Abstract
As mediators of cytokine and growth factor signaling, signal transducers and activators of transcription (STATs) transmit signals from the membrane and cytoplasm to the nucleus. While Y699 phosphorylation is required for STAT5b transcriptional activity, our previous studies show that mutation of two tyrosines in the transactivation domain of STAT5b (Y740/743F) increases Y699 phosphorylation leading to increased transcriptional activity and DNA synthesis in breast cancer cells [1]. In many instances, phosphorylation of serines in the transactivation domain also modulates STAT5b activity. Here, we demonstrate for the first time that EGF stimulation enhances S731 phosphorylation. Furthermore, we show that the increased activity of the Y740/743F STAT5b mutant requires S731. As STAT5b is implicated in several cancers, understanding how its activity is regulated through tyrosine and serine phosphorylation is vital for the development of potential novel cancer therapeutics.
Keywords: STAT5b, serine phosphorylation, tyrosine phosphorylation, breast cancer
Introduction
Signal transducers and activators of transcription (STATs) are involved in cytokine and growth factor signaling pathways including those involved in cell differentiation, development, proliferation, and survival [2]. While the STAT family consists of seven members (STAT 1, 2, 3, 4, 5a, 5b, and 6), a basic paradigm exists for all STAT activation [3]. STAT activation occurs via phosphorylation of a conserved tyrosine residue located in the carboxy-terminus. This phosphorylation can be mediated by receptor tyrosine kinases, such as the epidermal growth factor receptor (EGFR), or non-receptor tyrosine kinases, such as c-Src or Janus kinases (JAKs) [4]. Tyrosine phosphorylation subsequently results in phosphotyrosine-SH2 domain mediated dimerization. The STAT dimer then translocates to the nucleus, binds to consensus DNA sequences, and recruits additional transcription machinery to initiate specific gene transcription [5].
STAT5b is activated by a variety of factors, including growth hormone (GH), prolactin (Prl), and epidermal growth factor (EGF), resulting in phosphorylation of Y699 [6–8]. Phosphorylation of this tyrosine is required for dimerization, DNA binding, and transcriptional activity, such that mutation of this tyrosine results in a transcriptionally inactive STAT5b [6, 9]. We have previously identified and characterized three additional tyrosine phosphorylation sites (Y725, Y740, Y743) located in the transactivation domain of STAT5b (Figure 1A) [1, 10]. By tryptic digest and site directed mutagenesis, these tyrosines were identified as being phosphorylated upon EGF, but not GH treatment [10]. They are positive (Y725) or negative (Y740 and Y743) regulators of STAT5b activity, such that mutation of Y740 and Y743 results in a basally active STAT5b [1].
Figure 1. EGF induced STAT5b phosphorylation.
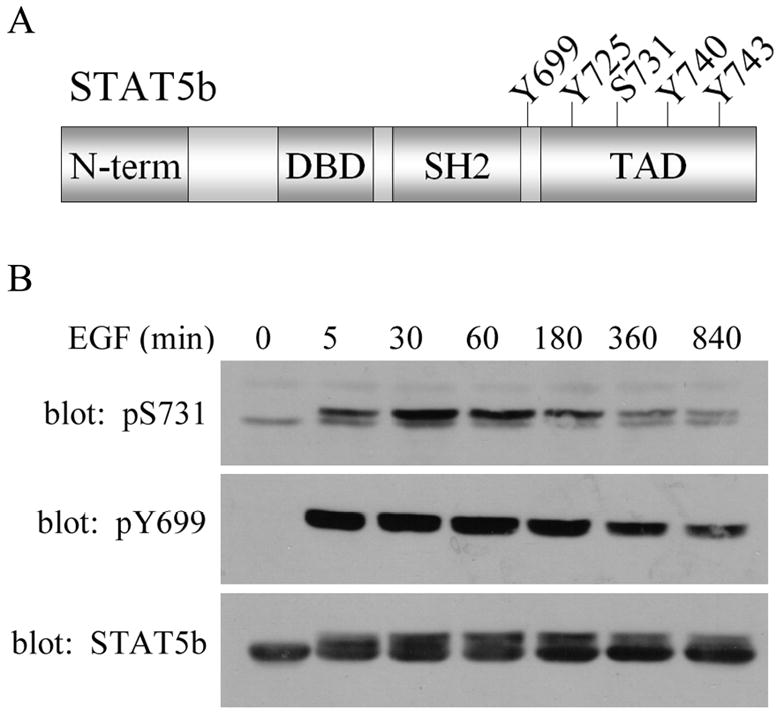
(A) Schematic of STAT5b structure illustrating the conserved domains of the STAT proteins: amino-terminus (N-term), DNA binding domain (DBD), Src homolog domain 2 (SH2), transactivation domain (TAD). The Y and S indicate where tyrosine and serine phosphorylation sites are located. (B) SKBr3 cells stably expressing His-wtSTAT5b were treated with 100ng/mL EGF for the times indicated. Whole cell lysates were analyzed by immunoblotting with antibodies directed against phospho-S731 STAT5b (top), phospho-Y699 STAT5b (middle) and total STAT5b (bottom).
