Abstract
The filamentous, heterocystous cyanobacterium Nostoc sp. strain PCC 7120 (Anabaena sp. strain PCC 7120) possesses an uptake hydrogenase and a bidirectional enzyme, the latter being capable of catalyzing both H2 production and evolution. The completely sequenced genome of Nostoc sp. strain PCC 7120 reveals that the five structural genes encoding the bidirectional hydrogenase (hoxEFUYH) are separated in two clusters at a distance of approximately 8.8 kb. The transcription of the hox genes was examined under nitrogen-fixing conditions, and the results demonstrate that the cluster containing hoxE and hoxF can be transcribed as one polycistronic unit together with the open reading frame alr0750. The second cluster, containing hoxU, hoxY, and hoxH, is transcribed together with alr0763 and alr0765, located between the hox genes. Moreover, alr0760 and alr0761 form an additional larger operon. Nevertheless, Northern blot hybridizations revealed a rather complex transcription pattern in which the different hox genes are expressed differently. Transcriptional start points (TSPs) were identified 66 and 57 bp upstream from the start codon of alr0750 and hoxU, respectively. The transcriptions of the two clusters containing the hox genes are both induced under anaerobic conditions concomitantly with the induction of a higher level of hydrogenase activity. An additional TSP, within the annotated alr0760, 244 bp downstream from the suggested translation start codon, was identified. Electrophoretic mobility shift assays with purified LexA from Nostoc sp. strain PCC 7120 demonstrated specific interactions between the transcriptional regulator and both hox promoter regions. However, when LexA from Synechocystis sp. strain PCC 6803 was used, the purified protein interacted only with the promoter region of the alr0750-hoxE-hoxF operon. A search of the whole Nostoc sp. strain PCC 7120 genome demonstrated the presence of 216 putative LexA binding sites in total, including recA and recF. This indicates that, in addition to the bidirectional hydrogenase gene, a number of other genes, including open reading frames connected to DNA replication, recombination, and repair, may be part of the LexA regulatory network in Nostoc sp. strain PCC 7120.
Cyanobacteria are capable of synthesizing two functionally different hydrogenases: an uptake hydrogenase and a bidirectional enzyme (18, 49). So far, the cyanobacterial uptake hydrogenase has been, in all known cases, found only in nitrogen-fixing strains and has the evident function of recycling the H2 produced by the nitrogen-fixing enzyme, nitrogenase. However, the soluble or loosely membrane-associated bidirectional hydrogenase is an enzyme present in both nitrogen-fixing and non-nitrogen-fixing cyanobacteria (49), and it can catalyze both H2 uptake and evolution.
The physiological role of the cyanobacterial bidirectional hydrogenase is not fully understood, and several functions have been proposed. It was thought to function predominantly in anaerobic or microaerobic environments catalyzing an uptake of hydrogen produced by other microorganisms (19, 20). It was also proposed as a mediator in the release of excess of reducing equivalents during fermentative growth (47, 51). However, the low Km for H2 (2.3 μM) suggests that the enzyme normally operates in the uptake direction (19, 20), and as the proton gradient is directed outward in cyanobacteria, the enzyme might reside at the periplasmic face of the cytoplasmic membrane and allocate electrons to the respiratory chain (22, 42). The coupling of the bidirectional hydrogenase activity to the respiratory chain has also been emphasized by the homology to subunits of the NADH-ubiquinone oxidoreductase (complex I) (2, 10, 39). In cyanobacteria, only an incomplete version of complex I, containing 11 out of the 14 subunits that are strictly conserved in other prokaryotes like Escherichia coli, can be found. However, studies of respiration in Nostoc punctiforme, a species naturally lacking the bidirectional hydrogenase, demonstrated comparable rates to cyanobacteria having the enzyme (8). In addition, hoxU mutants of Synechococcus sp. strain PCC 6301 (10) and hoxEF mutants of Synechocystis sp. strain PCC 6803 showed nonimpaired respiratory O2 uptake while being affected in H2 evolution (21). Several strains naturally lack the bidirectional hydrogenase completely, and it seems that it does not play an essential role in strains in which it is present, since inactivation of hoxH in both Synechocystis sp. strain PCC 6803 and Nostoc sp. strain PCC 7120 resulted in only a small decrease in the growth rate compared to that of the wild type (3, 26). In the unicellular, non-nitrogen-fixing Synechocystis sp. strain PCC 6803, this enzyme has been suggested to act as an electron valve for low-potential electrons generated during the light reaction of photosynthesis, thus preventing a slowing down of electron transport (3).
Five subunits have been demonstrated to form the bidirectional hydrogenase in cyanobacteria (41, 42). The enzyme consists of a diaphorase part, encoded by hoxEFU, and a hydrogenase part, encoded by hoxYH. The physical organizations of the structural genes encoding the bidirectional hydrogenase have strong similarities among different strains. In Anabaena variabilis ATCC 29413, Nostoc sp. strain PCC 7120, Synechococcus sp. strain PCC 6301, and Synechocystis sp. strain PCC 6803, one or several additional open reading frames (ORFs) between some of the structural genes have been identified. The intergenic region between hoxF and hoxU in Synechococcus sp. strain PCC 6301 and that in Nostoc sp. strain PCC 7120 are longer (333 kb and 8.8 kb, respectively) than those in other strains (49). Transcriptional studies using reverse transcription-PCR (RT-PCR) indicate that the structural hox genes form a single transcript together with two ORFs of unknown function in Anabaena variabilis ATCC 29413 (9) and forms a single transcript together with three additional ORFs in Synechocystis sp. strain PCC 6803 (30). In contrast, the hox genes in the unicellular Synechococcus sp. strain PCC 6301 are located on two different transcripts. hoxEF forms one transcript, and hoxUYH is part of a second transcript together with hoxW, hypA, and hypB (9). In Synechococcus sp. strain PCC 7942, hoxEF and hoxUYHW are located on two different transcripts (40). hoxUYHW may be polycistronic, with a second promoter located between hoxH and hoxW (40). The differences in structural gene organization and transcription units may imply a difference in transcriptional regulation between different strains.
The activity of the bidirectional hydrogenase has been investigated previously for both unicellular and filamentous cyanobacteria. In several studies, this activity was demonstrated to be induced under anaerobic conditions (19, 20, 39, 44). The bidirectional hydrogenase in Nostoc sp. strain PCC 7120 is present and active in both vegetative cells and heterocysts in aerobically grown filaments. When the cells were transferred to anaerobic conditions, the level of activity of the bidirectional hydrogenase increased by about two orders of magnitude, with roughly equal specific activities in both cell types (19, 20). Similar results were observed for Anabaena variabilis ATCC 29413 (44). However, the activity of the bidirectional hydrogenase in the unicellular Gloeocapsa alpicola and Chroococcidiopsis thermalis is not directly dependent on oxygen (45, 51). Relatively little is known about the transcriptional regulation of the bidirectional hydrogenase in cyanobacteria. The relative abundances of hoxY and hoxH transcript levels in Gleocapsa alpicola did not change significantly under nitrogen-limiting conditions (46), while the activity of the enzyme increased considerably (46, 51). The hoxH transcript level was not affected by a shift from ammonium-grown cells to N2-fixing conditions or the addition of hydrogen for Nostoc muscorum (6, 7).
