Abstract
In recent years the kinesin superfamily has become so large that several different naming schemes have emerged, leading to confusion and miscommunication. Here, we set forth a standardized kinesin nomenclature based on 14 family designations. The scheme unifies all previous phylogenies and nomenclature proposals, while allowing individual sequence names to remain the same, and for expansion to occur as new sequences are discovered.
Kinesins constitute a superfamily of microtubule-based motor proteins that perform diverse functions, including the transport of vesicles, organelles, chromosomes, protein complexes, and RNPs; they also help to regulate microtubule dynamics (for review see Hirokawa, 1998). Individually identified kinesins are often named on the basis of their functional characteristics (McDonald et al., 1990). Systematically identified kinesins have different names (Stewart et al., 1991; Aizawa et al., 1992; Vernos et al., 1993). Kinesins also have been named by other criteria, including the position of the motor core within the protein (Vale and Fletterick, 1997), and their evolutionary relatedness to other kinesins (for reviews see Goodson et al., 1994; Sekine et al., 1994; Hirokawa, 1998; Kirchner et al., 1999). Early in the process of kinesin discovery, it was relatively simple to be familiar with the names and potential functional relationships of all known kinesins (Goodson et al., 1994). Today, however, there are literally hundreds of kinesins being named by diverse criteria, and inconsistencies are emerging that cause genuine confusion (for specific examples, see “Problems with Previous Nomenclature” at http://www.proweb.org/kinesin/Nomenclature_Details.html).
The revised nomenclature
To address this and other confusion that have arisen in recent classifications, a special interest subgroup meeting was held at the 2003 meeting of the American Society for Cell Biology. As a result of this meeting and other feedback from kinesin researchers worldwide (available for review as an “Archived Web Discussion” at http://www.proweb.org/kinesin/Nomenclature_Details.html), we determined that a single nomenclature must be adopted to facilitate communication among researchers. Here, we propose a standardized nomenclature for all kinesin families (Table I). To minimize confusion, the names of individual sequences will remain unchanged. The formal kinesin nomenclature is shown in Table I and was constructed using the following logic and rules:
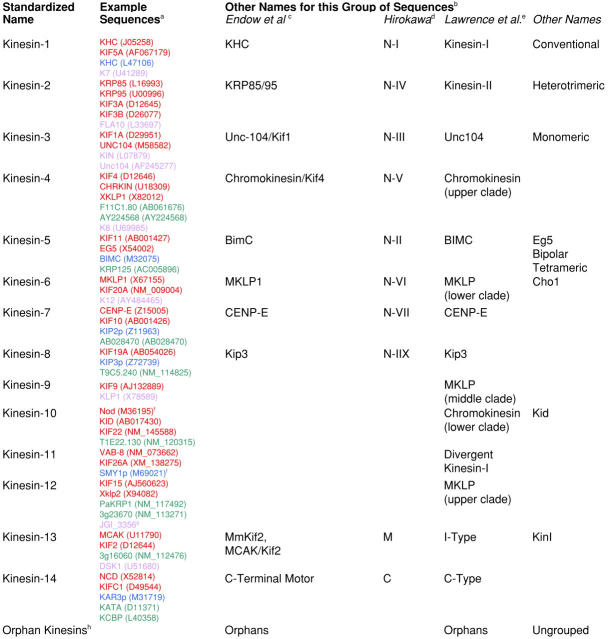
Table I.
Sequences are listed by common name followed by their GenBank ID in parentheses and are color-coded by kingdom: protist sequences are purple; plant sequences are green; animal sequences are red; and those from fungi are blue.
A list of kinesin sequences and their respective family classification (if known) is actively maintained and exists online at http://www.proweb.org/kinesin//MotorSeqTable.html.
Dagenbach and Endow, 2004; Goodson et al., 1994; Kim and Endow, 2000; Moore and Endow, 1996.
Hirokawa, 1998.
Lawrence et al., 2002.
