Abstract
With more than 1.2 million copies, Alu elements are one of the most important sources of structural variation in primate genomes. Here, we compare the chimpanzee and human genomes to determine the extent of Alu recombination-mediated deletion (ARMD) in the chimpanzee genome since the divergence of the chimpanzee and human lineages (∼6 million y ago). Combining computational data analysis and experimental verification, we have identified 663 chimpanzee lineage-specific deletions (involving a total of ∼771 kb of genomic sequence) attributable to this process. The ARMD events essentially counteract the genomic expansion caused by chimpanzee-specific Alu inserts. The RefSeq databases indicate that 13 exons in six genes, annotated as either demonstrably or putatively functional in the human genome, and 299 intronic regions have been deleted through ARMDs in the chimpanzee lineage. Therefore, our data suggest that this process may contribute to the genomic and phenotypic diversity between chimpanzees and humans. In addition, we found four independent ARMD events at orthologous loci in the gorilla or orangutan genomes. This suggests that human orthologs of loci at which ARMD events have already occurred in other nonhuman primate genomes may be “at-risk” motifs for future deletions, which may subsequently contribute to human lineage-specific genetic rearrangements and disorders.
Author Summary
The recent sequencing of a number of primate genomes shows that small segments of DNA known as Alu elements are found repeatedly along all chromosomes, and indeed comprise ∼10% of the human genome. Although older Alu elements that have been in the genome for a long time accumulate some random mutations, overall these elements retain high levels of sequence identity among themselves. The presence of many near-identical Alu elements located close to each other makes primate genomes prone to DNA recombination events that generate genomic deletions of varying sizes. Here, by scanning the chimpanzee genome for such deletions, we determined the role of the Alu recombination-mediated deletion process in creating structural differences between the chimpanzee and human genomes. Using a combination of computational and experimental techniques, we identified 663 deletions, involving the removal of ∼771 kb of genomic sequence. Interestingly, about half of these deletions were located within known or predicted genes, and in several cases, the deletions removed coding exons from chimpanzee genes as compared to their human counterparts. Alu recombination-mediated deletion shows signs of being a major sculptor of primate genomes and may be responsible for generating some of the genetic differences between humans and chimpanzees.
Introduction
Mobile elements are a major source of genetic diversity in mammals [1,2]. Alu elements, a family of short interspersed elements (SINEs), emerged ∼65 million y ago (Mya) and have successfully proliferated in primate genomes with >1.2 million copies [2–5]. Alu elements consist of a left monomer and a right monomer [2,6]. Each of these monomers independently evolved from 7SL-RNA [7] and subsequently fused into the dimeric Alu element in the primate lineage [6]. Alu elements are known to be associated with primate-specific genomic alterations by several mechanisms, including de novo insertion, insertion-mediated deletion, and unequal recombination between Alu elements [8–11]. The Alu family consists of a number of subfamilies, which maintain high sequence identity among themselves (70%–99.7%) [12–15].
Mispairing between two Alu elements has been shown to be a frequent cause of deletion or duplication in the host genome [10,11,16]. A recent study of human-specific Alu recombination-mediated deletion (ARMD) reported a significant number of events associated with Alu elements [10]. An ARMD may arise through either interchromosomal recombination by mismatch of sister or nonsister chromatids during meiosis [17] or by intrachromosomal recombination between two Alu elements on the same chromosome. Previously, Sen et al. [10] found 492 human-specific ARMD events responsible for ∼400 kb of deleted genomic sequence in the human lineage [10]. Here, we report 663 chimpanzee-specific ARMD events identified from comparative analysis of the chimpanzee and human genomes. The chimpanzee-specific ARMD events deleted a total of ∼771 kb of genomic sequence in chimpanzees, including exonic deletions in six genes, sometime after the divergence of the human and chimpanzee lineages (∼6 Mya). ARMD events in the chimpanzee genome have generated large deletions (up to ∼32 kb) relative to human-specific ARMD events. Taking deletions in both the human and chimpanzee lineages into account, we suggest that ARMD events may have contributed to genomic and phenotypic diversity between humans and chimpanzees.
Results
A Genome-Wide Analysis of Chimpanzee-Specific ARMD Events
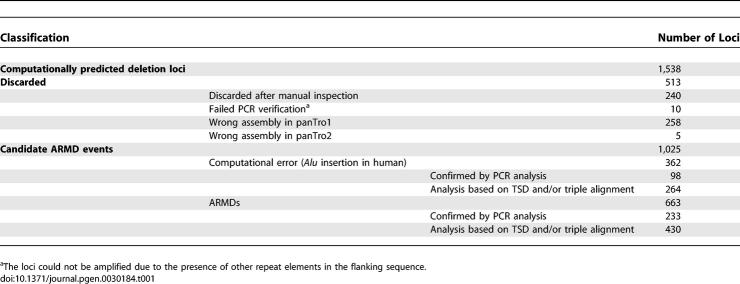
To investigate chimpanzee-specific ARMD loci, we first computationally compared the chimpanzee (panTro1) and human (hg17) genome reference sequences. A total of 1,538 ARMD candidates were initially retrieved using panTro1. These loci were converted to panTro2 (March 2006), which, due to the better quality of the sequence assembly, allowed us to eliminate a number of loci that mimicked authentic ARMD loci. Through a comparison of panTro1 and panTro2, we discarded 258 of the 1,538 loci (Table 1). The remaining 1,280 loci were manually inspected using the repetitive DNA annotation utility RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). In terms of local sequence architecture, human-specific mobile element insertions between two preexisting adjacent Alu elements could be computationally confused with a chimpanzee-specific deletion. Because the consensus sequences of the human-specific mobile elements (e.g., AluYb8, AluYa5, SVA, and L1Hs) have been well established in RepeatMasker, we were able to identify and eliminate from our analysis 189 human-specific insertion loci, including processed pseudogenes. The remaining 1,091 candidate ARMD loci were inspected using triple alignments of human (hg18), chimpanzee (panTro2), and rhesus macaque (rheMac2) sequences at each locus, and also on the basis of their target site duplication (TSD) structures (see Materials and Methods). After manual inspection, 342 of the candidate ARMD loci were examined by PCR to verify their status as authentic ARMD loci. Finally, combining computational and experimental results, 663 loci were confirmed as bona fide chimpanzee-specific ARMD loci (Table 1 and Dataset S1).