Recently, it was demonstrated that the transcription factor LexA interacts with the regulatory promoter region of the hox operon in Synechocystis sp. strain PCC 6803 (30), suggested to function as a transcription activator (16). Large-scale analyses demonstrated that the transcriptions of many genes are affected in a Synechocystis sp. strain PCC 6803 lexA-depleted mutant, i.e., it was possible to identify numerous genes whose expression was either activated or repressed in response to LexA depletion (14). In addition, Synechocystis sp. strain PCC 6803 LexA was recently suggested to function as a general regulator of redox-responsive gene expression (1, 33), a concept that discards the classical model of LexA as a DNA binding protein directly involved in the SOS response.
In the present study, the transcription of the structural genes encoding the bidirectional hydrogenase, the hox genes, in the filamentous, heterocystous cyanobacterium Nostoc sp. strain PCC 7120 was investigated. The regulation of the hox operons was examined during a transfer from aerobic conditions to anaerobic conditions. In addition, transcription start sites have been determined, and specific interactions between the transcriptional regulator LexA and the hox promoter regions have been observed.
MATERIALS AND METHODS
Organisms and growth conditions.
The filamentous heterocystous cyanobacterium Nostoc sp. strain PCC 7120 (= Anabaena sp. strain PCC 7120) was routinely grown in BG110 liquid medium (37) at 23°C under a continuous illumination of 40 μmol of photons m−2 s−1 and by sparging with air. Dark, anaerobic conditions were achieved by replacing the air with 100% argon and covering the culture with aluminum foil. All cultures were mixed with a magnetic stirrer to obtain a homogeneous suspension. Escherichia coli strains were grown in LB medium or on agar plates at 37°C.
Nucleic acid extraction.
Genomic DNA was isolated from Nostoc sp. strain PCC 7120 cultures grown with air. Cells were harvested (4,500 × g for 10 min) at room temperature, and DNA was extracted as described previously (48) and resuspended in water. Plasmid DNA was isolated from E. coli by using the GenElute plasmid miniprep kit (Sigma-Aldrich). For the isolation of total RNA, Nostoc sp. strain PCC 7120 cells were harvested at room temperature and washed once with 2 ml RNAlater (Ambion) followed by centrifugation at 20,000 × g for 60 seconds. The cells were disrupted in a Fast-Prep FP 120 BIO 101 homogenizer (Savant) at full speed for 1 min together with 0.6 g acid-washed glass beads (0.6-mm diameter) and 1 ml TRI Reagent (Molecular Research Center, Inc.), followed by a quick cooling on ice. After incubation at room temperature for 8 min, the suspension was transferred to a 2-ml Phase Lock Gel tube (Eppendorf) to which 100 μl BCP (1-bromo-3-chloropropane) and 100 μl water were added. Phases were separated by centrifugation at 12,000 × g for 5 min at 4°C. The upper aqueous layer was taken out, and the RNA was precipitated with 250 μl isopropanol and 250 μl RNA precipitation buffer (0.8 M sodium citrate and 1.2 M NaCl) for several hours at −20°C. The RNA was collected by centrifugation at 12,000 × g for 15 min, washed twice with 75% ethanol, and resuspended in RNA resuspension solution (1 mM sodium citrate, pH 6.4). RNA samples were further treated with DNase I according to the instructions of the manufacturer (Fermentas).
Agarose gel electrophoresis, PCR, DNA recovery, and sequencing.
Agarose gel electrophoresis was used to separate and analyze DNA and RNA according to standard procedures (38). PCRs were carried out in a thermal cycler (Gene Amp PCR system 2400; Applied Biosystems) with either Taq DNA polymerase (Fermentas) as described previously (48) or Phusion DNA polymerase (Finnzymes) according to guidelines provided from the supplier. All the oligonucleotides that were used are listed in Table 1. Obtained DNA fragments were isolated from agarose gels with the GFX PCR DNA and Gel Band Purification kit (Amersham Biosciences) according to the manufacturer's instructions. Probes used for electrophoretic mobility shift assay (EMSA) and Northern blotting were analyzed by sequencing before use. Sequencing reactions were performed at Macrogen Inc.
TABLE 1.
Oligonucleotides used in this studya
| Gene, ORF, or vector in indicated expt | Sense primer
|
Antisense primer
|
Size (bp) | Reference/source | ||
|---|---|---|---|---|---|---|
| Name | Sequence (5′→3′) | Name | Sequence (5′→3′) | |||
| RT-PCR and Northern blot analysis | ||||||
| alr0750 | Aalr0750F | AGTTGCATTGGATCGCTCCGATAT | Aalr0750R | CTCTGCCCAAATCATGCACTATATG | 467 | 5 |
| hoxE | AhoxEF | GTGGTGTGTACAGGTACGGC | AhoxER | TTACCTAAGACTTTGCCATC | 188 | 5 |
| hoxF | AhoxFF | ACATTGGCAGACAAGAACGC | AhoxFR | TTGCTGACGCATCTTCAGGC | 270 | 5 |
| alr0760 | Aalr0760F | TACCCTTCCAGTTCTCCAATCTTC | Aalr0760R | ATCTATCAACTTCATCTAAGCCG | 482 | 5 |
| alr0761 | Aalr0761F | ACAGACAATTTCATGTTACAAGC | Aalr0761R | TATCAACTTGGATTTGATATCTAC | 424 | This work |
| hoxU | AhoxUF | ACGACCAACTCATTAGCGC | AhoxUR | AGCCACGCAAACAGAAC | 290 | 5 |
| alr0763 | Aalr0763F | GACAAGCAACAACCAGA | Aalr0763R | CTGGTGCTTCATCTAGGAA | 417 | This work |
| hoxY | AhoxYF | TGAAGTTAGCAACAGTATGG | AhoxYR | AAGACTGATTCAGCACTACC | 307 | This work |
| alr0765 | Aalr0765F | GCCACGAACAAGATGCT | Aalr0765R | TTTCGGCTCGTTGGTGT | 417 | This work |
| hoxH | AhoxHF | GACTATTGCCTTGGATGCT | AhoxHR | GACAGAATATCTGGGTCGTT | 513 | This work |
| rnpB | rnpBF | GACCAAACTTGCTGGATAACG | rnpBR | TTGCGAGGGCAGTTATCTATC | 331 | This work |
| lexA | LexAF | TCGTCAGATGATGCAGGCGATGAA | LexAR | AGTGATGCGATCGCCACTTCGATA | 424 | This work |
| Cloning—lexA | AlexAF2 | GGATCCGAACGCCTAACAGAAGCGCA | AlexAR2 | AAGCTTTCACATATAACCGCGCCACA | 618 | This work |
| AlexAF3 | GAATTCATGGAACGCCTAACAGAAGCG | AlexAR3 | GGATCCTCACATATAACCGCGCCACAC | 618 | This work | |
| Identification of TSPs | ||||||
| alr0750 | Aalr0750R3 | CTCTGCCCAAATCATGCACTATA | This work | |||
| Aalr0750R4 | GTATTACCTGCGGTTGTT | This work | ||||
| hoxU | AhoxUR | AGCCACGCAAACAGAAC | This work | |||
| EMSA | ||||||
| alr0750 fragment | Aalr0750F2 | AAGTAAGCTAGAAGGCGCTTGC | Aalr0750R2 | ATATCGGAGCGATCCAATGCAACT | 499 | This work |
| hoxU fragment | Aalr0761F3 | GCATCTGGTTTAAATGGTTAC | AhoxUR2 | GCGCTAATGAGTTGGTCGT | 535 | This work |
| pQE-30 | pQEF | CCCGAAAAGTGCCACCTG | pQER | GTTCTGAGGTCATTACTGG | 334 | This work |
The underlined base pairs correspond to restriction sites.
