhibitory than ADP in both screening assays (quadrant III, black circles) can be discarded as “inactive”. Of higher interest are those nucleotide analogs that are more inhibitory than ADP in the ATPase assay (quadrant II, blue squares). Although these compounds do not perturb RecA-DNA filament assembly, they inhibit the ATP turnover that is required for RecA’s recombinational motor activities and may allow the motor-like and signaling functions of RecA to be separated (see Fig. 1).
Nucleotide analogs that were more inhibitory than ADP in both screening assays (quadrant I, red squares) suppressed all RecA activities, including both RecA-DNA filament assembly and ATP hydrolysis. These compounds prevent the assembly of active RecA-DNA filaments altogether - presumably by stabilizing the inactive RecA conformer - and would thereby abrogate both signaling and processive recombinational activities of RecA.
Nucleotide analogs that were more inhibitory than ADP in the filament assembly assay but not the ATPase assay (quadrant IV, red-and-white squares) provide additional insight. Although such compounds apparently prevent RecA-DNA filament assembly without inhibiting the subsequent ATP hydrolysis, this cannot be the true interpretation because RecA-DNA filament assembly is obligatory for ATP hydrolysis. Consideration of the ATP concentration in the different screening assays is important for unraveling this apparent conundrum. In particular, the ATPase assay is conducted in the presence of 0.5 mM ATP, while the filament assembly assay is conducted with only sub-stoichiometric ATPγS (2 μM). We conclude that nucleotide analogs found in quadrant IV are capable of selectively stabilizing the inactive RecA conformer, but can be competed off by ATP at elevated concentrations. This further suggests that such compounds are likely to be weakly-binding competitive inhibitors, whether they bind the ATP-binding or allosteric sites. Although compounds in this group will not be good candidates for in vivo inhibition, their properties will be important for the elucidation of structure-activity relationships among ligands for the inactive RecA conformation.
Conclusions
In summary, we have developed two rapid and robust microplate screening assays suitable for identifying inhibitors of E. coli RecA. We envision that the coupled pair of assays will allow maximum throughput at the early stages of library screening while providing for maximum characterization of first-assay hits. Indeed, the assays should reveal complementary sets of agents that can either selectively suppress only the ATP-driven motor activities of the RecA-DNA filament or prevent assembly of active RecA-DNA filaments altogether. Results from screening a larger collection of nucleotide analogs and a model suitable for interpreting the activities of compounds in these two sets will be described elsewhere (Wigle & Singleton, manuscript in preparation). Although current results are limited to validation assays conducted using NTPs, the screening assays can be readily configured for use in future automated HTS projects to discover inhibitors in other structural classes. We envision that such inhibitors may be developed into novel adjuvants for antibiotic chemotherapy that moderate the development and transmission of antibiotic resistance genes and increase the antibiotic therapeutic index.
Acknowledgments
We are grateful to Prof. Matthew Redinbo (UNC-CH Department of Chemistry) for use of the fluorescence polarization plate reader, and to Dr. Eric Ortlund of the Redinbo laboratory for expert assistance with the instrument’s use. This work was supported by a grant to S.F.S. by the National Institutes of Health (GM58114).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Matic I, Taddei F, Radman M. Survival versus maintenance of genetic stability: a conflict of priorities during stress. Research in Microbiology. 2004;155:337–41. doi: 10.1016/j.resmic.2004.01.010. [DOI] [PubMed] [Google Scholar]
- [2].Friedberg EC, Walker GC, Siede W. SOS responses and DNA damage tolerance in prokaryotes, DNA Repair and Mutagenesis. ASM Press; Washington, D.C.: 1995. pp. 407–464. [Google Scholar]
