Abstract
Complete, life-long exclusion of gluten containing foods from the diet is the only available treatment for celiac sprue, a widespread immune disease of the small intestine. Investigations into the molecular pathogenesis of celiac sprue have identified the major histocompatibility complex protein HLA-DQ2 and the multi-functional enzyme transglutaminase 2 as potential pharmacological targets. Based upon the crystal structure of HLA-DQ2, we rationally designed an aldehyde-functionalized, gluten peptide analogue as a tight-binding HLA-DQ2 ligand. Analogous aldehyde-bearing gluten peptide analogues were designed as high-affinity, reversible inhibitors of transglutaminase 2. By varying the side-chain length of the aldehyde-functionalized amino acid, we found that the optimal transglutaminase 2 inhibitor was 5 methylene units in length, 2 carbon atoms longer than its natural glutamine substrate.
Keywords: Celiac sprue, transglutaminase, HLA-DQ2, inhibition
1. Introduction
Celiac sprue is an inflammatory disease of the small intestine triggered by the dietary exposure to wheat, barley, or rye containing foods in genetically susceptible individuals.1, 2 Wheat, barley, and rye each contain a class of storage proteins called prolamines (or gluten) rich in the amino acids proline (15%) and glutamine (35%).3 The high proline content of gluten gives it an unusual level of digestive resistance against the gastro-pancreatic and brush border membrane proteases and peptidases that typically hydrolyze food into tripeptides, dipeptides, or single amino acids in the small intestine.4-6 Therefore, peptides of sufficient length to trigger a T cell-mediated immune response (greater than 9 amino acids long) persist a considerable distance into the lumen of the small intestine. Prior to causing inflammation, these metastable peptides are regiospecifically deamidated by the enzyme transglutaminase 2 (TG2).7 Deamidated gluten peptides have a considerably increased affinity for the major histocompatibility complex (MHC) protein HLA-DQ2,8, 9 which (together with DQ8) is the only known genetic risk factor for developing the disease and is expressed by more than 90% of patients.1 Small intestinal T cells recognize the gluten peptide-DQ2 complex as a foreign pathogen and orchestrate the ensuing small intestinal inflammatory response.1, 2 The chronic inflammation found in celiac patients on a gluten-containing diet eventually flattens the small intestinal villi thereby significantly decreasing the absorptive surface area of the small intestine. The inability to absorb sufficient levels of nutrients causes many of the common symptoms experienced by celiac patients, such as chronic diarrhea, fatigue, iron-deficient anemia, and osteoporosis, and leads to an increase in the mortality rate compared to the general population.1, 2, 10
The current treatment for celiac sprue involves complete, life-long exclusion of gluten containing foods from the diet. Although this treatment is effective, it is difficult to maintain and lowers the overall quality of life for patients.11, 12 Hence, there is an urgent need for non-dietary therapies to help treat this common disorder (approximately 0.5−1% of the population has celiac sprue).1 The disease-relevant proteins TG2 and HLA-DQ2 represent reasonable therapeutic targets due to their contribution to disease pathogenesis.
Peptidomimetic HLA-DQ2 blockers based upon highly inflammatory gluten peptides have been synthesized and shown to inhibit antigen presentation in HLA-DQ2 homozygous antigen presenting cells (APCs).13 Although these blocking peptides were fairly effective at competing with gluten peptide antigens for HLA-DQ2 occupancy in fixed APCs, the reduction in their ability to inhibit antigen presentation in non-fixed APCs required higher blocker concentrations for comparable levels of inhibition. Further, design of these blockers was based upon a gluten peptide itself. Therefore, the affinity of the HLA-DQ2 blocking peptides is comparable to certain unmodified gluten peptides thereby limiting their blocking potency. Formation of a covalent bond between the peptide ligand and HLA-DQ2 has the potential of greatly increasing the occupancy of the HLA-DQ2 binding pocket by a synthetic blocker. Several irreversible TG2 inhibitors that form stable covalent bonds with the active site cysteine are described in the literature.14 However, few reversible inhibitors have been described,15 and no reversible active site directed inhibitors have been reported. Because TG2 has been implicated to participate in many diverse biological functions, such as extracellular matrix stabilization, cell adhesion, cell migration, phagocytosis and cell signaling to name a few,16, 17 irreversible inhibition may cause undesirable side-effects.
All previous attempts to study the glutamine substrate side-chain length tolerance of TG2 have utilized irreversible inhibitors, which all inhibit the enzyme once a critical side-chain length is exceeded.18, 19 Therefore, structure-activity data concerning the side-chain length tolerance of TG2 may help with the design of more potent irreversible inhibitors without the inevitable loss of specificity caused by overextending the side-chain length.
In this report, we harness structural insights from the previously described crystal structure of HLA-DQ220 and available structure-activity data regarding TG2 regiospecificity7 to design novel aldehyde-bearing peptide-based inhibitors of these proteins. We found that substitution of an appropriately positioned α-amido aldehyde functional group in the DQ2-αI epitope dramatically increased its half-life in the HLA-DQ2 binding pocket compared to the unsubstituted DQ2-αI peptide. Moreover, the aldehyde bearing DQ2-αI epitope more effectively stimulated celiac patient T cell lines than its unsubstituted analogue. However, attempts to design longer HLA-DQ2 blockers containing multiple aldehyde functional groups decreased T cell stimulation. Substitution of an aldehyde functional group onto the amino acid side-chain of a TG2-targeted glutamine residue resulted in reversible inhibition. Variation of the aldehyde side-chain length showed that the optimal length for reversible inhibition was 5 carbon units long, 2 methylene units longer than its natural glutamine substrate.