In addition to tyrosines, there are several serines located in the transactivation domain of STAT proteins, although their functional implications are both STAT- and cell type-specific [11]. While serine phosphorylation of STAT1 and STAT3 has been shown to be necessary for full transcriptional activity, the regulation and importance of serine phosphorylation of STAT5b is poorly understood [11]. In some cell models, STAT5b is constitutively phosphorylated on S731, and this phosphorylation is enhanced upon Prl or GH stimulation [12–14]. Although the kinase that mediates phosphorylation of S731 has yet to be identified, mutation of S731 influences STAT5b transcriptional activity in response to PRL or GH [13–18]. For example, transient transfection of the S731A STAT5b mutant into HepG2 liver cells decreases the transcriptional activity of the GH-induced NTCP-promoter luciferase reporter compared to the corresponding wild type (wt) STAT5b [14]. In contrast, transient transfection of the S731A STAT5b mutant in COS-1 cells increases the transcriptional activity of the β-casein promoter luciferase reporter compared to wtSTAT5b [14]. These data suggest that the effect of serine phosphorylation on STAT5b function may be cell type- and gene-specific. In summary, it is apparent that the role of serine phosphorylation in STAT5b function is more complex than originally appreciated.
STAT5b has developed into a promising target for cancer therapeutics in part due to the ability of STAT5b to regulate the transcription of genes involved in cellular proliferation and survival [2, 19]. STAT5b has a fundamental role in the proliferation of breast, head and neck, and prostate cancers [1, 20–22]. Additionally, STAT5b is activated by tyrosine kinases that are frequently overexpressed or demonstrate increased activity in these cancers, such as the EGFR, HER2, c-Src, and Brk [1, 8, 10, 23–25]. The inhibition of STAT5b activity either by dominant negative constructs or anti-sense oligonucleotides impairs the transcriptional activity and transforming ability of STAT5b [22, 26].
Due to the involvement of STAT5b in cancer, a better understanding of how STAT5b activity is regulated through tyrosine and serine phosphorylation is of the utmost importance. Our previous studies revealed how tyrosines in the transactivation domain either positively or negatively regulate STAT5b activity [1, 10]. Since S731 is located between the positive (Y699 and Y725) and negative (Y740 and Y743) regulating tyrosines, we investigated its potential role in modulating STAT5b activity (Figure 1A).
Material and Methods
Cell lines and transient transfections
The human breast cancer cell line, SKBr3, was obtained from ATCC (Manassas, VA). Cells were passaged twice per week and maintained in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal calf serum (FCS). SKBr3 cells stably transfected with the His-wtSTAT5b expression vector were generated and maintained in DMEM plus 10% FCS with 50mg/mL gentamicin. Transient transfections of SKBr3 cells with His-tagged STAT5b constructs were performed using LipofectAMINE Plus (Invitrogen) per manufacturer’s directions as previously described [10].
Reagents
Recombinant human epidermal growth factor (rhEGF), molecular weight standards, and all tissue culture reagents were from Invitrogen. The polyclonal STAT5b-specific antibody was developed in our laboratory, as previously described [10]. The monoclonal anti-phospho-STAT5a/5b (Y694/Y699) antibody and anti-phospho-STAT5a/b (S726/S731) were acquired from Upstate Biotechnology. The protease cocktail inhibitor was from Calbiochem, and the acrylamide was obtained from Bio-Rad. Other reagents were of either reagent or molecular biological grade from Sigma.