Drosophila melanogaster Nod and Saccharomyces cerevisiae SMY1p do not always group with their respective families. The placement of these two sequences is difficult to attain without performing in-depth, careful phylogenetic analyses, and their family placement should be considered tenuous.
Chlamydomonas reinhardtii sequence from the DOE Joint Genome Institute (http://genome.jgi-psf.org/). JGI_3356 is a locus on scaffold 135 at positions 184041–190631.
Goldstein, 1993.
(1) Each family bears the name “Kinesin.” This was the name given to the first described superfamily member (from the Greek “kinein,” to move; Brady, 1985; Vale et al., 1985). Its use will avoid past confusion encountered by having families called “kin” (which can be confused with kinase), “kif” (an acronym for “kinesin superfamily”), and KLP (an acronym for kinesin-like protein).
(2) To designate each family, an Arabic number has been used. This avoids the problems with using Roman numerals for database searches, and helps us to steer clear of the structural and functional misinterpretations that could continue to arise if letters of the alphabet (e.g., C, N, I, and M) were to be used.
(3) Large subfamilies will be referred to as individual entities by appending a letter to the family name. For example, the Kinesin-14 family is made up of two large subfamilies, designated Kinesin-14A and Kinesin-14B (previously referred to as C-I and C-II, respectively; Lawrence et al., 2002).
(4) Wherever possible, new number designations are based on past names or known functional characteristics. Examples include Kinesin-1 (the first kinesin discovered was a member of this family), Kinesin-2 (the holoenzyme for these family members has been referred to as “kinesin II” in past publications; Scholey, 1996), and Kinesin-4 (named for mouse KIF4, the family's founding member; Sekine et al., 1994). The numbers associated with Hirokawa's class designations (1998) have been retained for Kinesin-1, -3, -6, -7, and -8.
(5) At this time, there are 14 recognized kinesin families. This number was derived by determining the consensus monophyletic groups conserved among past phylogenetic analyses (Moore and Endow, 1996; Hirokawa, 1998; Kim and Endow, 2000; Miki et al., 2001; Lawrence et al., 2002; Dagenbach and Endow, 2004) in conjunction with examining phylogenetic trees generated by S.C.D. using a dataset that includes many protistan kinesin sequences (manuscript in preparation) and a phylogenetic tree generated by H. Miki, Y. Okada, and N. Hirokawa using a dataset made up of 608 publicly available kinesin sequences (which can be viewed as a part of the “Archived Web Discussion” at http://www.proweb.org/kinesin/Nomenclature_Details.html). It is important to note that the naming system outlined here is based primarily on molecular systematic analysis, as are the accepted systems for other cytoskeletal gene families (e.g., the myosin and actin gene families; Cheney et al., 1993; Goodson and Spudich, 1993; Goodson and Hawse, 2002).
(6) To gain the status of a recognized kinesin family or subfamily, the group in question must be made up of sequences from at least two kingdoms. This criterion prevents classification of small groups of sequences from individual species or closely related groups of organisms as families or subfamilies, and it will help keep the number of recognized families small enough to be manageable. Sequences not grouping consistently within a family (based on measures of phylogenetic consistency such as high bootstrap support) will be called “orphan kinesins” (Goldstein, 1993).
It should be noted that in order for new groups of kinesins to become recognized as a new kinesin family, the group of sequences must conform to all rules set forth herein.
Referring to kinesin families in publications
In practice, there are two acceptable ways to identify the family to which a sequence belongs. The first uses the formal name of the appropriate family followed by one of the family's former names in parentheses (and a reference to an appropriate publication). The second uses only the formal name. Examples of acceptable usage follow.
(1) Human Eg5, a member of the Kinesin-5 family (previously referred to as BimC by Dagenbach and Endow, 2004), is involved in establishing the bipolar spindle.
(2) Human Eg5, a member of the Kinesin-5 family, is involved in establishing the bipolar spindle.
We recommend that such a statement be included within the introduction of any publication. However, it is not necessary to call the sequence itself by the name of the family to which it belongs: individual sequence names will remain unchanged to reduce needless confusion.