2. Results
2.1. Design and biochemical characterization of HLA-DQ2-binding aldehyde peptides
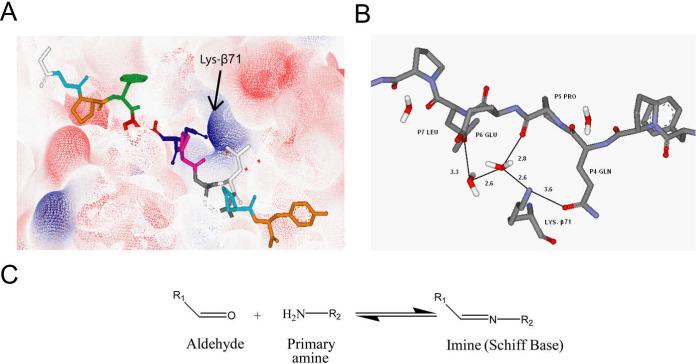
Like all class II MHC proteins, HLA-DQ2 is composed of an α and β chain bound together via non-covalent interactions to form a peptide binding groove.21 However, unlike other HLA proteins, DQ2 contains a lysine residue at position 71 of its β chain which protrudes into the binding groove and can interact with peptide anchor side-chain residues at the P4 and P6 positions via salt-bridge formation and hydrogen bonding (Figure 1A).20, 22 Indeed, examination of the crystal structure of the DQ2-αI peptide bound to HLA-DQ2 reveals hydrogen bonding interactions of Lys-β71 with the P4 glutamine and P6 glutamate side-chains of the DQ2-αI peptide (Figure 1B).20 The positive charge of Lys-β71 at least partially explains the preference HLA-DQ2 has for binding negatively charged peptides.
Figure 1.
Rationale behind the design of aldehyde containing gluten peptide analogues. (A) Electron density map showing the positively charged Lys-β71 in the HLA-DQ2 binding groove. (B) Lys-β71 directly forms hydrogen bonds with the P4 glutamine of the DQ2-αI peptide and forms a hydrogen bonding network to interact with the DQ2-αI P6 glutamate (numbers show the distance between corresponding atoms in Å). (C) Aldehydes react with primary amines in a reversible manner to create covalent imine bonds.
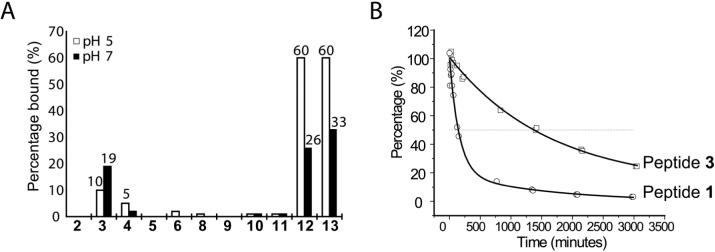
To take advantage of the structural insights provided by the crystal structure of HLA-DQ2 as well as the reactivity of the primary ε-amino group of Lys-β71, a series of peptides were synthesized with an α-amido aldehyde functional group substituted at the P4 or P6 positions of the DQ2-αI and DQ2-αII peptides (Figure 2). Aldehydes can reversibly react with primary amines to form imines (or Schiff bases), which we hoped would stabilize the peptide-DQ2 interaction (Figure 1C). In fact, when the DQ2-αI peptide P4 glutamine side chain was substituted with an aldehyde functional group of the appropriate length (peptide 3), improved binding affinity with recombinant human HLA-DQ2 was observed (Figure 3). Intriguingly, the affinity of the aldehyde substituted DQ2-αI peptide was higher at pH 7 than at pH 5, an unprecedented finding for gluten peptide binding to HLA-DQ2 (Figure 3A). This shift in the pH binding profile is likely due to the preferential imine formation at pH 7 rather than pH 5. Aldehyde peptide 3 also had an HLA-DQ2 dissociation half-life more than 8 times longer than its non-aldehyde analogue, peptide 1, again suggesting the formation of a stabilizing imine bond between the aldehyde peptide and HLA-DQ2 Lys-β71 (Figure 3B). Efforts to directly identify the Schiff's base adduct by sodium borohydride reduction were unsuccessful, presumably due to the small quantities of HLA-DQ2 protein.
Figure 2.
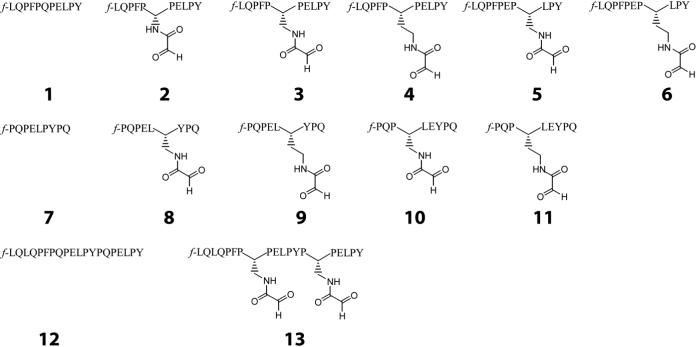
A panel of peptides bearing aldehyde functional groups was synthesized based upon the DQ2-αI (1) and DQ2-αII (7) gluten peptides. Substitutions were made at both the P4 and P6 HLA-DQ2 binding positions of the peptides. Further, the side-chain length was varied to find the binding optimum. Based upon the binding results of peptides 1−11, putatively optimal aldehyde substitutions were made in the 20mer gluten peptide (12). All peptides are labeled at the N-terminus with the fluorophore 5(6)-carboxyfluorescein (designated f).