Transcription analysis.
RT of 2 μg total RNA extracted from cells grown under dark, anaerobic conditions for 24 h were carried out with the Revert Aid first-strand cDNA synthesis kit (Fermentas), according to the instructions of the manufacturer. The antisense primers used for the cDNA synthesis were AhoxFR and AhoxHR (Table 1). PCR amplification of the hox cluster genes (Fig. 1) were performed using corresponding primers (Table 1). Genomic DNA from Nostoc sp. strain PCC 7120 was used as a positive control. Negative controls included the omission of reverse transcriptase in the RT reaction and a PCR to which no template was added. For Northern blotting, probes were obtained by PCR using Nostoc sp. strain PCC 7120 genomic DNA as the template and primer pairs listed in Table 1. RNA separation, transfer and hybridization were done according to the Hybond-N+ protocol (Amersham Biosciences), making use of probes labeled with [α-32P]dCTP. The even loading of the total RNA aliquots was controlled by verification of equal abundance of the rRNA bands and of the constitutive RNA component of the ribozyme RNase P (52). The relative positions of the isotope were visualized using a BAS-2000II bioimaging analyzer (Fuji Film) or a Pharos FX Plus molecular imager (Bio-Rad).
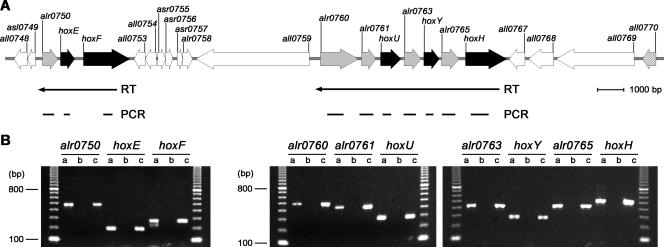
FIG. 1.
(A) Physical map of the hox gene clusters (depicted in black) in Nostoc sp. strain PCC 7120. ORFs present on the same transcript as the hox genes are depicted in gray, and the ORF corresponding to the putative bidirectional-hydrogenase-specific C-terminal endopeptidase, hoxW (all0770), is striped. (B) RT-PCR analysis of the two hox gene clusters. Total RNA was isolated from cells grown under dark, anaerobic conditions for 24 h. The RT reactions were performed with primers against hoxF, followed by PCRs for alr0750, hoxE, hoxF, and hoxH, followed by PCRs for alr0760, alr0761, hoxU, alr0763, hoxY, alr0765, and hoxH (primers are listed in Table 1). Lanes: a, RT-PCR product; b, negative control without reverse transcriptase in the RT reaction prior to the PCR; c, PCR-positive control; marker, 100-base-pair ladder (Amersham Biosciences) where the lowest visible band corresponds to 100 base pairs.
Identification of TSPs.
Transcription start points (TSPs) were localized with the a system for rapid amplification of cDNA ends (5′ RACE; Invitrogen). First-strand cDNA synthesis was performed using 3 μg of total RNA under aerobic and dark, anaerobic conditions together with random hexamer primers (Fermentas). PCR products obtained with the gene-specific primers listed in Table 1 were cloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions before being sequenced.
Cloning of lexA and purification of the gene product.
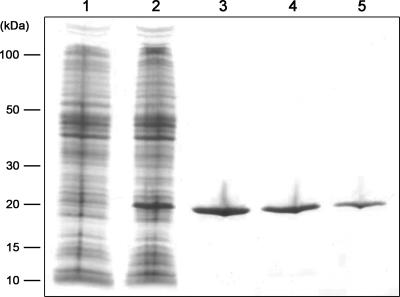
Nostoc sp. strain PCC 7120 lexA was amplified from genomic DNA by using the primers AlexAF2 and AlexAR2 (Table 1). The obtained PCR product was cloned into the pCR 2.1-TOPO (Invitrogen) vector and confirmed by sequencing. The Nostoc sp. strain PCC 7120 lexA was further subcloned into the pQE-30 (QIAGEN) vector by using the restriction enzymes BamHI and HindIII and introduced into M15 (pREP4) cells (QIAGEN). After DNA sequencing confirmed that no mutations had been introduced, Nostoc sp. strain PCC 7120 LexA was overexpressed by induction with IPTG (isopropyl-β-d-thiogalactopyranoside) and purified using Ni-nitrilotriacetic acid Superflow resin (QIAGEN) according to the instructions of the manufacturer. The obtained Nostoc sp. strain PCC 7120 LexA His-tagged protein was analyzed by Coomassie blue staining of a sodium dodecyl sulfate-polyacrylamide gel (see Fig. 4). Synechocystis sp. strain PCC 6803 His-tagged LexA was purified as described previously (30).
FIG. 4.
Overexpression and purification of Nostoc sp. strain PCC 7120 LexA. Lanes: 1, noninduced crude extract; 2, induced crude extract; 3 through 5, eluates of purified Nostoc sp. strain PCC 7120 LexA.
EMSA.