Table 1.
Summary of Chimpanzee-Specific ARMD Events
In this study, we combined computational data mining and wet-bench experimental verification, an approach that is optimal for identifying lineage-specific insertions and deletions [10]. Whereas Sen et al. [10] computationally compared the human and chimpanzee genomes, in our analysis, the draft version of the rhesus macaque genome sequence was used as an outgroup when filtering computational output for false positives (see Materials and Methods). This allowed us to eliminate 215 candidate ARMD loci prior to wet-bench verification, minimizing the cost and time needed to confirm authentic chimpanzee-specific ARMD events, as compared with the previous human-specific ARMD study.
Genomic Deletion Through Chimpanzee-Specific ARMD Events
Since the human-chimpanzee divergence ∼6 Mya, chimpanzee-specific ARMD events have occurred 1.3 times as often as their human-specific counterparts (663 chimpanzee-specific versus 492 human-specific events). The total amount of genomic DNA deleted by ARMD events from the chimpanzee genome is estimated to be 771,497 bp. However, when we consider that the average indel divergence between the human and chimpanzee genomes has been estimated at 5.07% [18], the precise amount of DNA deleted through ARMDs in the chimpanzee genome could be anywhere between ∼733 and ∼811 kb (±5.07% of ∼771 kb). The size distribution of DNA sequences deleted through chimpanzee-specific ARMD events ranged from 111 to 31,861 bp, with 1,164 bp average and 615 bp median ARMD sizes. Similar to the pattern observed in human-specific ARMD events [10], a histogram of the size distribution of chimpanzee-specific ARMDs is skewed toward deletions of shorter size, with ∼68% (449 of 663) of the deletion events shorter than 1 kb (Figure 1). As expected, about 70% of the deleted genomic DNA sequences are composed of repetitive elements (Table 2), of which Alu element sequences account for ∼64% (338 kb of 528 kb). Interestingly, the amount of sequence deleted through the ARMD process from the chimpanzee genome is twice as much as that from the human genome during the same period of time. Ten chimpanzee-specific ARMD events were found to have each deleted >7.3 kb of sequence (Figure 1); ARMD sizes this large were not observed in the human-specific study. Among these, the largest deleted sequence is 31,861 bp in length, within which only the SLC9A3P2 pseudogene and two intergenic regions are found in the ancestral sequence (i.e., human ortholog).
Figure 1. Size Distribution of Chimpanzee-Specific ARMD Events.
Size distribution of chimpanzee-specific ARMD events (red bars) compared with that of human-specific ARMD events (blue bars), displayed in 200-bp bin sizes.
Table 2.
Classification of Genomic DNA Deleted by ARMDs in Chimpanzee Lineage

To examine the possible effects of the removal of ancestral genomic sequences during the 663 chimpanzee lineage-specific ARMD events, we retrieved the pre-recombination sequences (i.e., unaltered orthologs) from the human genome. About 46% (305 of 663) of the ARMD events were located within known or predicted RefSeq genes (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=9606), and five ARMD events generated 13 exonic deletions in six genes annotated as either demonstrably or putatively functional in the human genome. Among them, two ARMD events deleted exons from demonstrably functional genes in the NBR2 (neighbor for BRCA1 [breast cancer 1] gene 2) and HTR3D (5-hydroxytryptamine [serotonin] receptor 3 family member D) genes. While no alternative pre-mRNA spliced forms exist for the NBR2 gene, the HTR3D gene shows three alternative pre-mRNA spliced forms in the human according to the ECR Browser (http://ecrbrowser.dcode.org). Among them, one of the HTR3D isoforms does not contain exon 3, which was deleted from the chimpanzee genome. Thus, chimpanzees could produce a similar protein to the HTR3D isoform mentioned above, because the ARMD event deleted the entire exon 3 and portions of some introns in the chimpanzee genome. However, we cannot rule out that the ARMD event has produced cryptic splicing sites causing either nonfunctionalization or subfunctionalization of HTR3D. The remaining three chimpanzee ARMD events generated exonic deletions in four putative human genes of unknown function (LOC339766, LOC127295, LOC729351, and LOC645203).
To further analyze the genomic sequences lost due to the ARMD process in the chimpanzee genome, we used the National Center for Biotechnology Information's (NCBI) UniGene utility (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene) to look at the orthologous loci in the human genome, which contained sequences that would have been present in the chimpanzee genome if the ARMD events had not occurred. UniGene indicated that 164 ARMD events had caused deletions of coding sequence on the basis of expressed sequence tags (ESTs), although this number decreased to 94 when a high threshold indicating protein similarities (≥98% ProtEST) was selected (Table S1). This number is much higher than the exonic deletions in six genes generated by ARMD events reported above when RefSeq annotation was used instead.