Figure 3.
Aldehdye peptide 3 forms a reversible imine covalent bond with Lys-β71 of HLA-DQ2. (A) Among the DQ2-αI and DQ2-αII aldehyde peptides, peptide 3 had the highest affinity for HLA-DQ2. Moreover, the affinity of peptide 3 for HLA-DQ2 was higher at pH 7 than pH 5. There was no change in the pH 5 affinity of the 20mer peptide (12) upon addition of two aldehyde functional groups (13), although there was a slight improvement at pH 7. (B) HLA-DQ2 dissociation kinetics of aldehyde containing peptide 3 were much slower than the unmodified parent peptide 1 (t1/2app (3) = 1400 min.; t1/2app (1) = 170 min.).
2.2. Aldehyde functionalization can enhance antigen presentation
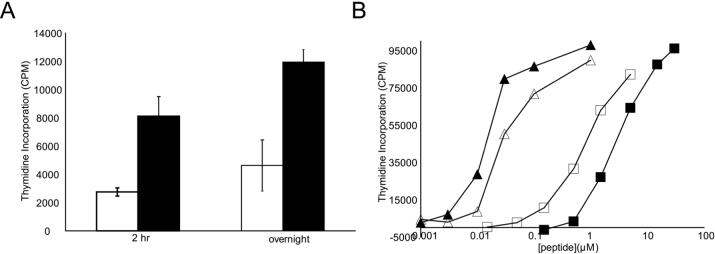
Because of the encouraging kinetic and equilibrium binding data (Figure 3) of aldehyde peptide 3, we wished to study the peptide 3-HLA DQ2 interaction in a more biologically relevant system. Therefore, peptide 3 and its parental peptide 1 were each incubated with γ-irradiated HLA-DQ2 homozygous B-lymphoblastoid VAVY cells for 2 hours or overnight to allow for antigen processing and presentation before being washed away by centrifugation. The polyclonal T cell line P28 TCL1, which contains a population of DQ2-αI responsive T cells, was then used to detect the quantity of αI peptide-DQ2 complexes on the surface of the VAVY cells. Consistent with the recombinant HLA-DQ2 binding data, peptide 3 caused more than twice as much T cell proliferation as peptide 1 at both time points (Figure 4A).
Figure 4.
Enhanced antigen presentation of peptide 3 relative to peptide 1. (A) Peptides 1 (□) and 3 (■) (10 μM) were incubated with γ-irradiated VAVY cells for either 2 hours or overnight before being removed. Polyclonal T cell line P28 TCL1 proliferation was used to assess the amount of peptide-DQ2 complexes on the surface of the VAVY cells. (B) Titration of peptides 1, 3, 12, and 13 using P28 TCL3 T cells. Although aldehyde peptide 3 showed enhanced antigen presentation relative to non-aldehyde peptide 1, di-aldehyde peptide 13 showed a decreased level of antigen presentation compared to its parent peptide 12. Peptide 1 ■ (EC50 = 3.3 μM); Peptide 3 □ (EC50 = 1.0 μM); Peptide 12 ▲ (EC50 = 0.016 μM); Peptide 13 △ (EC50 = 0.039 μM)
The HLA-DQ2 binding properties of peptide 12 have been shown to be nearly equivalent to one of the most potent naturally occurring gluten peptides studied thus far.13 The high affinity interaction of peptide 12 was previously exploited to create HLA-DQ2 blocking peptides capable of decreasing the T cell stimulation caused by unmodified, antigenic gluten peptides.13 In an effort to create a more potent blocking peptide, two aldehyde functional groups were substituted into peptide 12 at the same relative location (the DQ2-αI and DQ2-αIII epitopes) and with the same side-chain length used to create peptide 3 (see peptide 13, Figure 2). As shown in Figure 3A, peptide 12 and peptide 13 have equivalent affinity for HLA-DQ2 at pH 5 while peptide 13 shows an incremental improvement in affinity at pH 7.
In order to more quantitatively assess the HLA-DQ2 occupancy of peptides 1, 3, 12, and 13 on the surface of antigen presenting cells (APC), the peptides were incubated with γ-irradiated VAVY cells for 10 hours before being washed away. A polyclonal T cell line biased towards recognizing the DQ2-αI epitope (P28 TCL3) was used to assess APC presentation of each peptide. Although aldehyde peptide 3 showed a 3-fold improvement in EC50 over parental peptide 1, di-aldehyde peptide 13 showed a 2.5-fold worse EC50 relative to peptide 12 (Figure 4B). Similar results were obtained utilizing a monoclonal T cell line (P26 TCC1) specific for recognizing the DQ2-αII epitope (peptide 7) (data not shown).