Cell treatment and analysis of STAT5b
Cells were preincubated overnight in DMEM containing 0.1% bovine serum albumin (BSA). Following preincubation, cells were treated either with media alone (control) or 100ng/mL rhEGF at 37°C for the time indicated. After incubation, cells were washed twice in phosphate-buffered saline (PBS). Detergent lysates were prepared as previously described [1]. The His-tagged expressed proteins were isolated using nickel-NTA-agarose magnetic beads (Qiagen) as previously described [1] and analyzed via immunoblotting. Isolated proteins were separated on a 7.5% polyacrylamide gel and analyzed as previously described [1].
Site-directed mutagenesis
The serine point mutations of STAT5b were constructed by designing primers in which the serine would be mutated to an alanine in the nucleotide sequence using the QuikChange site-directed mutagenesis kit (Stratagene).
Luciferase Assay
SKBr3 cells were transiently transfected with the STAT5 specific Spi 2.1-containing luciferase reporter plasmid. Forty-eight hours post transfection, lysates were prepared, and luciferase activity was measured as previously described [10]. The luciferase values (arbitrary units), as measured by a Berthold luminometer, were normalized to protein amount.
DNA synthesis assay
SKBr3 cells were transiently transfected using Lipofectamine PLUS (Invitrogen) with His-tagged STAT5b constructs according to manufacturer’s recommendations [10]. Twenty-four hours after transfection, cells were serum starved for 18 hrs, and then incubated with 100uM of 5-bromodeoxyuridine (BrdU) for an additional 8 hours. Cells were fixed and stained with fluorescent antibodies directed against the His-tag and BrdU as previously described [1]. Expression of the His-tagged STAT5b construct and BrdU incorporation into DNA were visualized using a Leica DM RBE Fluorescence microscope (model# RS232C).
Results and Discussion
EGF stimulation increases S731 phosphorylation
To investigate basal and EGF-induced S731 phosphorylation of STAT5b, the human breast cancer cell line, SKBr3, was stably transfected with the His-wtSTAT5b expression vector and stimulated with EGF from 5 to 840 minutes (14 hours). Whole cell lysates were analyzed by immunoblotting with antibodies specific for phospho-S731 STAT5b or phospho-Y699 STAT5b (Figure 1B). While a low level of basal S731 phosphorylation was detectable, EGF stimulation significantly increased this phosphorylation as early as 5 minutes of treatment (top panel). As expected, Y699 phosphorylation was seen only upon EGF stimulation (middle panel). Phosphorylation of Y699 results in an upshift of the STAT5b band, as seen in the STAT5b immunoblot (lower panel). Similar band shifts of STAT5b upon phosphorylation have been reported by others [27–29]. Both the serine and tyrosine phosphorylation of STAT5b are sustained through 3 hours (180 minutes), at which point both phosphorylations begin to decrease. These results provide the first evidence of EGF-stimulated S731 phosphorylation in the transactivation domain of STAT5b. Also of importance is that this phosphorylation occurs in a breast cancer cell line that ovexpresses the EGFR and HER2 tyrosine kinases.
Effect of Y740/743F on S731 phosphorylation of STAT5b
As noted previously, S731 is located between the activating tyrosine (Y699) and negative regulatory tyrosines (Y740, Y743) in the transactivation domain of STAT5b (Figure 1A). Thus, we investigated the level of S731 phosphorylation in the Y740/743F STAT5b mutant as well as the potential impact of S731 on Y699 phosphorylation. SKBr3 cells were transfected with wtSTAT5b, Y740/743F, S731A, or S731A/Y740/743F and then treated for 15 minutes with or without EGF. Following isolation of the STAT5b constructs by nickel-NTA magnetic agarose beads, the S731 phosphorylation was analyzed by immunoblotting with the anti-phospho-S731 specific antibody (Figure 2). Specificity of the phospho-S731 STAT5b antibody was demonstrated by the lack of a detectable band for the S731A STAT5b mutant (Figure 2, top panel). Interestingly, the Y740/743F STAT5b mutant demonstrated increased basal and EGF-induced S731 phosphorylation compared to wtSTAT5b. As previously shown, the Y740/743F STAT5b mutant also demonstrated increased basal phosphorylation of Y699 compared to wtSTAT5b (middle panel). In contrast, the S731A STAT5b mutant demonstrated a level of basal and EGF-induced phosphorylation of Y699 similar to that seen with wtSTAT5b, suggesting that mutation of this serine site does not impact the phosphorylation of the activating tyrosine. However, mutation of S731 in the context of the Y740/743F STAT5b mutant (S731A/Y740/743F) not only abrogated the increased basal Y699 phosphorylation seen with the Y740/743F mutant, but also decreased the EGF-induced Y699 phosphorylation (middle panel). These results suggest that the S731 phosphorylation is necessary for the increased basal Y699 phosphorylation of the Y740/743F mutant.