Structural Features of ARMD Events
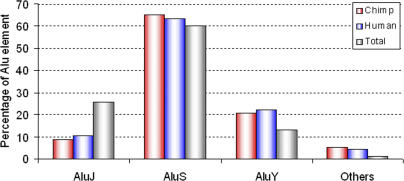
Ten different Alu subfamilies are associated with chimpanzee-specific ARMD events: AluJo, AluJb, AluSx, AluSq, AluSp, AluSg, AluSg1, AluSc, AluY, and AluYd8. Their composition and ratio in chimpanzee-specific ARMD events are remarkably similar to those in human-specific ARMD events (Figure 2). The Alu subfamily analysis shows that the number of elements from each Alu subfamily involved in the ARMD process is proportional to the genome-wide copy number of each Alu subfamily in the chimpanzee genome. For example, the AluS subfamily has contributed the most to chimpanzee-specific ARMD events because it is the most successful Alu subfamily in the primate genome in terms of copy number. However, we found one exception to this rule; the AluJ subfamily is more ubiquitous than the AluY subfamily in both the chimpanzee and human genomes (Figure 3), but more members of the AluY subfamily were found to be involved in the ARMD process. The major expansion of the AluJ subfamily in primate genomes occurred ∼60 Mya, whereas the AluY subfamily expanded only ∼24 Mya [14,19,20]. On the basis of these ages, the individual members of the AluJ subfamily have likely accumulated more point mutations than those of the AluY subfamily. As a result, AluY copies have more sequence identity among them than do the AluJ copies, which results in increased involvement in ARMD events. In addition, we investigated intra-Alu subfamily recombination-mediated deletions for both the AluJ and AluY subfamilies. Of the 103 events involving at least one AluJ element in the ARMD event, only 15 (14.6%) involved recombination between two AluJ elements. The AluY subfamily shows a higher rate of intra-subfamily recombination than the AluJ subfamily, with 219 loci in which at least one AluY element was involved in the recombination event, and 57 (26%) that were between two AluY elements. This suggests that the rate of recombination between AluY elements is 1.8 times higher than that between AluJ elements. Taken together, this suggests that, in addition to the copy number of each Alu subfamily, the level of sequence identity between the individual Alu elements in the genome is also an important variable influencing ARMD events.
Figure 2. Alu Subfamily Composition in ARMD Events.
Proportion of all Alu elements involved in chimpanzee- and human-specific ARMD events (red and blue bars, respectively) that belong to each Alu subfamily as noted.
Figure 3. Comparison of Alu Subfamilies Involved in ARMD Events.
Proportion of Alu elements involved in chimpanzee-specific (red bars) and human-specific (blue bars) ARMD events versus proportion of total Alu elements in each subfamily in the chimpanzee genome (gray bars).
From a mechanistic viewpoint, four different types of recombination may occur between two Alu elements. An Alu element consists of left and right monomers. In the first type, comprising about 88% (583 of 663) of the ARMD events in our study, the recombination occurred between the same monomers of the two Alu elements. A second type of recombination occurred between two Alu elements in which one had previously integrated into the middle of the other. Such insertions are commonly found in both the chimpanzee and human genomes because each Alu element bears two endonuclease cleavage sites (5′-TTTT/A-3′) between its two monomers. About 8% (51 of 663) of the ARMD events in the chimpanzee genome are products of this second type of recombination. The third type of recombination, seen in 25 of the 663 events (∼4%), involved recombination between the left and right monomers on two separate Alu elements. The last type occurred between oppositely oriented Alu elements. Instances of this type of ARMD are very rare, found only in four of the 663 cases (0.6%). This style of recombination is likely to be uncommon because the stretch of sequence identity between two Alu elements oriented in opposite directions to one another is too short to frequently generate unequal homologous recombination. Instead, these two Alu elements are more likely to cause Alu recombination-mediated inversions or A-to-I RNA editing through the posttranscriptional modification of RNA sequences [21].
Analysis of the ARMD “Hotspots”
To analyze the frequency of recombination at different positions along the length of the Alu elements (which we refer to as “recombination breakpoints”) at our ARMD loci, we aligned the two intact human Alu elements involved in each recombination event with the single chimeric Alu element from the chimpanzee genome (Figure S1). The windows between the two Alu elements range in size from 1 to 116 bp, with a mean of 20 bp and a mode of 22 bp. In general, the ARMD loci generated by intra-Alu subfamily recombination, as well as the recombination events between relatively young Alu elements, show longer stretches of sequence identity than others. Through this analysis, we identified a recombination “hotspot” on the Alu consensus sequence (5′-TGTAATCCCAGCACTTTGGGAGG-3′), located between positions 24 and 45 (Figure 4). This recombination hotspot is congruent with previous studies of gene rearrangements in the human LDL-receptor gene involving Alu elements [22], and with the pattern of recombination found in the 492 human-specific ARMD events [10]. Of these studies, the former suggested that the hotspot sequence (therein called the “core sequence”) might induce genetic recombination because it subsumes the prokaryotic chi sequence (the pentanucleotide motif CCAGC), which is known to stimulate recBC-dependent recombination [23]. We searched for and found the CCAGC motif at four places (positions 31–35, 85–89, 166–170, and 251–255) along the Alu consensus sequences. The percentages of breakpoints found at these positions are 0.00886%, 0.00336%, 0.00406%, and 0.00372%, respectively. Among these, the percentages of breakpoints found at the latter three positions are similar to the average percentage of breakpoints across the entire length of the Alu elements (0.0035%) in our ARMD events. The only spot where the motif is found that showed a substantially higher percentage of breakpoints is the one located at positions 31–35, which is within our proposed hotspot. Therefore, this motif may invoke, but does not seem to be essential for the generation of ARMD events.
Figure 4. Recombination Breakpoints during Chimpanzee-Specific ARMD Events.
Percentage of ARMD events found to have breakpoints at different positions along an Alu consensus sequence. The “hotspot” region is represented by a conserved 22-bp nucleotide sequence found in 634 ARMD loci (the first and second types of ARMD events) using WebLogo analysis (http://weblogo.berkeley.edu). The dashed line represents the average percentage (0.0035%) of breakpoints across the entire length of the Alu consensus sequence.
Interestingly, the 22-bp hotspot sequence contains no CpG dinucleotides. These CpG dinucleotides have been shown to mutate approximately six times faster than other dinucleotides in Alu elements [24] due to cytosine methylation and subsequent deamination [25]. In addition, when we aligned the consensus sequences of the 10 different Alu subfamilies involved in ARMDs, we found that the hotspot sequence is located within the longest stretch of their conserved regions. Furthermore, using the software utility WebLogo [26], we confirmed that this 22-bp sequence is the most conserved region among Alu elements involved in ARMD events (Figure 4). Therefore, the recombination hotspot that we have identified, by virtue of having an increased level of conservation among the Alu subfamilies involved in the ARMDs in our study, has potentially allowed frequent recombination between Alu repeats from different Alu subfamilies to occur.