Criteria for kinesin classification
Finally, for publication there should be an accepted protocol for identifying the family to which an unclassified kinesin sequence belongs. How this determination was made should be explicitly stated in the manuscript within the methods section. Our suggestion is to first do a BLAST search (Altschul et al., 1990) using the full-length protein sequence for the kinesin of interest as the query. If all top hits are members of a single kinesin family, the kinesin of interest is probably also a member of that family. To confirm the BLAST results and also to classify sequences where BLAST results do not clearly indicate assignment to a particular family, we recommend that the researcher download a published kinesin motor core alignment such as one of the Lawrence et al. (2002) alignments or the alignment available through the kinesin home page (http://www.proweb.org/kinesin/KinesinAlign.html), and add their own kinesin's motor core to that alignment (by hand or using an alignment tool such as Clustal; Higgins et al., 1996). (Lawrence et al., 2002 alignments ALIGN_000356, ALIGN_000357, and ALIGN_000358 are available online from EMBL at ftp://ftp.ebi.ac.uk/pub/databases/embl/align.) Next, use a simple method for building a phylogenetic tree (such as neighbor joining; Saitou and Nei, 1987) to find the family most closely related to the kinesin of interest. We also recommend that bootstrap resampling (easily performed by phylogenetic analysis programs such as Clustal) be used to determine whether a kinesin should be assigned to a particular family or remain an ungrouped “orphan kinesin.” These alignment and treebuilding methods are available together on a webserver at http://www.ebi.ac.uk/clustalw. Analysis of nonmotor regions can help to confirm family assignments (Dagenbach and Endow 2004), and as previously noted by Hirokawa (1998) and Vale (2003), specific domains or motifs should be used as secondary criteria for classification. For instance, many Kinesin-3 family members can be identified by the presence of both a fork head homology (FHA) domain COOH-terminal to the motor and a conserved insertion present within the third loop (Vale, 2003). Although the relatedness of families to one another varies among published phylogenies, the members of each family are relatively consistent among published trees, making it possible to use any published phylogeny as a guide to classification.
Acknowledgments
Research groups supporting the use of this system for kinesin nomenclature include the following: Victoria Allan, Linda Amos, Peter Baas, Steven Block, George Bloom, Konrad Böhm, Olaf Bossinger, Scott Brady, W. Zacheus Cande, Karen Christie, Don Cleveland, Douglas Cole, Duane Compton, Roger Cooke, Robert Cross, R. Kelly Dawe, Scott Dawson, Arshad Desai, Sharyn Endow, Anne Ephrussi, Jacek Gaertig, Volodya Gelfand, Joseph Gindhart, Lawrence Goldstein, Holly Goodson, Steven Gross, David Hackney, William Hancock, Scott Hawley, Steven Henikoff, Nobutaka Hirokawa, Keiko Hirose, Andreas Hoenger, Peter Hollenbeck, Jonathon Howard, Kenneth Johnson, Frank Jülicher, F. Jon Kull, Carolyn Lawrence, Bo Liu, Russell Malmberg, Eckhard Mandelkow, Richard McIntosh, Edgar Meyhöfer, Harukata Miki, David Mitchell, Timothy Mitchison, Virgil Muresan, Berl Oakley, Yasushi Okada,Elizabeth Raff, Krishanu Ray, A.S.N. Reddy, Thomas Reese, Mark Rose, Joel Rosenbaum, Douglas Ruden, Ted Salmon, Peter Satir, William Saunders, William Saxton, Manfred Schliwa, Bruce Schnapp, Jonathan Scholey, David Sharp, Hernando Sosa, Ann Sperry, Gero Steinberg, Susan Strome, Kazuo Sutoh, Ronald Vale, Isabelle Vernos, Claire Walczak, Richard Walker, Matthew Welch, Linda Wordeman, and Tim Yen.
Carolyn J. Lawrence's present address is Department of Genetics, Development and Cell Biology, Iowa State University, Ames, IA 50011.