- [3].Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. Journal of Molecular Biology. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- [4].Goodman MF. Coping with replication ‘train wrecks’ in Escherichia coli using Pol V, Pol II and RecA proteins. Trends in Biochemical Sciences. 2000;25:189–95. doi: 10.1016/s0968-0004(00)01564-4. [DOI] [PubMed] [Google Scholar]
- [5].Courcelle J, Ganesan AK, Hanawalt PC. Therefore, what are recombination proteins there for? Bioessays. 2001;23:463–70. doi: 10.1002/bies.1065. [DOI] [PubMed] [Google Scholar]
- [6].Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Progress in Nucleic Acid Research and Molecular Biology. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- [7].Casjens S. Prophages and bacterial genomics: what have we learned so far? Molecular Microbiology. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- [8].Kline KA, Sechman EV, Skaar EP, Seifert HS. Recombination, repair and replication in the pathogenic Neisseriae: the 3 R’s of molecular genetics of two human-specific bacterial pathogens. Molecular Microbiology. 2003;50:3–13. doi: 10.1046/j.1365-2958.2003.03679.x. [DOI] [PubMed] [Google Scholar]
- [9].Bisognano C, Kelley WL, Estoppey T, Francois P, Schrenzel J, Li D, Lew DP, Hooper DC, Cheung AL, Vaudaux P. A recA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. Journal of Biological Chemistry. 2004;279:9064–71. doi: 10.1074/jbc.M309836200. [DOI] [PubMed] [Google Scholar]
- [10].Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–31. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- [11].Hersh MN, Ponder RG, Hastings PJ, Rosenberg SM. Adaptive mutation and amplification in Escherichia coli: two pathways of genome adaptation under stress. Research in Microbiology. 2004;155:352–9. doi: 10.1016/j.resmic.2004.01.020. [DOI] [PubMed] [Google Scholar]
- [12].Foster PL. Stress responses and genetic variation in bacteria. Mutation Research. 2005;569:3–11. doi: 10.1016/j.mrfmmm.2004.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–4. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- [14].Hastings PJ, Rosenberg SM, Slack A. Antibiotic-induced lateral transfer of antibiotic resistance. Trends in Microbiology. 2004;12:401–4. doi: 10.1016/j.tim.2004.07.003. [DOI] [PubMed] [Google Scholar]
- [15].Lee AM, Ross CT, Zeng BB, Singleton SF. A Molecular Target for Suppression of the Evolution of Antibiotic Resistance: Inhibition of the Escherichia coli RecA Protein by N(6)-(1-Naphthyl)-ADP. J. Med. Chem. 2005;48:5408–11. doi: 10.1021/jm050113z. [DOI] [PubMed] [Google Scholar]
- [16].Lee AM, Singleton SF. Inhibition of the Escherichia coli RecA protein: zinc(II), copper(II) and mercury(II) trap RecA as inactive aggregates. J. Inorg. Biochem. 2004;98:1981–6. doi: 10.1016/j.jinorgbio.2004.08.018. [DOI] [PubMed] [Google Scholar]
- [17].Wigle TJ, Lee AM, Singleton SF. Conformationally Selective Binding of Nucleotide Analogues to Escherichia coli RecA: A Ligand-Based Analysis of the RecA ATP Binding Site. Biochemistry. 2006;45:4502–13. doi: 10.1021/bi052298h. [DOI] [PubMed] [Google Scholar]
- [18].Little JW, Edmiston SH, Pacelli LZ, Mount DW. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc. Natl. Acad. Sci. USA. 1980;77:3225–9. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu. Rev. Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- [20].Nishinaka T, Ito Y, Yokoyama S, Shibata T. An extended DNA structure through deoxyribose-base stacking induced by RecA protein. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6623–6628. doi: 10.1073/pnas.94.13.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].VanLoock MS, Yu X, Yang S, Lai AL, Low C, Campbell MJ, Egelman EH. ATP-mediated conformational changes in the RecA filament. Structure (Camb) 2003;11:187–96. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- [22].Ellouze C, Selmane T, Kim HK, Tuite E, Norden B, Mortensen K, Takahashi M. Difference between active and inactive nucleotide cofactors in the effect on the DNA binding and the helical structure of RecA filament dissociation of RecA--DNA complex by inactive nucleotides. Eur. J. Biochem. 1999;262:88–94. doi: 10.1046/j.1432-1327.1999.00357.x. [DOI] [PubMed] [Google Scholar]