Genomic Environment of ARMD Events
Most Alu elements located in the primate genomes that have been sequenced (e.g., human, chimpanzee, and rhesus macaque) exist in high-GC content regions [3–5], and also have high GC content (an average of ∼62.7%). Moreover, it has also been previously reported that human-specific ARMD events preferentially occur in areas of high GC content (∼45% GC content, on average) [10]. To analyze the genomic environment of chimpanzee-specific ARMD events, we estimated the GC content of 20 kb (±10 kb in either direction) of neighboring sequence for each ARMD locus. Our results indicate that the chimpanzee-specific ARMDs are similar to human-specific ARMDs in having a tendency to occur in GC rich regions (45.2% GC content, on average). This preference is correlated with the distribution of Alu elements involved in ARMDs (Figure 3) because the genomic distribution of ARMD events would in effect have an a priori dependence on the preferred locations of Alu elements after insertion of the different Alu subfamilies. About 74% of chimpanzee-specific ARMDs are associated with the older Alu subfamilies, AluJ and AluS. Although young Alu subfamilies are found in AT-rich, gene-poor regions, the older Alu subfamilies are most often found in GC-rich, gene-rich regions [3]. This could account for the preferential occurrence of ARMD events in GC-rich regions. Moreover, the local rate of genomic recombination has been shown to be positively correlated with GC content [27], which may further explain the observed distribution of ARMD events. About 44% of genomic DNA deleted through ARMD events were Alu sequences in the human ortholog. This could indicate that regions of high local Alu element density within chromosomes are more likely to provide increased opportunities for local recombination, a trend previously noticed during analysis of the global genomic distribution of human lineage-specific ARMD events [10].
To further characterize the genomic environment of chimpanzee-specific ARMD events, we estimated the gene density of the genomic regions flanking each chimeric Alu element resulting from the process by extracting 4 Mb of flanking genomic sequences (±2 Mb in either direction), and counting the number of known or predicted chimpanzee RefSeq genes. The gene density of the flanking regions of chimpanzee-specific ARMD events is estimated to be, on average, one gene per 60.7 kb, which is similar to that of human-specific ARMD events (one gene per 66 kb). This indicates that the global distribution of chimpanzee-specific ARMD events is biased towards gene-rich regions, since the global average gene density in the chimpanzee genome is approximately one gene per 112 kb. To test for any relationship between the size of an ARMD and its flanking gene density or GC content, we performed a correlation test. While the r-values for both tests were negative, as would be expected given the danger of large deletions in gene-rich areas, the low p-values indicate that no significant correlation exists between the two variables in either test (gene density: r = −0.028; p = 0.472; GC content: r = −0.065; p = 0.095).
Chimpanzee-Specific ARMD Polymorphism
In order to estimate the polymorphism rates in chimpanzees, we analyzed and amplified a total of 50 chimpanzee-specific ARMD loci on a panel composed of genomic DNA from 12 unrelated chimpanzee individuals (see Materials and Methods). Our results show that the polymorphism level of chimpanzee-specific ARMDs (28%) is about two times higher than the polymorphism rate of human-specific ARMD events (15%) [10], which is in general agreement with the polymorphism levels from previous studies of chimpanzee- or human-specific retrotransposons (e.g., Alu and L1 elements) [28,29].
Incomplete Lineage Sorting and Parallel Independent ARMDs
About 32% of the ARMD candidates were found to have ambiguous TSD structures and a triple alignment that proved too complex to assign ARMD status to the locus solely on the basis of our computational output. These loci were verified experimentally using PCR (see Materials and Methods) to determine the authenticity of the chimpanzee-specific ARMDs and identify false positives in the computational data, which were usually caused by human-specific Alu insertions. However, 16 ambiguous loci were identified at which human-specific Alu insertions were not present. In 11 of these loci, the human and gorilla genomes appear to have two Alu elements, while the chimpanzee and orangutan genomes have only one element at the orthologous position. DNA sequence analysis of the PCR products classified five of these 11 loci as chimpanzee-specific ARMDs, with the second of the two recombining Alu elements having integrated into the host genome after the divergence of orangutan and the common ancestor of humans, chimpanzees, and gorillas (Figure 5A). Four out of the 11 loci show a pattern consistent with incomplete lineage sorting, in which the ARMD event occurred before the divergence of great apes and was still polymorphic at the time of speciation. Subsequently, the chimeric Alu elements produced by these ARMD events became fixed in the chimpanzee and orangutan lineages while the two original Alu elements involved in the ARMDs were fixed in the human and gorilla genomes (Figure 5B). Incomplete lineage sorting has been reported in cases of retrotransposon insertion polymorphism involving closely related species [28,30]. In cases where the time between any genomic event and a subsequent speciation is very short, incomplete lineage sorting can easily occur. The remaining two of the 11 ambiguous loci were identified as parallel independent ARMD events in separate primate genomes by aligning the pre-recombination sequence and chimeric Alu elements (Figure 5C). These events suggest that orthologous loci may experience two independent lineage-specific ARMDs at different times (i.e., chimpanzee-specific ARMDs and orangutan-specific ARMDs).
Figure 5. Incomplete Lineage Sorting and Parallel Independent ARMD Events.
The DNA template used in each reaction is listed on top of the gel chromatograph (M, 100-bp ladder; H, human; C, chimpanzee; G, gorilla; O, orangutan). The large and small sizes of PCR products indicate two Alu elements and one Alu element, respectively. The thunderbolts represent recombination events between two Alu elements, causing ARMDs. Possible scenarios that explain the observed chromatograph: (A) chimpanzee-specific ARMDs, (B) incomplete lineage sorting of an ARMD event, and (C) parallel independent ARMD events.