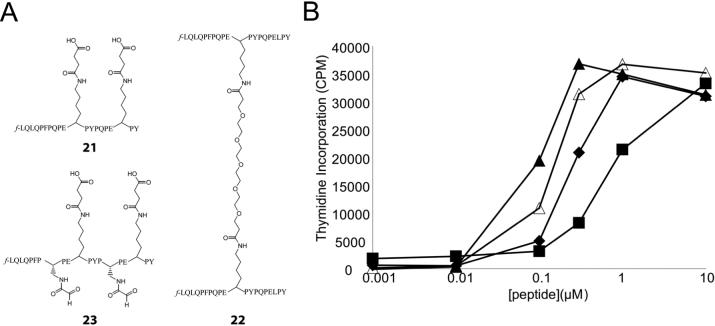
While peptide 13 is less effective at stimulating T cells than peptide 12, it is possible that the aldehyde functional groups on peptide 13 alter the normal conformation of 12 when bound to HLA-DQ2 such that it is not as well recognized by T cell receptors (TCRs). Thus, it cannot be ruled out that peptide 13 actually occupies a larger fraction of cell surface HLA-DQ2 molecules but is not as efficient at T cell stimulation as peptide 12. To test this hypothesis, blocker peptide 23 was synthesized (Figure 5A). The design of peptide 23 was based upon a previously synthesized HLA-DQ2 blocking peptide (peptide 21) that gave low intrinsic background T cell stimulation and showed effective inhibition of gluten peptide antigen presentation.13 More specifically, two solvent exposed leucine residues in peptide 13 pointing away from the DQ2 binding groove towards the TCR were mutated to lysine residues and modified with a bulky succinyl moiety to block TCR recognition. This novel DQ2 blocker was then compared head-to-head with two previously designed DQ2 blockers, peptides 21 and 22 (Figure 5A), in a standard T cell stimulation assay using fixed VAVY cells and peptide 12 as the stimulating antigen. However, peptide 23 was the least efficient DQ2 blocking peptide among the three suggesting that the addition of two aldehyde functional groups onto peptide 21 decreases its HLA-DQ2 occupancy on APCs (Figure 5B).
Figure 5.
Design and biological evaluation of aldehyde DQ2 blocker peptide 23. (A) The addition of bulky side-chains at the 11 and 18 positions of di-aldehyde peptide 13 creates a DQ2 blocker peptide similar to the previously studied peptide 21.13 The structure of the high affinity dimer peptide 22 is also shown. (B) Comparison of the HLA-DQ2 blocking efficiency of peptides 21, 22, and 23. Peptides 21, 22, and 23 (20 μM) were co-incubated with gluten peptide antigen 12 overnight in the presence of fixed APCs (VAVY). The peptides were washed away, and P28 TCL2 T cell proliferation was used to evaluate the blocking efficiency of each peptide. No blocker (▲) (EC50 = 0.08 μM); Peptide 21 (◆) (EC50 = 0.25 μM); Peptide 22 (■) (EC50 = 0.77 μM); Peptide 23 (△) (EC50 = 0.14 μM)
2.3. Design of transglutaminase 2 aldehyde peptide inhibitors
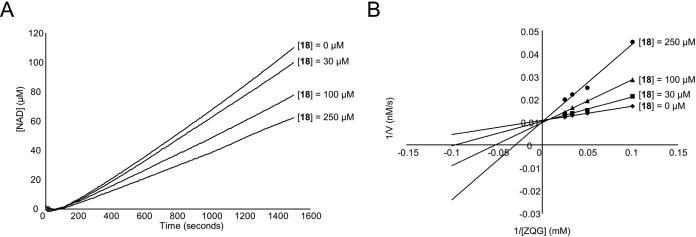
Encouraged by the success of functionalizing small gluten peptides with an α-amido aldehyde group to enhance HLA-DQ2 affinity, we decided to see if a similar strategy could yield TG2 inhibitors that react reversibly with the nucleophilic TG2 active site cysteine residue, presumably through the formation of a hemithioacetal adduct between the aldehyde group of the peptide and the thiol group of the TG2 active site cysteine. Previous studies suggest that recombinant human TG2 optimally recognizes the QXP tripeptide.23, 24 Further, examination of the regiospecificity of TG2 towards the α-gliadin peptide PQPQLPYPQPQLPY showed that it preferentially deamidates the underlined glutamine residues.7 Therefore, peptides based upon the α-gliadin peptide sequence PQPQLPF were mutated at the TG2 targeted 4-position glutamine into an α-amido aldehyde functionalized side-chain. Kinetic evaluation of TG2 catalyzed progress curves revealed a dose dependent decrease in the linear reaction velocities with increasing concentration of aldehyde inhibitors suggesting reversible inhibition (Figure 6A). Lineweaver-Burk analysis revealed competitive inhibition, because the y-intercept was constant at varying concentrations of inhibitor (Figure 6B).
Figure 6.