The different DNA fragments were obtained by PCR using Nostoc sp. strain PCC 7120 genomic DNA or pQE-30 vector (QIAGEN) as the template (for details, see Table 1 and Fig. 3). All fragments were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas) as described (31). Various amounts of purified LexA from Nostoc sp. strain PCC 7120 or Synechocystis sp. strain PCC 6803 were incubated with 20 fmol of each fragment alone or together with 20 fmol of labeled unrelated DNA. For binding competition experiments, an excess of either specific or nonspecific unlabeled fragments was included. LexA from Nostoc sp. strain PCC 7120 was used in assays carried out as described previously (28), except that 1 μg of salmon sperm DNA was included in the incubation buffer. Assay mixtures were incubated at 30°C for 30 min before separation on a 6% (wt/vol) nondenaturing polyacrylamide gel at 200 V. The assays performed with Synechocystis sp. strain PCC 6803 LexA were carried out as described previously (30), and the reaction mixtures were separated by electrophoresis on a 10 to 15% (wt/vol) gradient native polyacrylamide gel by using the PhastSystem (Amersham Biosciences). All gels were visualized using a BAS-2000II bioimaging analyzer (Fuji Film).
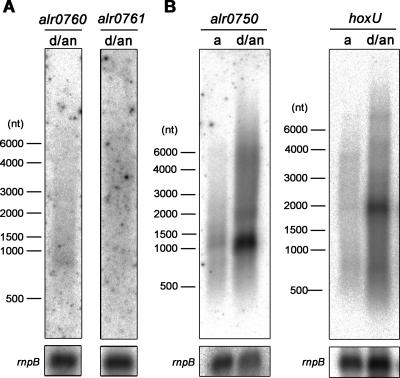
FIG. 3.
Northern blot analysis of alr0760 and alr0761 (A) and alr0750 and hoxU (B) in Nostoc sp. strain PCC 7120 under aerobic (a) and dark, anaerobic (d/an) conditions. Probes for the Northern blot hybridization were obtained by PCR using Nostoc sp. strain PCC 7120 genomic DNA as the template and primer pairs Aalr0760F-Aalr0760R and Aalr0761F-Aalr0761R (A) and Aalr0750F-Aalr0750R and AhoxUF-AhoxUR (B) (see Table 1). Total RNA was isolated from Nostoc sp. strain PCC 7120 cultures grown in air or from cultures induced in dark, anaerobic conditions for 24 h, and a total amount of 10 μg RNA was used for each lane. The rnpB gene was used to standardize the loading and transfer of the RNA. Numbers on the left side of the panels indicate sizes in nucleotides (nt).
In vivo bidirectional hydrogenase activity measurement.
The activity of the bidirectional hydrogenase was assayed by determining the evolution of H2 from methyl viologen reduced by sodium dithionite as described previously (48), using a Clarus 500 gas chromatograph with a Molecular Sieve 5A 60/80 mesh column (Perkin Elmer) and Ar as the carrier gas.
RESULTS
Characterization of the hox transcription units.
The hox genes encoding the bidirectional hydrogenase in the filamentous, nitrogen-fixing cyanobacterium Nostoc sp. strain PCC 7120 (= Anabaena sp. strain PCC 7120) are located in two different gene clusters separated by approximately 8.8 kb (Fig. 1A). To investigate the transcription units of the hox genes, RT reactions were carried out using total RNA extracts from Nostoc sp. strain PCC 7120. The two primers AhoxFR and AhoxHR (Table 1) were used as hoxF- and hoxH-specific antisense primers, respectively, and the resulting cDNA was used as the template in PCR amplifications. From the RT reaction originating from hoxF, it was possible to obtain PCR products for hoxF, hoxE, and the ORF alr0750, suggesting that all three genes are located on at least one single transcript (Fig. 1B). From the RT reaction originating from hoxH, PCR products were obtained from all six upstream genes, including the four ORFs located before and between the hox genes, suggesting a larger transcript harboring alr0760, alr0761, hoxU, alr0763, hoxY, alr0765, and hoxH. However, when using probes against genes in the two putative operons and Northern blot hybridizations, the transcript levels of the two ORFs located upstream from hoxU (alr0760 and alr0761) were significantly lower than those for any other genes (see Fig. 3A, for cells with induced hydrogenase activity, and see below).
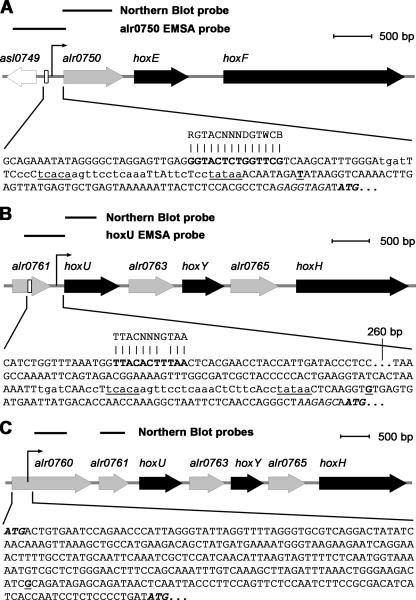
Attempts to identify TSPs upstream from alr0750, alr0760, and hoxU were performed by using the 5′ RACE system. Although no TSP could be found upstream from alr0760, a TSP was identified within the annotated ORF, 244 bp downstream from the translation start codon. Interestingly, it was possible to find additional ORFs within the annotated alr0760 corresponding to shorter versions, since the putative translation start points are found in the same frame as in the original alr0760. One of those additional ORFs starts 321 nucleotides downstream from the annotated ATG start codon and 78 bp downstream from the putative TSP identified in this work by use of 5′ RACE. The N-terminal region of Alr0760, in particular, the first 80 amino acids, shows significantly lower similarity to this TSP than the remaining part of the protein in BLAST searches. This may indicate that the identified TSP could be real and located within a falsely annotated gene. The presence of multiple TSPs also cannot be excluded, and this occurrence has been reported previously, e.g., ntcA of Nostoc sp. strain PCC 7120 shows multiple transcripts with different 5′ ends depending on nitrogen availability (35). However, no significant sequence motifs could be found either upstream from the identified TSP within alr0760 or upstream from the translation start codon of the annotated ORF. TSPs were also identified upstream from alr0750 and hoxU, located 66 bp and 57 bp upstream from the respective start codons. The 5′ RACE experiments were performed using RNA extracted from cells of Nostoc sp. strain PCC 7120 grown aerobically and from cells induced in dark, anaerobic conditions for 24 h with identical results. Furthermore, when analyzing the different promoter regions, it was possible to identify putative sigma70 promoter sequences and ribosomal binding sites upstream from alr0750 and hoxU (Fig. 2). The positions and characters of the −10 and −35 elements show similarity to known promoters from other genes in cyanobacteria (13). Interestingly, the alr0750 and hoxU promoter regions have segments of 40 nucleotides that share a high degree of homology, suggesting a similar regulation of the two different promoter regions. No significant sequence motifs could be found upstream from the identified TSP within the annotated ORF alr0760. By use of Northern hybridizations, accumulations of transcripts of approximately 1,200 and 1,900 nucleotides were observed when using alr0750 and hoxU as probes, respectively (Fig. 3B). The 1,200-nucleotide transcript, starting from the identified TSP, would cover alr0750 and hoxE, ending in the intergenic region between hoxE and hoxF. However, no TSP upstream from hoxF could be identified (5). The 1,900-nucleotide transcript may cover hoxU, alr0763, and part of hoxY.