Figure 2. Increased S731 phosphorylation of Y740/743F.
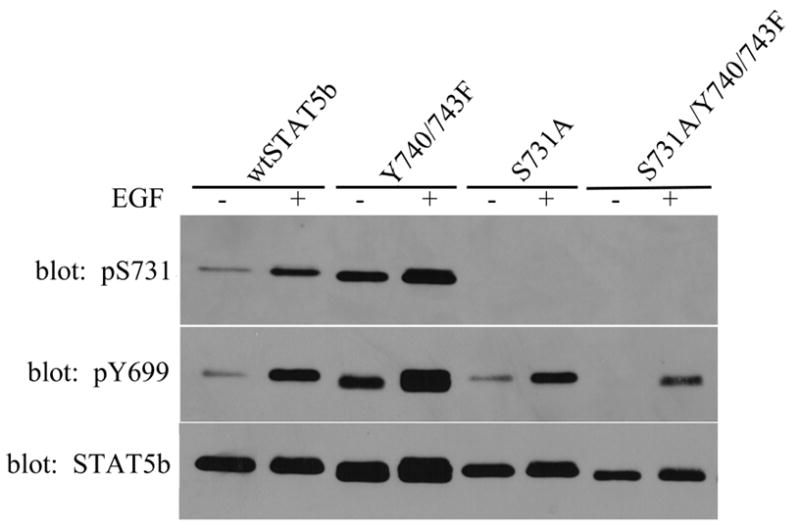
After transfection with His-STAT5b constructs, SKBr3 cells were either unstimulated (−) or stimulated (+) with 100ng/mL EGF for 15 minutes. STAT5b constructs were isolated using nickel-NTA magnetic agarose beads and analyzed by immunoblotting with anti-phospho-S731 STAT5b (top), anti-phospho-Y699 STAT5b (middle), or anti-STAT5b antibodies (bottom). Averages ± S.E. for wtSTAT5b and Y740/743F were calculated from densitometric analysis of three experiments to quantitate fold increase of Y740/743F con over wtSTAT5b con. The values of S731 phosphorylation per STAT5b are: wtSTAT5b con (1.00 ± 0.00); Y740/743F con (3.73 ± 0.97). Student’s t test was utilized to determine statistical significance between wtSTAT5b con and Y740/743F con. *, p=0.0475.
Modulation of transcriptional activity by S731
To determine whether S731 phosphorylation affects the transcriptional activity of STAT5b, particularly the increased basal seen with the Y740/743F STAT5b mutant, the constructs were tested in a luciferase reporter assay with a STAT5 specific response element (Spi2.1-luciferase). As previously shown, the Y740/743F STAT5b mutant increased the transcriptional activity of the STAT5 response element in SKBr3 cells (Figure 3) [1]. In contrast, the mutation of S731 did not alter basal transcriptional activity from that seen with wtSTAT5b (Figure 3), correlating with no change in Y699 phosphorylation (Figure 2). Most significant, the S731A/Y740/743F STAT5b mutant decreased basal transcriptional activity not only compared to the Y740/743F STAT5b mutant, but also compared to wtSTAT5b. Again, these data correlate with the decreased Y699 phosphorylation seen in Figure 2. Thus, mutation of S731 negates the enhanced basal transcriptional activity of the Y740/743F STAT5b mutant. Together, these studies suggest that the increased basal transcriptional activity of the Y740/743F STAT5b mutant requires S731. In fact, S731 phosphorylation plays a more important role in the activity of the Y740/743F STAT5b than it does with wtSTAT5b.