Kinetic analysis of TG2 inhibition using aldehyde inhibitors. (A) Example of the reaction progress curves generated using a previously described continuous spectrophotometric assay for tracking TG2 activity.7 Straight lines were fit to the linear region of the data to determine the reaction rates. Increasing the concentration of peptide 18 decreased the linear reaction velocity of 15 mM ZQG in a dose dependent fashion. (B) Lineweaver-Burk plot indicating that TG2 inhibition using aldehyde inhibitor 18 is competitive. TG2 reaction velocities were measured at 10, 20, 30, and 40 mM ZQG in the presence of 0, 30, 100, or 250 μM inhibitor 18.
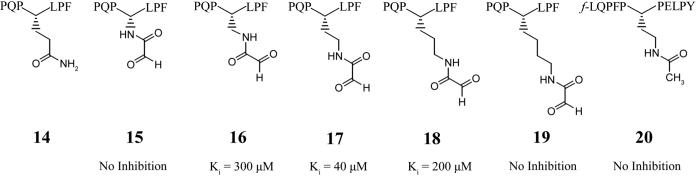
In order to study the side-chain length tolerance of TG2, a series of aldehyde inhibitor peptides sequentially extended by one methylene subunit were synthesized (Figure 7). Inhibition constants (Ki) were calculated for each inhibitor using a competitive inhibition model. Interestingly, the inhibitor with the optimum side-chain length (peptide 17) was two methylene units longer than the natural glutamine substrate (peptide 14). To ensure that the aldehyde functional group was responsible for the observed inhibition, a peptide having the optimal side-chain length was synthesized with an acetamido group in place of the α-amido aldehyde (peptide 20). This peptide, peptide 20, showed no inhibition.
Figure 7.
TG2 aldehyde inhibitors and their competitive inhibition parameter, Ki. The maximum concentration of inhibitor tested for compounds 15, 19, and 20 was 750, 500, and 100 μM, respectively.
3. Discussion and conclusions
Development of small molecule, peptide, or peptidomimetic inhibitors of protein-protein interactions has been extensively investigated due to the ubiquitous nature of protein-protein interactions in almost every cellular process and in the pathology of most diseases.25-28 Despite research efforts, the limited number of successful therapeutics based upon the disruption of protein-protein interactions has precluded the development of standardized methods to design and biologically evaluate such therapeutics.25-28 Class II major histocompatibility complex (MHC) proteins provide a unique opportunity for the development of protein-protein interaction blockers since 1) the crystal structures of a number of MHC proteins with bound ligands have been solved21; 2) MHC proteins have a structurally defined binding pocket (or groove) into which the ligand binds as opposed to many other protein-protein interactions between two large, relatively flat and undefined protein surface interfaces; 3) the MHC ligand is a peptide rather than a large protein; and 4) the MHC groove can only accommodate 9−10 amino acids thereby limiting the number of protein-peptide contacts that form.
A key challenge in the design of effective blockers of HLA-DQ2 is the development of high-affinity, proteolytically stable DQ2 ligands. The challenge is especially severe in the context of celiac sprue, where an orally administered ligand is expected to survive the exceptionally harsh proteolytic environment of the upper gastrointestinal tract. Naturally occurring gluten antigens have been selected for their intrinsic proteolytic resistance5; however, their affinity for HLA-DQ2 is not as high as compared to benchmark ligands of this class II MHC.9 In this study, we attempted to enhance the HLA-DQ2 affinity of a synthetic peptide ligand through formation of a Schiff's base between an aldehyde group in the peptide and a signature Lys residue in the HLA-DQ2 binding pocket.
Guided by the crystal structure of HLA-DQ220, a panel of gluten peptide analogues containing an α-amido aldehyde moiety positioned to react with the polymorphic Lys-β71 residue unique to HLA-DQ2 was synthesized. The highest affinity DQ2-αI peptide (peptide 3) displayed the unusual behavior of preferentially binding to recombinant HLA-DQ2 at pH 7 rather than pH 5, probably because imine bond formation is favored at pH 7 more so than pH 5 within the HLA-DQ2 peptide binding groove (Figure 3A). Further, peptide 3 showed a 3-fold decrease in the T cell stimulation EC50 relative to parent peptide 1 proving its effectiveness in a more biologically complex, and disease relevant, system. The generality of this potential solution for enhancing DQ2 affinity is unclear, in light of the results obtained for peptide 23.
Aldehyde functionalization was also used to create a class of reversible transglutaminase 2 (TG2) inhibitors. Surprisingly, variation of the side-chain length showed that the optimal TG2 inhibitor was two methylene units longer than its natural glutamine substrate. Previous results using irreversible TG2 inhibitors indicated that all inhibitors with reactive functional groups exceeding a minimum side-chain length were able to irreversibly inhibit TG2 activity.18,19 Our data using reversible α-amido aldehyde inhibitors shows that TG2 does indeed have a chain length preference at this side-chain. One possible explanation for this difference may be that the reactive functional groups installed on irreversible inhibitors simply need access to the TG2 active site cysteine thiol in order to form a covalent bond. Thus, substrate specificity probably cannot be revealed for irreversible inhibitors with relatively long side-chains. In contrast, the aldehyde functional group may require accurate positioning to react with the active cysteine thiol group to reversibly form a hemithioacetal. Consequently, these inhibitors show chain length specificity. We also synthesized a panel of PQPXLPF peptides containing a bromomethylketone functional group at this position with varying side-chain lengths. Consistent with the previously reported results18,19, all bromomethylketone inhibitors with side-chain lengths of 5 methylene units or longer were able to irreversibly inhibit TG2 (data not shown).