FIG. 2.
Schematic representation of the Nostoc sp. strain PCC 7120 hox gene clusters and the regulatory promoter regions of the two operons. Positions of the DNA probes used for Northern blot hybridizations (Fig. 3) and DNA fragments used in EMSAs (Fig. 5 and 6) are depicted as lines above the respective maps. The identified TSPs are indicated by arrows and depicted in boldface and underlined in the sequence. Putative −10 and −35 regulating sequences are underlined, and the segments of 40 nucleotides that show a high degree of homology in the two promoter regions of alr0750 and hoxU are indicated with the matching nucleotides shown in lowercase. The ribosomal binding site is shown in italics, and the start codons of hoxE (A), hoxU (B), and alr0760 (C) are shown in boldface and italics. The putative LexA binding sites are indicated with a white box in the map and depicted in boldface in the sequences. In addition, the suggested consensus LexA binding sites (11, 27) are shown above the Nostoc sp. strain PCC 7120 sequences.
Anaerobic induction of the two hox gene clusters.
It has been demonstrated that anaerobic conditions induce higher transcription levels of hoxH transcript in the filamentous, heterocystous Nostoc muscorum and Anabaena variabilis ATCC 29413 (6, 46). In the present study, when cells of Nostoc sp. strain PCC 7120 were transferred to anaerobic conditions and the relative amounts of alr0750 and hoxU transcript were monitored by Northern blot hybridization, significantly higher transcript levels were observed (Fig. 3B) with a simultaneous increase in the enzyme activity (from 3.29 ± 1.75 nmol H2 evolved·h−1·μg chlorophyll a−1 under aerobic conditions to 32.83 ± 2.50 nmol H2 evolved·h−1·μg chlorophyll a−1 under dark, anaerobic conditions), in agreement with earlier physiological studies (19, 20). This also suggests that the regulations of the hox genes, located on two different operons, are similar with respect to O2.
LexA interacts with the promoter regions of the hox genes.
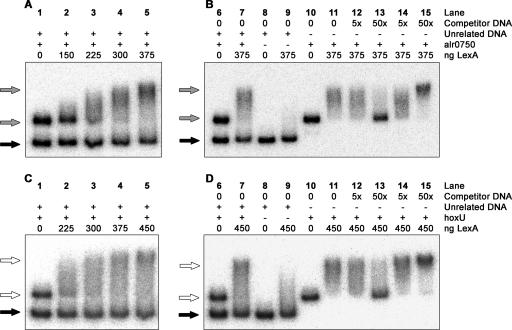
The sequence GGTACTCTGGTTCG found 63 to 76 bp upstream from the TSP of alr0750 (Fig. 2A) corresponds to the previously defined motif RGTACNNNDGTWCB, described as a LexA box in cyanobacteria (27). Recently, LexA was shown to interact with the promoter region of the bidirectional hydrogenase in Synechocystis sp. strain PCC 6803 (30). Additional studies were therefore carried out in order to clarify if this novel role of LexA is specific for Synechocystis sp. strain PCC 6803 or if its role can be broadened to other cyanobacteria. A 499-bp fragment containing part of the upstream region of alr0750 (herein referred to as the alr0750 fragment), harboring the putative binding site, was used in EMSAs with purified His-tagged LexA from Nostoc sp. strain PCC 7120 (Fig. 4). Increasing amounts of LexA produced a retardation which increased in intensity in relation to the amount of protein, while an unrelated DNA fragment produced no retardation (Fig. 5A). To stress the specificity in the binding, competition assays were carried out with either unlabeled specific fragments or unlabeled nonspecific fragments: it was clearly shown that a nonspecific fragment does not compete with the fragment harboring the LexA binding site (Fig. 5B). Since the five hox genes in Nostoc sp. strain PCC 7120 are separated on two independent transcripts, it would be expected that they are regulated in a similar manner. However, the LexA box located upstream from alr0750 could not be found upstream from hoxU. In a recent report, the LexA-binding sequence of Bdellovibrio bacteriovorus was identified, and it was further suggested that the cyanobacterial LexA is probably a common ancestor of the B. bacteriovorus LexA (11). In fact, EMSAs using Nostoc sp. strain PCC 7120 LexA showed that it could recognize the LexA box of B. bacteriovorus (11). The consensus sequence TTACNNNGTAA used in the same work to identify additional LexA binding sites in the genome of B. bacteriovorus (11) shows similarity to a stretch of 11 bp upstream from hoxU (Fig. 2B). A 535-bp fragment harboring this site (herein referred to as the hoxU fragment) was therefore used to elucidate whether LexA interacts with the promoter region of the second cluster as well. The results clearly demonstrate a specific interaction between the hoxU fragment and LexA (Fig. 5C and D), and we conclude that LexA can in fact interact with the promoter regions of the two hox clusters.
FIG. 5.
EMSAs with purified LexA from Nostoc sp. strain PCC 7120 and the two regulatory hox operon regions. The alr0750 (A and B) and hoxU (C and D) DNA fragments were generated by PCR using genomic DNA from Nostoc sp. strain PCC 7120, and the unrelated DNA fragment was generated by using pQE-30 vector (QIAGEN). The positions of the DNA fragments relative to the regulatory regions of the hox operons are shown in Fig. 2. EMSAs were first carried out with the specific alr0750 (A) and hoxU (C) fragments, incubated together with the unrelated DNA fragment without protein (lane 1) or together with increasing amounts of LexA (lanes 2 through 5). The unrelated DNA is indicated with a black arrow and the alr0750 and hoxU fragments and their retardations are indicated with gray and white arrows, respectively. EMSAs were carried out to demonstrate the specificities in the binding between Nostoc sp. strain PCC 7120 LexA and the alr0750 (B) and hoxU (D) fragments. The unrelated DNA was incubated with and without LexA, alone or together with the alr0750 or hoxU fragment (lanes 6 through 9). The alr0750 and hoxU fragments were also incubated together with a 5× or 50 × excess of either unlabeled specific (lanes 12 and 13) or nonspecific (lanes 14 and 15) DNA. The amount of LexA (in ng) used for each lane is indicated in the figure. A plus sign indicates that the fragment was included in the assay and a minus sign indicates that the fragment was excluded from the assay.