Figure 3. Increased transcriptional activity of Y740/743F is mediated through S731.
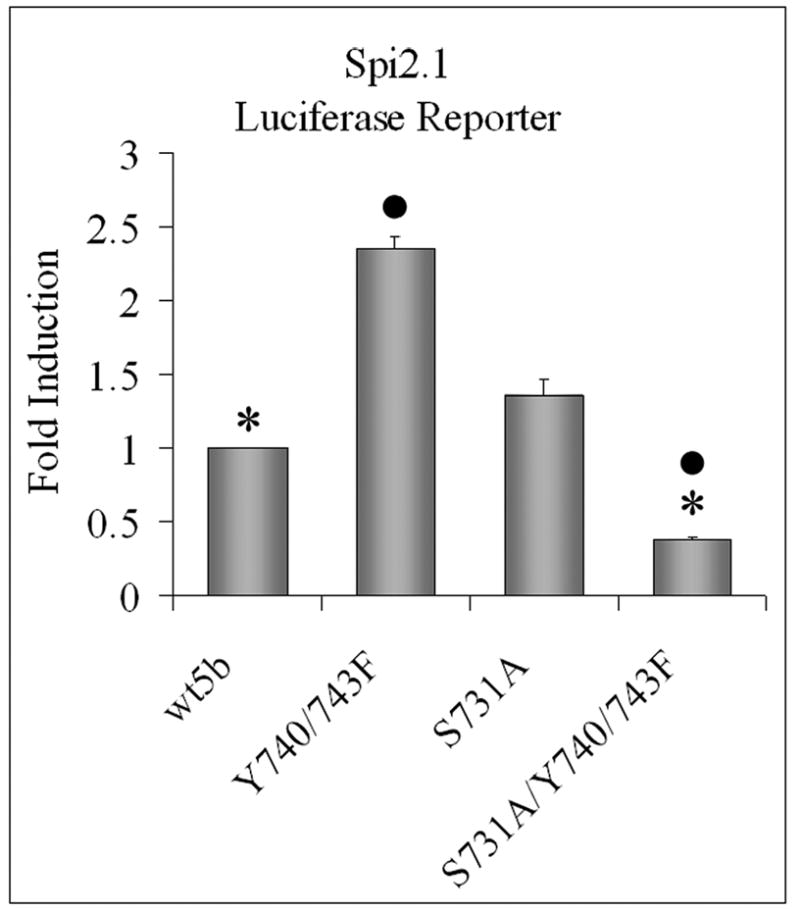
SKBr3 cells were transfected with the Spi 2.1 promoter luciferase construct and either wtSTAT5b, Y740/743F, S731A, or S731A/Y740/743F. Luciferase activity from unstimulated cells was measured as described in “Experimental Procedures.” The graph represents three independent experiments with each group in each experiment done in triplicate. Values are reported as fold induction over wtSTAT5b (con) ± S. E. They are: wtSTAT5b (1.00 ± 0.00); Y740/743F (2.36 ± 0.79); S731A (1.36 ± 0.12); S731A/Y740/743F (0.38 ± 0.02). Student’s t test was utilized to determine statistical significance between Y740/743F and S731A/Y740/743F in addition to wtSTAT5b and S731A/Y740/743F. *, p< 0.001 ●, p<0.001.
Implications of S731 on biological activity
To examine the effect of S731 phosphorylation on biological activity, DNA synthesis assays were performed. SKBr3 cells were transfected with STAT5b constructs, and bromodeoxyuridine (BrdU) incorporation was used as a measurement of DNA synthesis [1]. As shown before, the Y740/743F STAT5b mutant significantly increased basal DNA synthesis (Figure 4) [1]. The DNA synthesis of the S731A STAT5b mutant was comparable to wtSTAT5b, consistent with no change in Y699 phosphorylation or transcriptional activity (Figures 2 and 3). As reflected in the Y699 phosphorylation and transcriptional activity, the S731A/Y740/743F STAT5b mutant significantly decreased DNA synthesis compared to the Y740/743F STAT5b mutant as well as to wtSTAT5b. Thus, mutation of S731 is not only able to abrogate the increased basal activity of the Y740/743F STAT5b mutant, but also able to decrease the level of DNA synthesis below that seen with wtSTAT5b.