Our findings may have important implications for the future design of irreversible inhibitors of TG2. Indeed, one of the most potent reported TG2 inhibitors contains a reactive 6-diazo-5-oxo-norleucine (Don) functional group that reacts with the TG2 active site thiol at a position one methylene group further than its natural glutamine substrate.29 On the other hand, overextending the side-chain length of irreversible inhibitors may cause an increase in off-target binding due to the increased accessibility of the reactive moiety. Finally, it should be noted that the neighboring carbonyls of the α-amido aldehyde side-chains have a different electronic configuration compared to the natural glutamine side-chain, which contains only one carbonyl group. The increased molecular rigidity of the α-amido aldehyde group relative to the glutamine amide may itself alter the interactions between the TG2 active site and inhibitor thereby affecting the results. Thus, the results should be interpreted with this structural difference in mind.
4. Experimental
4.1. Peptide synthesis and purification
Peptides used in this study were synthesized using Boc/HBTU chemistry starting from N-α-t-Boc-L-aminoacyl-phenylacetamidomethyl (PAM) resin as previously described.9, 13 Lysine analogues, i.e. N-Boc-N'-Fmoc-diaminoacetic acid (DAA), Boc-Nβ-Fmoc-L-2,3-diaminopropionic acid (DAP), Boc-Nγ-Fmoc-L-2,4-diaminobutyric acid (DAB) (chemimpex) were used for synthesis of aldehyde containing peptides. While still attached to the resin, the Fmoc group was deprotected by 20% piperidine followed by coupling of Fmoc protected serine group to install a serine residue on the side chain of each lysine analogue.
Following cleavage of the peptidyl resin in trifluoroacetic acid/trifluoromethanesulfonic acid/thioanisole (TFA/TFMSA/thioanisole 10:1:1, v/v/v) for 4 h, the crude peptides were precipitated in cold ether and dissolved in 1:1 v/v acetonitrile/water. HPLC purified peptides with terminal serine residue were treated with 2−5 equivalents sodium periodinate in phosphate solution at pH 7 for 2 hours.30 The α-amido aldehyde containing peptide analogues were then purified by reverse HPLC. The identity and purity of the peptides and peptide analogues were confirmed by liquid chromatography coupled electrospray mass spectrometry (LC-ESMS). The peptides were lyophilized and stored at –20°C. Prior to use, peptide stock solutions were prepared by dissolving in PBS. The concentrations of the non-fluorescent peptide stocks were determined by UV spectrophotometry at 280 nm in pH 7.2 PBS using the absorption coefficient factor 1280 cm−1M−1 for every tyrosine residue, whereas the concentration of carboxyfluorescein labeled peptides were determined using the same method at 495 nm in pH 7.2 PBS with an absorption coefficient of 80,200 cm−1M−1.
4.2. Peptide exchange assay
Peptide exchange assays were conducted as previously described.9, 13 In brief, soluble recombinant DQ2 molecules with a gliadin epitope fused to the N-terminus of the β-chain were expressed and purified. Prior to use in exchange experiments, recombinant DQ2 molecules were treated with ∼2% w/w thrombin in pH 7.3 PBS at 0°C for 2 h to release the covalently linked epitope for peptide exchange measurements. Thrombin treated DQ2 was incubated with fluorescein-conjugated ligands in a 25:1 ratio (i.e. 4.7 μM DQ2 with 0.185 μM fluorescent peptide) at 37°C in a 1:1 mixture of PBS buffer (10 mM sodium phosphate, 150 mM NaCl, pH 7.3, supplemented with 0.02% NaN3) and McIlvaine's citrate-phosphate buffer (pH 5 or pH 7) such that the final pH was either 5.5 or 7.3, respectively. Peptide binding was measured by high performance size exclusion chromatography (HPSEC) coupled with fluorescence detection with excitation at 495 nm and emission at 520 nm. The DQ2-peptide 1:1 complex eluted at ∼8.5 min, with free peptides emerging ∼2 min later. When present, the 2:1 DQ2-peptide complex eluted ∼0.5 min before the 1:1 complex. Peak areas corresponding to the DQ2-peptide complex and the free peptide were used to calculate the fractional yield of the DQ2-fluoresceinated peptide complex. At least two independent measurements were conducted, with an error <5%.
4.3. Peptide dissociation assay
For dissociation experiments, DQ2-fluoresceinated peptide complexes were prepared by incubating thrombin treated DQ2 (3−5 μM) with 20-fold excess fluorescein-conjugated peptides in phosphate buffer at pH 7 for 25 hours. Excess free peptide was separated from the complex on a chilled spin column (Bio-Rad) packed with Sephadex G50 superfine medium and blocked with 1% BSA solution to minimize the binding of DQ2 to the column. Spin columns were pre-washed with pH 7.3 PBS buffer, and the fluorescein-conjugated peptide + DQ2 mixture was applied to the column. The DQ2-fluoresceinated peptide complex was eluted in a volume of ∼230 μl in pH 7.3 PBS buffer. 20 μM of a tight DQ2 binding peptide (AAIAAVKEEAF) was added to prevent the re-binding of dissociated fluorescent peptide to DQ2.9, 13 Kinetic measurements of ligand dissociation were performed at 37°C, and a time course was obtained by injecting 20 μl aliquots into HPSEC column.