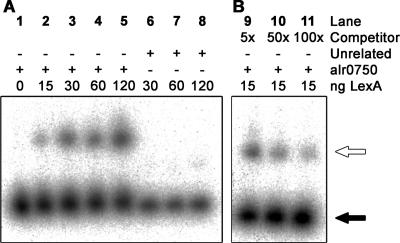
Additionally, LexA from the Synechocystis sp. strain PCC 6803, another cyanobacterial strain, was further used in EMSAs, making use of the same alr0750 and hoxU DNA fragments, to determine whether the identified LexA binding sites found in Nostoc sp. strain PCC 7120 can be recognized by a close phylogenetic relative. These experiments clearly demonstrated that there is indeed an interaction between LexA from Synechocystis sp. strain PCC 6803 and the promoter region of alr0750, the region containing the cyanobacterial LexA binding box (Fig. 6). Moreover, the signal decreased when increasing amounts of unlabeled alr0750 fragment were added, indicating a specific binding, and no shift was observed when an unrelated DNA fragment was used (Fig. 6). However, no shift was observed when the DNA fragment covering the promoter region of hoxU (data not shown) containing the LexA binding site similar to that of B. bacteriovorus was used.
FIG. 6.
EMSAs with purified LexA from Synechocystis sp. strain PCC 6803 and the Nostoc sp. strain PCC 7120 regulatory hox operon region covered by the alr0750 DNA fragment. The fragments used were generated by PCR using genomic DNA from Nostoc sp. strain PCC 7120 (lanes 1 through 5 and 9 through 11) or pQE-30 vector from QIAGEN (unrelated DNA, lanes 6 through 8). The position of the DNA fragment relative to the regulatory region of the hox operon is shown in Fig. 2. The assays were carried out using 20 fmol of each labeled fragment incubated with 0, 15, 30, 60, or 120 ng purified LexA (lanes 1 through 5). Competition experiments were carried out with 15 ng LexA in the presence of 5×, 50 ×, and 100 × molar excesses of the unlabeled alr0750 fragment as competitor DNA (lanes 9 through 11). White arrows indicate unbound DNA fragments, and black arrows indicate a shift, i.e., LexA bound to the DNA fragment.
The LexA box is widespread in the Nostoc sp. strain PCC 7120 genome.
The Nostoc sp. strain PCC 7120 LexA box RGTACNNNDGTWCB (27) was used in a whole-genome search, using a Perl script developed in-house, to identify other genes associated with the binding sequence. To broaden the search, two more variations of the sequence were included. The C in the first part of the motif was therefore exchanged for a T (RGTAC→RGTAT) to include the LexA binding site found upstream from recA in Nostoc sp. strain PCC 7120 (27). The first part of the motif was also altered by replacing the T with an A (RGTAC→RGAAC), a change made so the motif would resemble the closely related gram-positive LexA binding site CGAACRNRYGTTYC (55). LexA has previously been shown to interact with both these motifs in Nostoc sp. strain PCC 7120, although it interacts with the latter with a lower efficiency (27). A total number of 216 putative binding sites were found in the genome of Nostoc sp. strain PCC 7120; most of them, though, were located within annotated ORFs. A selected part of the identified putative binding sites found is listed in Table 2. Functional LexA binding sites have been previously reported to be located within ORFs (4, 33), and a few interesting putative binding sites located within ORFs are therefore also included in Table 2.
TABLE 2.
Putative LexA binding sites in the genome of Nostoc sp. strain PCC 7120
| Sequence | ORF | Positiona | Protein annotationb | Putative function/comment/reference |
|---|---|---|---|---|
| AGTACTTTTGTTCC | all0061 | −869 | Site-specific DNA-methyltransferase | Inside alr0063 and rbfA |
| AGTACTTATGTACT | alr0088 | −28 | Single-stranded DNA-binding protein | 27 |
| AGTACTTTTGTACT | asl0401 | −71 | Unknown protein | Probable nitroreductase |
| AGAACCAGAGTTCC | all0687N | −246 | [NiFe] uptake hydrogenase large subunit, N end fragment | Inside all0688 (hupS), located upstream from all0687N |
| GGTACTCTGGTTCG | alr0750 | −142 | Hypothetical protein | Encodes universal stress protein (Usp) |
| GGAACAGGAGTACT | all0998 | +630 | Putative catalase | Inside gene, protein contains manganese catalase domain |
| AGTACTTAAGTACT | alr1041 | −175 | Fructose-1,6-bisphosphatase | |
| AGAACTTTAGTTCT | alr1194 | −425 | Two-component response regulator | |
| GGTACTGTAGTACG | all1512 | −311 | Cytochrome b6/f-complex iron-sulfur protein PetC | Inside asl1513, encodes hypothetical protein |
| AGAACAATTGTACC | alr1855 | −150 | Unknown protein | Similar to DNA double-strand break repair rad50 ATPase gene |
| GGTACTGATGTTCC | alr1879 | +228 | Glycogen synthase | Inside gene |
| AGTACATAAGTACT | alr2104 | −59 | Probable methyltransferase | |
| AGAACTTTAGTTCT | alr2481 | −265 | Two-component sensor histidine kinase | |
| AGTACAGAAGTTCT | all3374 | +654 | DNA repair and genetic recombination protein RecF (encoded by recF) | Inside gene |
| AGTACTTAAGTACC | all3503 | −109 | Probable integrase | |
| AGTACTATTGTTCT | alr3716 | −84 | Excinuclease ABC subunit A (encoded by uvrA) | 27 |
| AGTACCAAAGTTCT | asl3860 | −46 | Glutaredoxin | |
| GGTACTTGTGTTCC | all3866 | −234 | Probable dioxygenase, Rieske iron-sulfur component | Inside alr3867, encodes putative aminotransferase |
| GGTACTTTTGTTCG | alr4239 | −241 | Toxin secretion ABC transporter ATP-binding protein | Upstream from alr4240, encodes RTX toxin transporter |
| GGTACAAATGTACG | all4790 | −45 | Hypothetical protein | Upstream from all4789, encodes DNA helicase Re cG |
| AGTACTAATGTTCT | alr4908 | −47 | SOS function regulatory protein, LexA repressor | Upstream from alr4909, similar to DNA repair gene rad25 (27) |
| AGTACTATGGTACG | asr4942 | −53 | Unknown protein | Upstream from alr4943, encodes putative 4Fe-4S cluster binding protein |
| GGTACTCCAGTACG | all0475 | +528 | Probable short-chain dehydrogenase | Inside gene |
| GGTATTTATGTACT | alr1044 | −472 | Transcriptional regulator | |
| GGTATCGCTGTACG | alr1095 | +687 | GAPDH (encoded by gap3) | Inside gene |
| GGTATCGTTGTACT | alr2780 | −276 | DNA topoisomerase I | |
| AGAATTAGTGTACC | all2951 | +33 | Probable helicase protein | Inside gene |
| AGTATTATTGTTCC | all5215 | −104 | Transcriptional-repair coupling factor | Helicase domain |
| GTATCCAAGTTCC | all3735 | −291 | Fructose-bisphosphate aldolase class I | Inside all3736, encodes unknown protein |
| AGTATATCTGTTCT | all3272 | −66 | Recombination protein RecA (encoded by recA) | 27 |
Position of the first base pair in the motif in relation to the TSP of the downstream gene.