Figure 4. Increased biological activity of Y740/743F is mediated through S731.
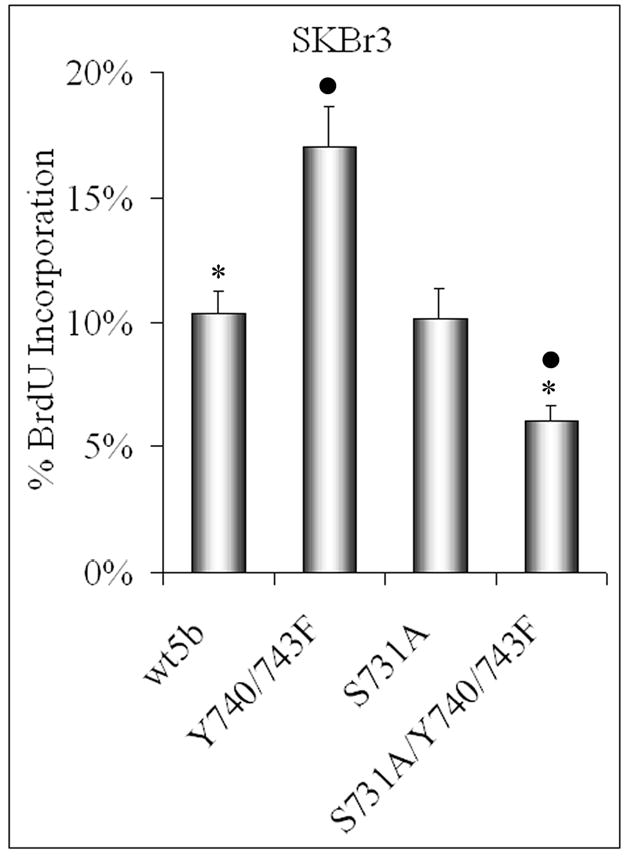
Twenty-four hours after SKBR3 cells were transfected with the STAT5b constructs, bromodeoxyuridine (BrdU) was added to the media for 8hrs. The cells were fixed and stained with fluorescent antibodies against the His-tag and BrdU. The cells that were transfected with the His-tagged construct and incorporated BrdU were counted and graphed. Between 80–120 cells were counted for each treatment group. Results are expressed as the mean percentage ± S.E. of transfected cells that were positive for BrdU incorporation. They are: wtSTAT5b (10.36 ± 0.85); Y740/743F (17.03 ± 1.61); S731A (10.17 ± 1.17); S731A/Y740/743F (6.07 ± 0.58). Student’s t test was utilized to determine statistical significance between transfected wtSTAT5b and S731A/Y740/743F in addition to Y740/743F and S731A/Y740/743F. *, p=0.0002; ●, p=0.0003.
In summary, we have demonstrated for the first time that EGF induces the phosphorylation of S731 in the transactivation domain of STAT5b in a human breast cancer cell line (Figure 1B). While mutation of S731 does not modulate the basal activity of wtSTAT5b, it dramatically inhibits the phosphorylation of Y699, transcriptional activity, and function of the basally active Y740/743F STAT5b mutant (Figures 2, 3, and 4). Thus, mutation of S731 inhibits the mechanism by which the Y740/743F STAT5b mutant increases Y699 phosphorylation and subsequently increases STAT5b activity. Our previous studies determined that Y699 phosphorylation is required for the increased activity of the Y740/743F STAT5b mutant [1]. Together, these data show that both Y699 and S731 are vital to the increased basal activity of the Y740/743F STAT5b mutant. In addition to Y699, these sites in the transactivation domain of STAT5b (S731, Y740, Y743) provide us with targets to mechanistically modulate STAT5b function in the context of a breast cancer cell. Ultimately, understanding the biological mechanisms by which STAT5b is regulated will contribute to the design of potential therapeutics for cancers with aberrant STAT5b signaling.