4.4. T cell proliferation assay
T cell proliferation assays were performed as previously described.9, 13 Briefly, HLA-DQ2 homozygous B-lymphoblastoid VAVY cells were γ-irradiated (12,000 rads) and resuspended in T cell media (Iscove's Modified Dulbecco Medium, 10% fetal bovine serum, 2% human serum, 100 U/ml penicillin, 100 μg/ml streptomycin) to 2*106 cells/ml. Sixty-five μl/well of VAVY cell suspension was added to a flat-bottom 96-well plate and peptides were added at the indicated concentration for the indicated amount of time. The peptides were then washed out by doubling the volume (65 μl/well), pipetting each well into an eppendorf tube, and centrifuging at 800g for 3 minutes at 4°C. The supernatant was aspirated and 130 μl of T cell media was added to the pellet to give 1*106 VAVY cell/ml. Fifty μl were added to a U-bottom 96-well plate to which 50 μl of T cells resuspended to 1*106 cells/ml were also added. The cells were incubated together for 48 hours before 1 μCi/well [methyl-3H] thymidine (Amersham, TRK120) was added for 18−24 hours. Cells were harvested using a Tomtec cell harvester and radioactivity counted using a Wallac 1205 Betaplate scintillation counter.
For the HLA-DQ2 blocking peptide experiment, VAVY cells were fixed with 1% paraformaldehyde in PBS for 10 minutes at room temperature. Blocking peptides 21, 22, or 23 were added at 20 μM to appropriate wells and antigenic 20mer peptide 12 was titrated against each blocker. After 12 hours, the peptides were aspirated following centrifugation and the typical T cell proliferation assay protocol was performed as described above.
4.5. Generation of DQ2-αI specific T cell line P28 TCL3
In order to evaluate the amount of αI peptide-DQ2 complexes on the surface of antigen presenting cells, a polyclonal T cell line recognizing the DQ2-αI epitope exclusive to the DQ2-αII epitope was created. VAVY cells were γ-irradiated with 12,000 rads, resuspended to 2*106 cells/ml and incubated overnight with 20 μM of DQ2-αI peptide in a 24-well plate. The VAVY cell volume was doubled to give 1*106 cells/ml and 1 ml/well was added to a 24 well plate. One ml of T cell line P28 TCL1 cells was added at 1*106 cells/ml to the VAVY cells to give 2 ml/well. The growth factor IL-2 was added at 50 U/ml on days 3, 5, 7, 10, and 12. Cells were tested for proliferation against the DQ2-αI and DQ2-αII peptides after each stimulation round. Cells went through 5 rounds of expansion against the DQ2-αI epitope to generate P28 TCL3.
All other T cell lines (P28 TCL1, P28 TCL2, P26 TCC1) were generated as described previously.13, 31
4.6. Continuous TG2 kinetic assay
Transglutaminase 2 reactions were tracked using a continuous spectrophotometric assay measuring TG2 deamidation rates as previously described7 with a few modifications. Briefly, a 5X MOPS buffer (200 mM MOPS, 5 mM CaCl2, 1 mM EDTA, 10 mM α-ketoglutarate pH 7.2) was prepared along with a 24 mg/ml stock of glutamate dehydrogenase (GDH), 20 mM stock of NADH, and a 300 mM stock of Z-Gln-Gly (ZQG) all dissolved in water. Aldehyde inhibitors were dissolved to 1 mM in water and recombinant human TG2 (rhTG2) was dissolved to 4 μM in water. Reactions were run in a 96-well plate (Greiner bio-one, 655801) by adding 76 μl water, 40 μl 5X MOPS, 20 μl GDH, 4 μl NADH, 20 μl ZQG dissolved to 10X, and 20 μl of inhibitors dissolved to 10X. To start the reaction, 20 μl of rhTG2 was added and the reaction was tracked using a Molecular Devices Spectramax 190 96-well spectrophotometric plate reader. Reaction velocities were determined by linear regression. Inhibition parameter, Ki, was determined by fitting the resulting reaction velocities (at a given inhibitor concentration) versus ZQG concentrations to the Michaelis-Menten equation by fixing Vmax to the value determined in the absence of inhibitor and using non-linear regression in Origin 6.0 to find KMapp. The following equation, based upon competitive inhibition, was used to calculate Ki.