- [23].Roca AI, Singleton SF. Direct evaluation of a mechanism for activation of the RecA nucleoprotein filament. J Am Chem Soc. 2003;125:15366–75. doi: 10.1021/ja0270165. [DOI] [PubMed] [Google Scholar]
- [24].Moreau PL, Carlier MF. RecA protein-promoted cleavage of LexA repressor in the presence of ADP and structural analogues of inorganic phosphate, the fluoride complexes of aluminum and beryllium. J. Biol. Chem. 1989;264:2302–6. [PubMed] [Google Scholar]
- [25].Lee JW, Cox MM. Inhibition of recA protein promoted ATP hydrolysis. 2. Longitudinal assembly and disassembly of recA protein filaments mediated by ATP and ADP. Biochemistry. 1990;29:7677–83. doi: 10.1021/bi00485a017. [DOI] [PubMed] [Google Scholar]
- [26].Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–95. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- [27].Yasuda T, Morimatsu K, Kato R, Usukura J, Takahashi M, Ohmori H. Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenesis. Embo J. 2001;20:1192–202. doi: 10.1093/emboj/20.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Singleton SF, Simonette RA, Sharma NC, Roca AI. Intein-mediated affinity-fusion purification of the Escherichia coli RecA protein. Protein Expression Purif. 2002;26:476–88. doi: 10.1016/s1046-5928(02)00571-5. [DOI] [PubMed] [Google Scholar]
- [29].Craig NL, Roberts JW. Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein-directed cleavage of phage lambda repressor. J Biol Chem. 1981;256:8039–44. [PubMed] [Google Scholar]
- [30].Defais M, Phez E, Johnson NP. Kinetic mechanism for the formation of the presynaptic complex of the bacterial recombinase RecA. Journal of Biological Chemistry. 2003;278:3545–51. doi: 10.1074/jbc.M204341200. [DOI] [PubMed] [Google Scholar]
- [31].Berger MD, Lee AM, Simonette RA, Jackson BE, Roca AI, Singleton SF. Design and evaluation of a tryptophanless RecA protein with wild type activity. Biochemical and Biophysical Research Communications. 2001;286:1195–203. doi: 10.1006/bbrc.2001.5525. [DOI] [PubMed] [Google Scholar]
- [32].Morrical SW, Lee J, Cox MM. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of recA protein and single-stranded DNA. Biochemistry. 1986;25:1482–94. doi: 10.1021/bi00355a003. [DOI] [PubMed] [Google Scholar]
- [33].Neet KE. Cooperativity in enzyme function: equilibrium and kinetic aspects. Methods Enzymol. 1980;64:139–92. doi: 10.1016/s0076-6879(80)64009-9. [DOI] [PubMed] [Google Scholar]
- [34].Menge KL, Bryant FR. ATP-stimulated hydrolysis of GTP by RecA protein: kinetic consequences of cooperative RecA protein-ATP interactions. Biochemistry. 1988;27:2635–40. doi: 10.1021/bi00407a055. [DOI] [PubMed] [Google Scholar]
- [35].Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. USA. 1992;89:4884–7. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nixon AE, Brune M, Lowe PN, Webb MR. Kinetics of inorganic phosphate release during the interaction of p21ras with the GTPase-activating proteins, p120-GAP and neurofibromin. Biochemistry. 1995;34:15592–8. doi: 10.1021/bi00047a026. [DOI] [PubMed] [Google Scholar]
- [37].Strasser F, Pelton PD, Ganzhorn AJ. Kinetic characterization of enzyme forms involved in metal ion activation and inhibition of myo-inositol monophosphatase. Biochem J. 1995;307(Pt 2):585–93. doi: 10.1042/bj3070585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wong C, Wu W, Obenshain SA, Diah SK, Faiola B, Kennelly PJ. High-molecular-weight polypeptide substrates for phospholysine phosphatases. Anal. Biochem. 1994;222:14–8. doi: 10.1006/abio.1994.1447. [DOI] [PubMed] [Google Scholar]
- [39].Wong C, Faiola B, Wu W, Kennelly PJ. Phosphohistidine and phospholysine phosphatase activities in the rat: potential protein-lysine and protein-histidine phosphatases? Biochem J. 1993;296(Pt 2):293–6. doi: 10.1042/bj2960293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang ZX, Cheng Q, Killilea SD. A continuous spectrophotometric assay for phosphorylase kinase. Anal. Biochem. 1995;230:55–61. doi: 10.1006/abio.1995.1437. [DOI] [PubMed] [Google Scholar]