Acknowledgments
This research was supported by National Institutes of Health (NIH)/NCI grants RO1-CA085462 and 5T32CA001909. We appreciate the expert technical assistance of Elise M. Branch. We thank Teresa M. Bernaciak and Dr. Sarah J. Parsons’ laboratory for insightful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weaver AM, Silva CM. Modulation of signal transducer and activator of transcription 5b activity in breast cancer cells by mutation of tyrosines within the transactivation domain. Molecular Endocrinology. 2006;20:2392–2405. doi: 10.1210/me.2005-0418. [DOI] [PubMed] [Google Scholar]
- 2.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nature Reviews Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 3.Levy D, Darnell J., Jr STATs: transcriptional control and biological impact. Nature Reviews Molecular Cell Biology. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 4.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–8023. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 5.Desrivieres S, Kunz C, Barash I, Vafaizadeh V, Borghouts C, Groner B. The biological functions of the versatile transcription factors STAT3 and STAT5 and new strategies for their targeted inhibition. Journal of Mammary Gland Biology and Neoplasia. 2006;11:75–87. doi: 10.1007/s10911-006-9014-4. [DOI] [PubMed] [Google Scholar]
- 6.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO Journal. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallego MI, Binart N, Robinson GW, Okagaki R, Coschigano KT, Perry J, Kopchick JJ, Oka T, Kelly PA, Hennighausen L. Prolactin, growth hormone, and epidermal growth factor activate STAT5 in different compartments of mammary tissue and exert different and overlapping developmental effects. Developmental Biology. 2001;229:163–175. doi: 10.1006/dbio.2000.9961. [DOI] [PubMed] [Google Scholar]
- 8.Kloth MT, Laughlin KK, Biscardi JS, Boerner JL, Parsons SJ, Silva CM. STAT5b, a Mediator of Synergism between c-Src and the Epidermal Growth Factor Receptor. Journal of Biological Chemistry. 2003;278:1671–1679. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- 9.Lin JX, Mietz J, Modi WS, John S, Leonard WJ. Cloning of human Stat5B. Reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells. Journal of Biological Chemistry. 1996;271:10738–10744. [PubMed] [Google Scholar]
- 10.Kloth MT, Catling AD, Silva CM. Novel activation of STAT5b in response to epidermal growth factor. Journal of Biological Chemistry. 2002;277:8693–8701. doi: 10.1074/jbc.M111884200. [DOI] [PubMed] [Google Scholar]
- 11.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita H, Nevalainen MT, Xu J, LeBaron M, Wagner K, Erwin RA, Harmon JM, Hennighausen L, Kirken RA, Rui H. Role of serine phosphorylation of Stat5a in prolactin-stimulated β-casein gene expression. Molecular and Cellular Endocrinology. 2001;183:151–163. doi: 10.1016/s0303-7207(01)00546-9. [DOI] [PubMed] [Google Scholar]
- 13.Wartmann M, Cella N, Hofer P, Groner B, Liu X, Hennighausen L, Hynes N. Lactogenic hormone activation of STAT5 and transcription of the β-casein gene in mammary epithelial cells is independent of p42 ERK2 mitogen-activated protein kinase activation. Journal of Biological Chemistry. 1996;271:31863–31686. doi: 10.1074/jbc.271.50.31863. [DOI] [PubMed] [Google Scholar]
- 14.Park S, Yamashita H, Rui H, Waxman DJ. Serine phosphorylation of GH-activated signal transducer and activator of transcription 5a (STAT5a) and STAT5b: impact on STAT5 transcriptional activity. Molecular Endocrinology. 2001;15:2157–2171. doi: 10.1210/mend.15.12.0746. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita H, Xu J, Erwin RA, Farrar WL, Kirken RA, Rui H. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. Journal of Biological Chemistry. 1998;273:30218–30244. doi: 10.1074/jbc.273.46.30218. [DOI] [PubMed] [Google Scholar]