References
- Aizawa, H., Y. Sekine, R. Takemura, Z. Zhang, M. Nangaku, and N. Hirokawa. 1992. Kinesin family in murine central nervous system. J. Cell Biol. 119:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Brady, S.T. 1985. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 317:73–75. [DOI] [PubMed] [Google Scholar]
- Cheney, R.E., M.A. Riley, and M.S. Mooseker. 1993. Phylogenetic analysis of the myosin superfamily. Cell Motil. Cytoskeleton. 24:215–223. [DOI] [PubMed] [Google Scholar]
- Dagenbach, E.M., and S.A. Endow. 2004. A new kinesin tree. J. Cell Sci. 1:3–7. [DOI] [PubMed] [Google Scholar]
- Goldstein, L.S. 1993. With apologies to scheherazade: tails of 1001 kinesin motors. Annu. Rev. Genet. 27:319–351. [DOI] [PubMed] [Google Scholar]
- Goodson, H.V., and W.F. Hawse. 2002. Molecular evolution of the actin family. J. Cell Sci. 115:2619–2622. [DOI] [PubMed] [Google Scholar]
- Goodson, H.V., S.J. Kang, and S.A. Endow. 1994. Molecular phylogeny of the kinesin family of microtubule motor proteins. J. Cell Sci. 107:1875–1884. [DOI] [PubMed] [Google Scholar]
- Goodson, H.V., and J.A. Spudich. 1993. Molecular evolution of the myosin family - relationships derived from comparisons of amino-acid-sequences. Proc. Natl. Acad. Sci. USA. 90:659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D., J. Thompson, and T. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383–402. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N. 1998. Kinesins and dynein superfamily proteins and the mechanism of organelle transport. Science. 279:519–526. [DOI] [PubMed] [Google Scholar]
- Kim, A., and S. Endow. 2000. A kinesin family tree. J. Cell Sci. 113:3681–3682. [DOI] [PubMed] [Google Scholar]
- Kirchner, J., G. Woehlke, and M. Schliwa. 1999. Universal and unique features of kinesin motors: insights from a comparison of fungal and animal conventional kinesins. Biol. Chem. 380:915–921. [DOI] [PubMed] [Google Scholar]
- Lawrence, C.J., R.L. Malmberg, M.G. Muszynski, and R.K. Dawe. 2002. Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 54:42–53. [DOI] [PubMed] [Google Scholar]
- McDonald, H., R. Stewart, and L.S.B. Goldstein. 1990. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell. 63:1159–1165. [DOI] [PubMed] [Google Scholar]
- Miki, H., S. Mitsutoshi, K. Kaneshiro, and N. Hirokawa. 2001. All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA. 98:7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J.D., and S.A. Endow. 1996. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays. 18:207–219. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 6:406–425. [DOI] [PubMed] [Google Scholar]
- Scholey, J. 1996. Kinesin-II, a membrane traffic motor in axons, axonemes, and spindles. J. Cell Biol. 133:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine, Y., Y. Okada, Y. Noda, S. Kondo, H. Alzawa, R. Takemura, and N. Hirokawa. 1994. A novel microtubule-based motor protein (KIF4) for organelle transport, whose expression is regulated developmentally. J. Cell Biol. 127:187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, R.G., P.A. Pesavento, D.N. Woerpel, and L.S. Goldstein. 1991. Identification and partial characterization of six members of the kinesin superfamily in Drosophila. Proc. Natl. Acad. Sci. USA. 88:8470-8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale, R. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480. [DOI] [PubMed] [Google Scholar]
- Vale, R., and R. Fletterick. 1997. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13:745–777. [DOI] [PubMed] [Google Scholar]
- Vale, R., T. Reese, and M. Sheetz. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 42:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernos, I., J. Heasman, and C. Wylie. 1993. Multiple kinesin-like transcripts in Xenopus oocytes. Dev. Biol. 157:232–239. [DOI] [PubMed] [Google Scholar]