4.7. Production and purification of recombinant human TG2
Production and purification of rhTG2 was described previously.7
Acknowledgements
This research was supported by a grant from the NIH (R21 DK065965). M. Siegel is a recipient of a predoctoral scholarship from the ARCS foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Sollid LM. Nat. Rev. Immunol. 2002;2(9):647–655. doi: 10.1038/nri885. [DOI] [PubMed] [Google Scholar]
- 2.Kagnoff MF. Gastroenterology. 2005;128(4 suppl 1):S10–18. doi: 10.1053/j.gastro.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Wieser H. Baillieres Clin. Gastroenterol. 1995;9(2):191–207. doi: 10.1016/0950-3528(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 4.Piper JL, Gray GM, Khosla C. J. Pharmacol. Exp. Ther. 2004;2004;311(1):213–219. doi: 10.1124/jpet.104.068429. [DOI] [PubMed] [Google Scholar]
- 5.Shan L, Molberg O, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Science. 2002;297(5590):2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 6.Arentz-Hansen H, McAdam SN, Molberg O, Fleckenstein B, Lundin KE, Jorgensen TJ, Jung G, Roepstorff P, Sollid LM. Gastroenterology. 2002;123(3):803–809. doi: 10.1053/gast.2002.35381. [DOI] [PubMed] [Google Scholar]
- 7.Piper JL, Gray GM, Khosla C. Biochemistry. 2002;41(1):386–393. doi: 10.1021/bi011715x. [DOI] [PubMed] [Google Scholar]
- 8.Qiao SW, Bergseng E, Molberg O, Jung G, Fleckenstein B, Sollid LM. J. Immunol. 2005;175(1):254–261. doi: 10.4049/jimmunol.175.1.254. [DOI] [PubMed] [Google Scholar]
- 9.Xia J, Sollid LM, Khosla C. Biochemistry. 2005;44(11):4442–4449. doi: 10.1021/bi047747c. [DOI] [PubMed] [Google Scholar]
- 10.Logan RF, Rifkind EA, Turner ID, Ferguson A. Gastroenterology. 1989;97(2):265–271. doi: 10.1016/0016-5085(89)90060-7. [DOI] [PubMed] [Google Scholar]
- 11.Zarkadas M, Cranney A, Case S, Molloy M, Switzer C, Graham ID, Butzner JD, Rashid M, Warren RE, Burrows V. J. Hum. Nutr. Diet. 2006;19(1):41–49. doi: 10.1111/j.1365-277X.2006.00659.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee A, Newman JM. J. Am. Diet Assoc. 2003;103(11):1533–1535. doi: 10.1016/j.jada.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Xia J, Siegel M, Bergseng E, Sollid LM, Khosla C. J. Am. Chem. Soc. 2006;128(6):1859–1867. doi: 10.1021/ja056423o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel M, Khosla C. Pharmacol. Ther. 2007 doi: 10.1016/j.pharmthera.2007.05.003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Case A, Stein RL. Biochemistry. 2007;46(4):1106–1115. doi: 10.1021/bi061787u. [DOI] [PubMed] [Google Scholar]
- 16.Lorand L, Graham RM. Nature Rev. Mol. Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 17.Zemskov EA, Janiak A, Hang J, Waghray A, Belkin AM. Front. Biosci. 2006;11:173–185. doi: 10.2741/1863. [DOI] [PubMed] [Google Scholar]
- 18.Marrano C, de Macedo P, Keillor JW. Bioorg Med Chem. 2001;9(7):1923–1928. doi: 10.1016/s0968-0896(01)00101-8. [DOI] [PubMed] [Google Scholar]
- 19.Marrano C, de Macedo P, Gagnon P, Lapierre D, Gravel C, Keillor JW. Bioorg. Med. Chem. 2001;9(12):3231–3241. doi: 10.1016/s0968-0896(01)00228-0. [DOI] [PubMed] [Google Scholar]
- 20.Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Proc. Natl. Acad. Sci. 2004;101(12):4175–4179. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph MG, Stanfield RL, Wilson IA. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 22.Quarsten H, Molberg O, Fugger L, McAdam SN, Sollid LM. Eur. J. Immunol. 1999;29(8):2506–2514. doi: 10.1002/(SICI)1521-4141(199908)29:08<2506::AID-IMMU2506>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Vader LW, de Ru A, van der Wal Y, Kooy YM, Benckhuijsen W, Mearin ML, Drijfhout JW, van Veelen P, Koning F. J. Exp. Med. 2002;195(5):643–649. doi: 10.1084/jem.20012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleckenstein B, Molberg O, Qiao SW, Schmid DG, von der Mulbe F, Elgstoen K, Jung G, Sollid LM. J. Biol. Chem. 2002;277(37):34109–34116. doi: 10.1074/jbc.M204521200. [DOI] [PubMed] [Google Scholar]
- 25.Arkin MR, Wells JA. Nat. Rev. Drug Discov. 2004;3(4):301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 26.Way JC. Curr. Opin. Chem. Biol. 2000;4(1):40–46. doi: 10.1016/s1367-5931(99)00049-6. [DOI] [PubMed] [Google Scholar]
- 27.Loregian A, Palu G. J. Cell Physiol. 2005;204(3):750–762. doi: 10.1002/jcp.20356. [DOI] [PubMed] [Google Scholar]
- 28.Yin H, Hamilton AD. Angew Chem. Int. Ed. Engl. 2005;44(27):4130–4163. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- 29.Hausch F, Halttunen T, Maki M, Khosla C. Chem. Biol. 2003;10(3):225–231. doi: 10.1016/s1074-5521(03)00045-0. [DOI] [PubMed] [Google Scholar]
- 30.Geoghegan KF, Stroh JG. Bioconjug. Chem. 1992;3(2):138–146. doi: 10.1021/bc00014a008. [DOI] [PubMed] [Google Scholar]
- 31.Siegel M, Bethune MT, Gass J, Ehren J, Xia J, Johannsen A, Stuge TB, Gray GM, Lee PP, Khosla C. Chem. Biol. 2006;13(6):649–658. doi: 10.1016/j.chembiol.2006.04.009. [DOI] [PubMed] [Google Scholar]