- 16.Haq R, Halupa A, Beattie BK, Mason JM, Zanke BW, Barber DL. Regulation of erythropoietin-induced STAT serine phosphorylation by distinct mitogen-activated protein kinases. Journal of Biological Chemistry. 2002;277:17359–17366. doi: 10.1074/jbc.M201842200. [DOI] [PubMed] [Google Scholar]
- 17.Kirken RA, Malabarba MG, Xu J, Liu X, Farrar WL, Hennighausen L, Larner AC, Grimley PM, Rui H. Prolactin stimulates serine/tyrosine phosphorylation and formation of heterocomplexes of multiple Stat5 isoforms in Nb2 lymphocytes. Journal of Biological Chemistry. 1997;272:14098–14103. doi: 10.1074/jbc.272.22.14098. [DOI] [PubMed] [Google Scholar]
- 18.Uddin S, Lekmine F, Sassano A, Rui H, Fish EN, Platanias LC. Role of Stat5 in type I interferon-signaling and transcriptional regulation. Biochemical and Biophysical Research Communications. 2003;308:325–330. doi: 10.1016/s0006-291x(03)01382-2. [DOI] [PubMed] [Google Scholar]
- 19.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clinical Cancer Research. 2002;8:945–954. [PubMed] [Google Scholar]
- 20.Ahonen TJ, Xie J, LeBaron M, Zhu J, Nurmi M, Alanen K, Rui H, Nevalainen MT. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. Journal of Biological Chemistry. 2003;278:27287–27292. doi: 10.1074/jbc.M304307200. [DOI] [PubMed] [Google Scholar]
- 21.Xi S, Zhang Q, Dyer KF, Lerner EC, Smithgall T, Gooding WE, Kamens J, Grandis JR. Src kinases mediate STAT growth pathways in squamous cell carcinoma of the head and neck. Journal of Biological Chemistry. 2003;278:31574–31583. doi: 10.1074/jbc.M303499200. [DOI] [PubMed] [Google Scholar]
- 22.Xi S, Zhang Q, Gooding WE, Smithgall TE, Grandis JR. Constitutive activation of Stat5b contributes to carcinogenesis in vivo. Cancer Research. 2003;63:6763–6771. [PubMed] [Google Scholar]
- 23.Weaver AM, Silva CM. Signal transducer and activator of transcription 5b (STAT5b): A new target of breast tumor kinase. 2007 doi: 10.1186/bcr1794. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabotyanski EB, Rosen JM, Kazansky AV. Signal transduction pathways regulated by prolactin and Src result in different conformations of activated Stat5b. Journal of Biological Chemistry. 2003;278:17218–17227. doi: 10.1074/jbc.M301578200. [DOI] [PubMed] [Google Scholar]
- 25.Olayioye M, Beuvink I, Horsch K, Daly JM, Hynes N. ErbB receptor-induced activation of Stat transcription factors is mediated by Src tyrsoine kinases. Journal of Biological Chemistry. 1999;274:17209–17218. doi: 10.1074/jbc.274.24.17209. [DOI] [PubMed] [Google Scholar]
- 26.de Groot R, Raaijamkers J, Lammers J, Jove R, Koenderman L. STAT5 activation by BCR-Abl contributes to transformation of K562 leukemia cells. Blood. 1999;94:1108–1112. [PubMed] [Google Scholar]
- 27.Friedrichsen BN, Galsgaard ED, Nielsen JH, Moldrup A. Growth hormone- and prolactin-induced proliferation of insulinoma cells, INS-1, depends on activation of STAT5 (signal transducer and activator of transcription 5) Molecular Endocrinology. 2001;15:136–148. doi: 10.1210/mend.15.1.0576. [DOI] [PubMed] [Google Scholar]
- 28.Gebert CA, Park S, Waxman DJ. Regulation of signal transducer and activator of transcription (STAT) 5b activation by the temporal pattern of growth hormone stimulation. Molecular Endocrinology. 1997;11:400–414. doi: 10.1210/mend.11.4.9904. [DOI] [PubMed] [Google Scholar]
- 29.Smit LS, Vanderkuur JA, Stimage A, Han Y, Luo G, Yu-Lee L, Schwartz J, Carter-Su C. Growth hormone-induced tyrosyl phosphorylation and deoxyribonucleic acid binding activity of Stat5A and Stat5B. Endocrinology. 1997;138:3426–3434. doi: 10.1210/endo.138.8.5332. [DOI] [PubMed] [Google Scholar]