In contrast, PCR analysis of the remaining five ambiguous loci (from the 16 referred to above) showed that humans and orangutans have two Alu elements, whereas chimpanzees and gorillas have only one at the orthologous position. Of these five loci, three showed a pattern suggesting incomplete lineage sorting events, while the other two were parallel independent ARMDs. For one of the loci displaying a parallel independent ARMD event, the structural characteristics of the two chimeric Alu elements resulting from independent recombination events are clearly different between the chimpanzee and gorilla genomes. The 574-bp chimpanzee genomic deletion occurred between the left monomer on the first Alu and the right monomer on the second Alu, whereas the 708-bp genomic deletion in the gorilla happened between the two left monomers of the two Alu elements.
These results indicate that at least ∼0.9% of chimpanzee-specific ARMD loci (2 of 233 loci which were analyzed by PCR) are shared by the gorilla genome and another ∼0.9% are shared by the orangutan genome, due to parallel independent ARMDs at two different time points in two separate primate genomes. As such, the presence of independently occurring ARMD events in both the human and chimpanzee genomes could lead to false negative events being missed during the previous analysis done by Sen et al. [10], although the frequency of such false negatives is likely to be very low. In addition, we believe that the human orthologs of the chimpanzee-specific ARMD loci represent sites predisposed for potential future ARMDs in the human genome that could generate human lineage-specific rearrangements and genetic disorders. Identifying putative ARMD hotspot genomic regions is not surprising based upon the frequency of Alu-mediated recombination events that have given rise to mutations in a number of different loci, including the LDLR and MLL1 genes [11,31–33].
Discussion
Differential Level of Lineage-Specific ARMD Events
Despite the high level of overall similarity between their genomes, humans and chimpanzees have subtly different genomic landscapes because of alterations such as insertions, deletions, inversions, and duplications after their divergence from a common ancestral primate [8–11,34,35]. Although from a mechanistic viewpoint, the chimpanzee-specific ARMD events are similar to the human-specific ones, the total number and size of deletions are substantially different between the two lineages. One reason for the observed differences between these two lineage-specific ARMD patterns may be the increased genetic diversity of the chimpanzee population as compared to the human population, which is known to have experienced a significant reduction in its effective population size after the divergence of humans and chimpanzees [36], leading to a consequent reduction in genetic diversity. These results are supported by the higher polymorphism level for chimpanzee-specific ARMDs than human-specific ARMDs.
Balance of Chimpanzee Genome Size
Alu elements as well as other retrotransposons can contribute to the size expansion of primate genomes by increasing their copy numbers and causing homology-mediated segmental duplications [37–39]. However, the retrotransposon-mediated increase in genome size is not unilateral, because several processes such as retrotransposon-mediated deletions and recombination-mediated deletions concurrently act in the opposite direction, causing reduction in genome size as well [8–10]. Retrotransposon-mediated negative control of genome size has been well documented in plants such as Arabidopsis and rice [40,41].
In this study, we analyzed the contribution of ARMDs to genome size regulation in the chimpanzee genome by estimating an Alu-mediated sequence turnover rate, which is the amount of sequence increase caused by chimpanzee-specific Alu insertions relative to the amount of reduction by the chimpanzee-specific ARMD process. The copy number of chimpanzee-specific Alu elements (i.e., those that inserted after the divergence of human and chimpanzee) is ∼2,340, accounting for ∼700 kb of inserted sequence in the chimpanzee lineage [3], while the amount of sequence deleted by chimpanzee-specific ARMDs is ∼771 kb. Therefore, within the past ∼6 million y, the genome size of chimpanzees has not expanded but rather has contracted by ∼71 kb, when considering the combined effects of Alu retrotransposition and recombination-mediated deletion (i.e., the Alu-mediated sequence turnover rate is more than 100% in the chimpanzee genome). This observation suggests that ARMD events efficiently counteract genomic expansion caused by novel Alu inserts in the chimpanzee genome when compared to the human genome. A previous analysis of human-specific ARMD events indicates that the Alu-mediated sequence turnover rate is ∼20% in the human genome [10]. This significantly different turnover rate between the two species could be explained by differences in the tempo of Alu amplification (i.e., higher Alu retrotransposition activity in the human genome) and rates of ARMD events (i.e., higher ARMD activity in the chimpanzee genome). Ultimately, it is worth noting that at least in the chimpanzee lineage, concurrent Alu insertion/ARMD mechanisms have balanced the gain and loss of sequences during Alu-mediated genomic alterations.
Retrotransposition of Chimeric Alu Elements
To investigate whether chimeric Alu elements are able to retrotranspose in the chimpanzee genome, we tried to find progeny of the 663 chimpanzee-specific chimeric Alu elements using the BLAST-Like Alignment Tool (BLAT) program (http://genome.ucsc.edu/cgi-bin/hgBlat). However, we failed to recover any such elements in the chimpanzee genome for one or more of a number of reasons. First, Alu elements involved in ARMD events are expected to be relatively old (i.e., more than 6 million y) because our comparative analysis detects only ARMD events involving Alu elements that were inserted into the genome before the divergence of humans and chimpanzees. Therefore, most of the ARMD-associated Alu elements probably lost their ability to retrotranspose before the Alu–Alu recombination process. In reality, the contribution of chimpanzee-specific young Alu elements to the ARMD process may be extremely limited due to their low copy number (∼2,000 copies) in the chimpanzee genome [3]. Indeed, ARMD events generated by the relatively young AluY subfamilies account for 0.19% of the total AluY elements in the chimpanzee genome. Second, only a few source genes are responsible for new Alu subfamily amplification through retrotransposition. Although some Alu subfamilies (e.g., AluYc1) are still active in the chimpanzee genome [3,29], it is improbable that their source gene(s) are involved in the Alu–Alu recombination events. Similarly during an earlier analysis [10], we investigated the retrotransposition ability of 492 human-specific ARMD-generated chimeric Alu elements and were unable to recover their progeny as well.
ARMD as an Endogenous Process Affecting Human and Chimpanzee Variation
Recently, the genomic relationship and genetic divergence between the human and chimpanzee genomes have been the subjects of extensive comparative genomic analyses on the basis of their respective draft genome sequences [3,35,42–44]. However, these studies have not focused on Alu-mediated genomic deletions in the chimpanzee lineage, aside from the 14 Alu retrotransposition-mediated deletions reported previously [9].