According to Cyanobase (http://www.kazusa.or.jp/cyano/cyano.html).
DISCUSSION
The results demonstrate that the hox genes, encoding the bidirectional hydrogenase, in Nostoc sp. strain PCC 7120 are located in at least two separate operons. Two of the three genes encoding the diaphorase part (hoxE and hoxF) form one operon together with the ORF alr0750. The third diaphorase subunit, hoxU, is thus separated from hoxE and hoxF and located on a second operon together with the two genes hoxY and hoxH, which encode the hydrogenase part. Interestingly, a similar genomic arrangement and transcription units of the hox genes are found in the unicellular cyanobacteria Synechococcus sp. strain PCC 7942 (40) and Synechococcus sp. strain PCC 6301 (10).
In order to study the transcription of the hox genes in Nostoc sp. strain PCC 7120, two different techniques were used: RT-PCR and Northern blotting. Both confirmed the existence of two separate operons and, in addition, due to different sensitivities, resulted in additional and complementary information. The very sensitive RT-PCR experiments demonstrated that alr0760 and alr0761 are part of a long transcript together with hoxU-alr0763-hoxY-alr0765-hoxH. However, the very low transcript levels of alr0760 and alr0761 observed by use of Northern blot hybridization (Fig. 3A) indicate that the alr0760-hoxH transcript is present only at a very low level, meaning that it is not the main contributor of the hoxUYH transcripts to the total mRNA pool. This suggestion is supported by the TSP upstream from hoxU. However, a careful examination of the Northern blot hybridizations revealed a rather complex transcription of the polycistronic hoxU-alr0763-hoxY-alr0765-hoxH transcription. In addition to the fainter signal in the form of a band that smears down on the lane (hoxU; Fig. 3B), matching to the large transcript, the predominance of shorter transcripts of approximately 1,900 nucleotides can be distinguished. Considering the size of the band and the TSP found upstream from hoxU, it is possible to predict that the band corresponds to the transcript consisting of hoxU, alr0763, and part of hoxY. A similar situation has been reported for Synechococcus sp. strain PCC 7942 by Schmitz et al. (40), who used reporter gene constructs and real-time RT-PCRs. In addition, they showed that the level of expression of hoxU is approximately six times higher than that of hoxH, even though hoxU and hoxH are clustered and belong to the same operon. In agreement, we suggest that the bands with stronger signals in our Northern blot hybridizations correspond to partial termination of the transcription and/or mRNA instability within the regions between hoxY and hoxH. Interestingly, a similar observation can be made for the operon alr0750-hoxE-hoxF: the 1,200-nucleotide strong band in the Northern blot hybridizations, occurring when using the alr0750 probe (Fig. 3B), may correspond to the alr0750-hoxE transcript. It is also important to notice that the Northern blot hybridizations in the present work are based on double-stranded DNA fragments, denatured at 95°C prior to the hybridization. As a consequence, any presence of antisense RNA, as has been described for the hox operons in Synechococcus sp. strain PCC 7942 (40), will result in hybridization patterns with more complex interpretations.
The roles of the ORFs transcribed together with the structural hox genes as well as the ORFs located around the two hox clusters are unclear. For most of them, no obvious connection to either the bidirectional hydrogenase or the hydrogen metabolism can be found. The three ORFs alr0750, alr0763, and alr0765, found on the same transcripts as the hox genes, are all physically located in the same position in Anabaena variabilis ATCC 29413 (42; see also the U.S. Department of Energy Joint Genome Institute website [http://genome.jgi-psf.org/finished_microbes/anava/anava.home.html]), although in this strain, the hox genes are located in one single cluster. Interestingly, alr0750 and alr0765 are also found in Nostoc punctiforme, a species lacking the bidirectional hydrogenase (48). Homologues of alr0760 are present in both Anabaena variabilis ATCC 29413 and Nostoc punctiforme. In Table 3, the annotations of all genes noted in Fig. 1A are listed, and for some of the genes, putative functions and characteristics are proposed. The first gene in the first hox cluster, alr0750, contains an UspA (universal stress protein) domain. UspA in E. coli has been reported to be stimulated in response to a large variety of conditions, including, e.g., carbon and nitrogen (25). Nachin et al. (29) proposed that Usp proteins have evolved different physiological functions to reprogram the cell to defense during cellular stress. They further showed that UspA is involved in regulating the capacity for the cells to withstand oxidative agents. Larger proteins in the Usp family in Archaea, cyanobacteria, and plants often contain other functional domains (25), and the domain found in alr0750 also includes a part of an Na+/H+ antiport motif. There are no obvious putative domains in alr0760, alr0761, or alr0763. Orf3 in Anabaena variabilis ATCC 29413 (9, 49), corresponding to alr0765 in Nostoc sp. strain PCC 7120, shows homology to a small protein named CP12 (34). This protein has been reported to oligomerize with two of the key enzymes in the Calvin cycle, phosphoribulokinase (PRK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Recent data suggest that under light and dark conditions, the oligomerization of CP12 with PRK and GAPDH regulates the activities of both these enzymes and, thus, the carbon flow from the Calvin cycle to the oxidative pentose phosphate cycle in the cyanobacterium Synechococcus sp. strain PCC 7942 (50). Furthermore, the discovery of genes for CP12 in mosses, green algae, and cyanobacteria, together with the demonstration of conserved PRK/CP12/GAPDH complex composition and function in Chlamydomonas and Synechocystis spp. suggests that the regulation of the Calvin cycle, via an NADPH-mediated dissociation of the complex, is conserved in all photosynthetic organisms (34, 54). Wedel and Soll (54) stated that the detection of proteins containing CP12, or parts of it, shows that these motifs have been used as evolutionary conserved modules for natural genetic engineering, allowing control of different enzymatic activities by NADP(H). The last step in maturation of NiFe enzymes involves the endoproteolytic cleavage of a C-terminal peptide of the large subunit precursor (12). The gene encoding the endopeptidase responsible for the cleavage in Nostoc sp. strain PCC 7120, the hoxW, was found to be located in close proximity to the hox gene clusters (Fig. 1A) (56). Four of the ORFs located in the 8.8-kb DNA region between the two hox clusters, all0753, all0754, asr0757, and alr0758 (Fig. 1A and Table 3), were recently identified as toxin-antitoxin loci (32). These loci seem to be abundant in bacterial and archaeal chromosomes (15), and suggested functions include nutritional stress response elements and/or quality control elements increasing the fitness of free-living prokaryotes (32). Interestingly, toxin-antitoxin loci are also found between the hox genes in Anabaena variabilis ATCC 29413 (U.S. Department of Energy Joint Genome Institute [http://genome.jgi-psf.org/finished_microbes/anava/anava.home.html]) and Synechocystis sp. strain PCC 6803 (32). In Synechocystis sp. strain PCC 6803, the two ORFs constituting the toxin-antitoxin locus are part of the hox operon (30).