- [41].Henkel RD, VandeBerg JL, Walsh RA. A microassay for ATPase. Anal Biochem. 1988;169:312–8. doi: 10.1016/0003-2697(88)90290-4. [DOI] [PubMed] [Google Scholar]
- [42].Kessen U, Schaloske R, Aichem A, Mutzel R. Ca(2+)/calmodulin-independent activation of calcineurin from Dictyostelium by unsaturated long chain fatty acids. Journal of Biological Chemistry. 1999;274:37821–6. doi: 10.1074/jbc.274.53.37821. [DOI] [PubMed] [Google Scholar]
- [43].Hausmann S, Shuman S. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. Journal of Biological Chemistry. 2002;277:21213–20. doi: 10.1074/jbc.M202056200. [DOI] [PubMed] [Google Scholar]
- [44].Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli. Steady state kinetic analysis of ATP hydrolysis. J Biol Chem. 1981;256:8845–9. [PubMed] [Google Scholar]
- [45].Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. Journal of Molecular Biology. 1985;181:281–95. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- [46].Lee JW, Cox MM. Inhibition of recA protein promoted ATP hydrolysis. 1. ATP gamma S and ADP are antagonistic inhibitors. Biochemistry. 1990;29:7666–76. doi: 10.1021/bi00485a016. [DOI] [PubMed] [Google Scholar]
- [47].Chabbert M, Cazenave C, Helene C. Kinetic studies of recA protein binding to a fluorescent single-stranded polynucleotide. Biochemistry. 1987;26:2218–25. doi: 10.1021/bi00382a022. [DOI] [PubMed] [Google Scholar]
- [48].Cox MM, McEntee K, Lehman IR. A simple and rapid procedure for the large scale purification of the recA protein of Escherichia coli. J. Biol. Chem. 1981;256:4676–8. [PubMed] [Google Scholar]
- [49].Cox MM, Soltis DA, Lehman IR, DeBrosse C, Benkovic SJ. ADP-mediated dissociation of stable complexes of recA protein and single-stranded DNA. J. Biol. Chem. 1983;258:2586–92. [PubMed] [Google Scholar]
- [50].McEntee K, Weinstock GM, Lehman IR. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1979;76:2615–9. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gourves AS, Defais M, Johnson NP. Equilibrium binding of single-stranded DNA to the secondary DNA binding site of the bacterial recombinase RecA. J. Biol. Chem. 2001;276:9613–9. doi: 10.1074/jbc.M004855200. [DOI] [PubMed] [Google Scholar]
- [52].Singleton SF, Roca AI, Lee AM, Xiao J.Probing the structure of RecA-DNA filaments. Advantages of a fluorescent guanine analog Tetrahedron 2006. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Volodin AA, Smirnova HA, Bocharova TN. Efficient interaction of RecA protein with fluorescent dye-labeled oligonucleotides. FEBS Lett. 1994;349:65–68. doi: 10.1016/0014-5793(94)00640-7. [DOI] [PubMed] [Google Scholar]
- [54].Wittung P, Ellouze C, Maraboeuf F, Takahashi M, Norden B. Thermochemical and kinetic evidence for nucleotide-sequence-dependent RecA-DNA interactions. Eur. J. Biochem. 1997;245:715–9. doi: 10.1111/j.1432-1033.1997.00715.x. [DOI] [PubMed] [Google Scholar]
- [55].Roca AI, Singleton SF. Direct evaluation of a mechanism for activation of the RecA nucleoprotein filament. Journal of the American Chemical Society. 2003;125:15366–75. doi: 10.1021/ja0270165. [DOI] [PubMed] [Google Scholar]
- [56].Singleton SF, Roca AI, Lee AM, Xiao J.Probing the structure of RecA-DNA filaments. Advantages of a fluorescent guanine analog Tetrahedron 2007. doi:10.1016/j.tet.2006.10.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lee AM, Singleton SF. Intersubunit Electrostatic Complementarity in the RecA Nucleoprotein Filament Regulates Nucleotide Substrate Specificity and Conformational Activation. Biochemistry. 2006;45:4514–29. doi: 10.1021/bi052296x. [DOI] [PubMed] [Google Scholar]
- [58].Stole E, Bryant FR. Reengineering the nucleotide cofactor specificity of the RecA protein by mutation of aspartic acid 100. J. Biol. Chem. 1996;271:18326–8. doi: 10.1074/jbc.271.31.18326. [DOI] [PubMed] [Google Scholar]
- [59].Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli. Characterization of ATP hydrolysis. J Biol Chem. 1981;256:8829–34. [PubMed] [Google Scholar]
- [60].Weinstock GM, McEntee K, Lehman IR. Interaction of the recA protein of Escherichia coli with adenosine 5′-O-(3-thiotriphosphate) J Biol Chem. 1981;256:8850–5. [PubMed] [Google Scholar]