Thus, our study forms the first comprehensive analysis of recombination-mediated genomic alteration by Alu elements in a nonhuman primate (chimpanzee) lineage. We found 305 chimpanzee-specific deletions within protein-coding genes as annotated by the RefSeq gene annotation database, 299 genes from which introns were deleted, and six genes in which thirteen exons were deleted. Remarkably, two chimpanzee-specific ARMD events deleted exons from genes demonstrably functional in the human lineage (NBR2 and HTR3D), providing direct proof that the ARMD process contributes to creating phenotypic differences between humans and chimpanzees. The NBR2 gene is located near the BRCA1 gene on Chromosome 17, which is responsible for tumor repressor activity in the human genome, and shares a common promoter for transcription, forming a bidirectional transcriptional unit with BRCA1. Although the complete NBR2 cDNA sequence is ∼1.3 kb, it has a short open reading frame (112 amino acids), and is subject to nonsense-mediated decay [45,46]. In humans, this gene is suppressed by a non–tissue-specific protein complex that binds to its first intron (i.e., the 18-bp repressor element) [47]. However, in the chimpanzee lineage, an ARMD event occurred between the third intron and the 3′ flanking region, causing an exonic deletion (Figure 6A). Thus, this ARMD event could potentially inhibit NBR2 gene expression in the chimpanzee genome, regardless of whether or not the repressor element is present. Although the exonic deletion of the NBR2 gene has been independently reported through a comparative analysis of cancer genes between the human and chimpanzee genomes, the previous analysis did not report what caused this genetic difference between human and chimpanzee genomes [48]. Our study of chimpanzee-specific ARMDs illuminates the underlying molecular mechanism for this deletion.
Figure 6. Exonic Deletions Caused by Two ARMD Events.
Black arrows represent the direction of transcription, and gray and black boxes indicate the noncoding exons and coding exons, respectively. Green and purple arrows indicate elements from two different Alu subfamilies, and dual-color arrows indicate chimeric Alus generated by ARMD events (map is not drawn to scale).
(A) An exonic deletion within the NBR2 gene. The AluSg and AluY elements are located within the third intron and the 3′ flanking sequence, respectively, in the human genome. The exon4 sequence is deleted due to an ARMD event in the chimpanzee lineage.
(B) An exonic deletion within the HTR3D gene. The AluSx and AluSq elements are located within the second and third introns, respectively, in the human genome. The exon3 sequence, which includes the initiation codon ATG, is deleted due to an ARMD event in the chimpanzee lineage.
A chimpanzee-specific ARMD event also deleted the first coding exon of HTR3D, a functional gene in humans (Figure 6B). This gene belongs to the 5-HT3 serotonin receptor-like gene family, which has been recently characterized [49]. The 5-HT3D subunit is not a functional receptor on its own (i.e., a homomeric receptor), but when it binds to the 5-HT3A subunit to form the heteroligomeric receptor, 5-HT, maximum response is significantly increased as compared to the homomeric 5-HT3A receptor [50]. HTR3D is primarily expressed in the gastrointestinal tract [50], where serotonin is synthesized extensively [51]. We speculate that the exonic deletion in this gene caused by the chimpanzee-specific ARMD event may lead to a reduction in serotonin levels in the chimpanzee lineage, and thus have an impact on physiological variation between the human and chimpanzee lineages.
The analyses using the RefSeq and UniGene annotations (see Results) indicate that ARMD events could have affected the expression of many genes. Moreover, intronic or intergenic deletions caused by ARMD events may also affect the levels of gene expression in both the human and chimpanzee genomes through alteration of splicing patterns and loss of transcription factor binding sites, further contributing to the divergence of the human and chimpanzee lineages. Additional studies of the functional genomics of the genes altered in both human and chimpanzee ARMD events will be instructive and provide new insight into the genetic and phenotypic differences between the two species.
Conclusion
Retrotransposon-mediated genomic rearrangement could be one of the major factors responsible for the lineage-specific changes in genomes that ultimately lead to speciation. Comparative investigations of the ARMD events apparent between the human and chimpanzee genomes indicate that this process plays an important role in the biological differences between humans and chimpanzees, and provides a reliable record of lineage-specific evolutionary histories due to the nearly homoplasy-free nature of these mutations. Moreover, in the chimpanzee lineage, the chimpanzee-specific ARMD process has completely counteracted the genomic expansion caused by new Alu inserts since the divergence of the chimpanzee and human lineages. The existence of parallel independent ARMD events found at the orthologous loci of some of the 663 chimpanzee-specific ARMD events suggest that other chimpanzee-specific ARMD orthologs in humans may be predisposed to undergo recombination between the two Alu elements in the future. These ARMD orthologous loci may be sites of unstable structure in humans as well as other apes, because they still preserve the pre-recombination structure that has proven itself susceptible to unequal recombination in the chimpanzee lineage.
Materials and Methods
Computational search and manual inspection of chimpanzee-specific ARMD loci.
To computationally screen the chimpanzee genome for potential ARMD loci, we used a technique previously described by Sen et al. [10] in a study of human lineage-specific ARMD events, with the distinction that, for this analysis, the query and target genomes were reversed. In summary, we extracted 400 bp of 5′ and 3′ flanking sequence for all chimpanzee Alu elements (PanTro1; November 2003 freeze) and joined the two 400 bp sequences to form a single “query” sequence. A best match for each query sequence was determined by using BLAT [52] against the reference human genome (hg17; May 2004 freeze). Then, the sequence in the human genome (the “hit”) found between the orthologs of the two 400 bp stretches of the query was extracted and aligned with the chimpanzee Alu element sequence initially used to design the query (the “query Alu”) using a local installation of the NCBI bl2seq utility.
One hallmark of de novo Alu insertion is the presence of TSDs flanking each side of the Alu element, generated by the target-site primed reverse transcription process [1,53–55]. However, the single chimeric Alu element created by an ARMD event lacks matching TSD structures in the chimpanzee because it is comprised of fragments from a pair of Alu elements with mutually unique TSDs at the orthologous ancestral locus [10]. If a potential ARMD locus exhibited the structures of a valid ARMD as described by Sen et al. [10], we accepted the computational detection as an authentic ARMD locus. In addition, we used the BLAT software utility [52] to compare the human, chimpanzee, and rhesus macaque genomes at each potential ARMD locus. If the two Alu elements in the human genome that are considered to be the pre-recombination Alu elements for an ARMD locus are shared with the rhesus macaque genome at orthologous loci, despite the presence or absence of TSDs, the single Alu element remaining at the orthologous chimpanzee locus is most likely a chimeric element generated an ARMD event. On the basis of these features, we manually inspected 1,538 potential ARMD loci retrieved by the computational data analysis. However, some loci displayed ambiguous TSD structure or remained ambiguous after analysis using the triple alignment. These loci were subjected to PCR analysis and, if necessary, DNA sequencing in order to confirm or eliminate each as being products of bona fide ARMD events.
PCR amplification and DNA sequence analysis.
PCR analysis was performed using four different primate species as templates. The cell lines used to isolate DNA samples corresponding the primate species are as follows: human (Homo sapiens) HeLa (CCL2; American Type Culture Collection [ATCC], http://atcc.org), common chimpanzee “Clint” (Pan troglodytes; NS06006B), gorilla (Gorilla gorilla; AG05251) and orangutan (Pongo pygmaeus; AG05252A). To evaluate polymorphism rates, we amplified 50 randomly selected ARMD loci on a common chimpanzee population panel composed of 12 unrelated individuals of unknown geographic origin obtained from the Southwest Foundation for Biomedical Research (San Antonio, Texas, United States).
Oligonucleotide primers for the PCR amplification of ARMD events were designed using the Primer3 utility (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The sequences of the oligonucleotide primers, annealing temperatures, and PCR product sizes are shown in Table S2. Each PCR amplification was performed in 25-μl reactions using 10–50 ng DNA, 200 nM of each oligonucleotide primer, 200 μM dNTPs in 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.4), and 2.5 U Taq DNA polymerase. Each sample was subjected to an initial denaturation step of 5 min at 95 °C, followed by 35 cycles of PCR at 1 min of denaturation at 95 °C, 1 min at the annealing temperature, and 1 min of extension at 72 °C, followed by a final extension step of 10 min at 72 °C. PCR amplicons were loaded on 1%–2% agarose gels, depending on the amplicon sizes, stained with ethidium bromide, and visualized using UV fluorescence. In cases where the expected size of the PCR product was greater than 1.5 kb, iTaq (Bio-Rad, http://www.bio-rad.com) or Ex Taq polymerase (TaKaRa, http://www.takara-bio.com) were used, following the manufacturer's suggested protocols.
When necessary, individual PCR amplicons were gel purified using the Wizard gel purification kit (Promega, http://www.promega.com) and cloned into vectors using the TOPO-TA Cloning kit (Invitrogen, http://www.invitrogen.com) according to the manufacturer's instructions. DNA sequencing was performed using dideoxy chain-termination sequencing [56] on an Applied Biosystems ABI3130XL automated DNA sequencer (Applied Biosystems, http://www.appliedbiosystems.com). Raw sequence reads were assembled using DNASTAR's Seqman program in the Lasergene version 5.0 software package (http://www.dnastar.com).
Analysis of flanking sequences.
For each chimpanzee-specific ARMD locus, 10 kb of flanking sequence upstream and downstream were collected using a combination of in-house Perl scripts and the nibFrag utility bundled with the BLAT software package. The GC content of the flanking regions of each ARMD locus was calculated by analyzing the combined 20 kb of flanking sequence using another in-house Perl script, which excluded Ns from the analysis. Gene density around individual ARMD loci was estimated using the NCBI Map Viewer utility, run on Build 2.1 of the Pan troglodytes genome (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=9598). The neighboring 2 Mb of sequence 5′ and 3′ to each chimeric chimpanzee Alu element was analyzed, and the number of genes found within this combined 4 Mb were noted. All computer programs used are available from the authors upon request.
Supporting Information
(2.2 MB TXT)
The chimeric chimpanzee Alu sequence is shown at the top. The sequences of the intact human AluSx and AluJb involved in the ARMD events are shown below. The dots below represent the same nucleotides as the chimeric chimpanzee Alu sequence, and the dashes represent the gaps. A yellow box on the sequences denotes the recombination window.
(49 KB DOC)
(41 KB XLS)
(69 KB XLS)
Accession Numbers
The gorilla and orangutan DNA sequences generated during the course of this study have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/Genbank) under accession numbers EF682150–EF682182. The GenBank accession numbers for the three HTR3D isforms discussed in this article are NM_182537, BC101090, and AJ437318.
Acknowledgments
We thank Dr. J. Kim for his useful comments during preparation of the manuscript and L. Song for technical assistance. We are especially grateful to J. A. Walker for her help throughout this project.
Abbreviations
- ARMD
Alu recombination-mediated deletion
- Mya
million y ago
- TSD
target site duplication
Footnotes
A previous version of this article appeared as an Early Online Release on September 10, 2007 (doi:10.1371/journal.pgen.0030184.eor).
Author contributions. KH and JL conceived and designed the experiments. KH, JL, and DS performed the experiments. JW, TJM, and PL performed the computational analysis. KH, JL, TJM, SKS, DS, and MAB analyzed the data. PL and MAB contributed reagents/materials/analysis tools. KH, JL, and MAB wrote the paper.
Funding. This research was supported by National Science Foundation BCS-0218338 (MAB) and EPS-0346411 (MAB), National Institutes of Health RO1 GM59290 (MAB), R03 CA101515 (PL), and P30 CA16056 (Roswell Park Cancer Institute), and the State of Louisiana Board of Regents Support Fund (MAB).
Competing interests. The authors have declared that no competing interests exist.
References
- Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Quentin Y. Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res. 1992;20:487–493. doi: 10.1093/nar/20.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Han K, Sen SK, Wang J, Callinan PA, Lee J, et al. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33:4040–4052. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Wang J, Herke SW, Garber RK, Liang P, et al. Alu retrotransposition-mediated deletion. J Mol Biol. 2005;348:791–800. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- Sen SK, Han K, Wang J, Lee J, Wang H, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Baron WF, Stout DB, Davidson EH. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci U S A. 1988;85:4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagel V, Flemington E, Traina-Dorge V, Bradshaw H, Deininger P. Clustering and subfamily relationships of the Alu family in the human genome. Mol Biol Evol. 1987;4:19–29. doi: 10.1093/oxfordjournals.molbev.a040422. [DOI] [PubMed] [Google Scholar]
- Jurka J, Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 1988;85:4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C, Maraia R. Transcriptional regulation and transpositional selection of active SINE sequences. Curr Opin Genet Dev. 1992;2:874–882. doi: 10.1016/s0959-437x(05)80110-8. [DOI] [PubMed] [Google Scholar]
- Hackenberg M, Bernaola-Galvan P, Carpena P, Oliver JL. The biased distribution of Alus in human isochores might be driven by recombination. J Mol Evol. 2005;60:365–377. doi: 10.1007/s00239-004-0197-2. [DOI] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, et al. Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet. 1994;3:223–228. doi: 10.1093/hmg/3.2.223. [DOI] [PubMed] [Google Scholar]
- Wetterbom A, Sevov M, Cavelier L, Bergstrom TF. Comparative genomic analysis of human and chimpanzee indicates a key role for indels in primate evolution. J Mol Evol. 2006;63:682–690. doi: 10.1007/s00239-006-0045-7. [DOI] [PubMed] [Google Scholar]
- Price AL, Eskin E, Pevzner PA. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14:2245–2252. doi: 10.1101/gr.2693004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger NS, Gregersen N, Kielland-Brandt MC. One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res. 1995;23:256–260. doi: 10.1093/nar/23.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl FW. Special sites in generalized recombination. Annu Rev Genet. 1979;13:7–24. doi: 10.1146/annurev.ge.13.120179.000255. [DOI] [PubMed] [Google Scholar]
- Xing J, Hedges DJ, Han K, Wang H, Cordaux R, et al. Alu element mutation spectra: Molecular clocks and the effect of DNA methylation. J Mol Biol. 2004;344:675–682. doi: 10.1016/j.jmb.2004.09.058. [DOI] [PubMed] [Google Scholar]
- Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Bernardo Carvalho A, Clark AG. Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol. 2001;18:1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- Lee J, Cordaux R, Han K, Wang J, Hedges DJ, et al. Different evolutionary fates of recently integrated human and chimpanzee LINE-1 retrotransposons. Gene. 2007;390:18–27. doi: 10.1016/j.gene.2006.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, et al. Differential alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Xing J, Salem AH, Batzer MA. SINEs of a nearly perfect character. Syst Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- Hess JL. MLL: A histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Purandare SM, Patel PI. Recombination hot spots and human disease. Genome Res. 1997;7:773–786. doi: 10.1101/gr.7.8.773. [DOI] [PubMed] [Google Scholar]
- Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, et al. Mutation in LDL receptor: Alu–Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhao S, Bailey JA, Sahinalp SC, Alkan C, et al. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–368. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA. Evolution of genome size: New approaches to an old problem. Trends Genet. 2001;17:23–28. doi: 10.1016/s0168-9525(00)02157-0. [DOI] [PubMed] [Google Scholar]
- Devos KM, Brown JK, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis . Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, MacDonald JR, Tang T, Carson AR, Li M, et al. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 2005;1:e56. doi: 10.1371/journal.pgen.0010056. doi: 10.1371/journal.pgen.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, et al. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Selfe J, Whitehouse C, Morris JR, Solomon E, et al. Structural evolution of the BRCA1 genomic region in primates. Genomics. 2004;84:1071–1082. doi: 10.1016/j.ygeno.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Xu CF, Brown MA, Nicolai H, Chambers JA, Griffiths BL, et al. Isolation and characterisation of the NBR2 gene which lies head to head with the human BRCA1 gene. Hum Mol Genet. 1997;6:1057–1062. doi: 10.1093/hmg/6.7.1057. [DOI] [PubMed] [Google Scholar]
- Suen TC, Tang MS, Goss PE. Model of transcriptional regulation of the BRCA1-NBR2 bi-directional transcriptional unit. Biochim Biophys Acta. 2005;1728:126–134. doi: 10.1016/j.bbaexp.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Puente XS, Velasco G, Gutierrez-Fernandez A, Bertranpetit J, King MC, et al. Comparative analysis of cancer genes in the human and chimpanzee genomes. BMC Genomics. 2006;7:15. doi: 10.1186/1471-2164-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesler B, Frank B, Kapeller J, Rappold GA. Cloning, physical mapping and expression analysis of the human 5-HT3 serotonin receptor-like genes HTR3C, HTR3D and HTR3E . Gene. 2003;310:101–111. doi: 10.1016/s0378-1119(03)00503-1. [DOI] [PubMed] [Google Scholar]
- Niesler B, Walstab J, Combrink S, Moeller D, Kapeller J, et al. Characterization of the novel human serotonin receptor subunits 5-HT3C, 5- HT3D, and 5-HT3E. Mol Pharmacol. 2007;72:8–17. doi: 10.1124/mol.106.032144. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Hasegawa H, Kaneko E, Ichiyama A. Gastrointestinal serotonin: Depletion due to tetrahydrobiopterin deficiency induced by 2,4-diamino-6-hydroxypyrimidine administration. J Pharmacol Exp Ther. 1991;256:773–779. [PubMed] [Google Scholar]
- Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(2.2 MB TXT)
The chimeric chimpanzee Alu sequence is shown at the top. The sequences of the intact human AluSx and AluJb involved in the ARMD events are shown below. The dots below represent the same nucleotides as the chimeric chimpanzee Alu sequence, and the dashes represent the gaps. A yellow box on the sequences denotes the recombination window.
(49 KB DOC)
(41 KB XLS)
(69 KB XLS)