TABLE 3.
Annotations of genes and the ORFs found in the vicinity of the hox genes (Fig. 1A) in Nostoc sp. strain PCC 7120
| Gene/ORF | Protein annotationa | Putative function/characteristic of protein | Reference |
|---|---|---|---|
| all0748 | Hypothetical protein | ||
| asl0749 | 30S ribosomal protein S15 | ||
| alr0750 | Hypothetical protein | Contains UspA domain | |
| hoxE | NADH dehydrogenase I chain E | 42 | |
| hoxF | Hydrogenase subunit; HoxF | 42 | |
| all0753 | Hypothetical protein | Toxin of toxin-antitoxin system (vapC) | 32 |
| all0754 | Unknown protein | Antitoxin of toxin-antitoxin system (vapB) | 32 |
| asr0755 | Hypothetical protein | ||
| asr0756 | Unknown protein | ||
| asr0757 | Unknown protein | Antitoxin of toxin-antitoxin system (mazE) | 32 |
| alr0758 | Hypothetical protein | Toxin of toxin-antitoxin system (mazF) | 32 |
| all0759 | WD repeat-containing protein | Contains WD-40 (Trp-Asp, ∼40 amino acids) repeat domains | |
| alr0760 | Hypothetical protein | ||
| alr0761 | Unknown protein | ||
| hoxU | Hydrogenase chain U | 42 | |
| alr0763 | Hypothetical protein | ||
| hoxY | Hydrogenase small subunit; HoxY | 42 | |
| alr0765 | Hypothetical protein | Contains CP12 domain | 32 |
| hoxH | Hydrogenase large subunit; HoxH | 42 | |
| all0767 | Hypothetical protein | Endoribonuclease domain | |
| all0768 | Hypothetical protein | Acetyltransferase | |
| all0769 | Acetyl coenzyme A synthetase | ||
| all0770 | Hypothetical protein | hoxW protein; hydrogenase-specific endopeptidase | 56 |
According to Cyanobase (www.kazusa.or.jp/cyano/cyano.html).
Looking carefully at the promoter regions of the two hox operons in Nostoc sp. strain PCC 7120, it is possible to observe that the positions and characters of the −10 and −35 boxes show similarity to known promoters from other genes in cyanobacteria (13). The segments of 40 nucleotides of the alr0750 and hoxU promoter regions that share a high degree of homology (Fig. 2) harbor the promoter recognition elements, suggesting a regulation common between the different transcripts. During searches for additional recognition motifs upstream from the transcriptional start site of alr0750 and hoxU, putative FNR (fumarate and nitrate reduction) binding sites at positions 218 to 231 bp and 208 to 221 bp upstream from the TSP, respectively, showing high similarity to the E. coli FNR consensus sequence could be found (43). FNR proteins are global transcription regulators that respond to fluctuations in environmental oxygen, and in E. coli, they regulate the response to the transition between aerobic growth and anaerobic growth (23). A putative FNR binding site has been previously reported to be upstream from hupSL genes encoding the uptake hydrogenase in Anabaena variabilis ATCC 29413 (17). The hyn operon of the purple sulfur bacterium Thiocapsa roseopersicina, encoding a membrane-associated [NiFe] hydrogenase, is up-regulated under anaerobic conditions, and it was recently demonstrated that an FNR homologue can bind to two proposed sites and initiate transcription (24). Although no obvious FNR protein-encoding gene can be found in the annotated genome of Nostoc sp. strain PCC 7120, it is possible to identify ORF proteins of interest; e.g., All4541 shows 22% amino acid sequence identity to FNR of E. coli and 31% identity to FNR of Bacillus subtilis. Furthermore, All4541 contains a conserved domain of the Fnr/Crp superfamily of transcription factors and has three cysteine residues that resemble the cluster of conserved cysteine residues coordinating the [4Fe-4S] cluster required for function (23, 36).
Interestingly, when cells of Nostoc sp. strain PCC 7120 were grown under dark, anaerobic conditions, the expression levels of the hox transcripts increased substantially (Fig. 3), a result which is in agreement with physiological data reported earlier (19, 20). It has long been demonstrated that microaerobic/anaerobic conditions influence the bidirectional hydrogenase activity in heterocystous cyanobacteria (6, 19, 20, 39, 44, 46). However, this is the first time that it is demonstrated that the anaerobically induced activity of the enzyme is in parallel with an increase in transcription of the two hox operons in Nostoc sp. strain PCC 7120.
The two identified LexA binding sites upstream from alr0750 and hoxU that interact with LexA purified from Nostoc sp. strain PCC 7120 (Fig. 5) suggest that this novel role of LexA described by Domain et al. (14) may not be restricted to Synechocystis sp. strain PCC 6803 but can possibly be expanded to other cyanobacterial strains. Interestingly, when using Synechocystis sp. strain PCC 6803 LexA in EMSAs, it was observed that it could interact only with the alr0750 promoter region (which harbors the previously described LexA box) but not with the hoxU DNA fragment. This fact is in agreement with the results reported by Mazon et al. (27), in which Nostoc sp. strain PCC 7120 LexA recognized the LexA box upstream from the Synechocystis sp. strain PCC 6803 lexA gene. However, here we show that although the LexA proteins from Synechocystis sp. strain PCC 6803 and Nostoc sp. strain PCC 7120 can both recognize the box RGTACNNNDGTWCB, they have different DNA recognition capabilities, which might be a consequence of the differences found on the amino-acidic level (14, 27, 30).
In E. coli, LexA is well characterized in the SOS response system, where it is accepted as a typical repressor (53). In cyanobacteria, microarray experiments carried out by Domain et al. (14) demonstrated that the expression of a number of genes in a LexA-depleted mutant of Synechocystis sp. strain PCC 6803 was either up- or down-regulated. Many of the identified genes are involved in carbon assimilation or controlled by carbon availability, although none connected to the SOS response system (14). Furthermore, Patterson-Fortin et al. (33) reported recently that the expression of the DEAD box RNA helicase, the crhR gene product, in Synechocystis sp. strain PCC 6803 is negatively regulated by LexA. Interestingly, the expression of crhR is regulated in response to conditions which elicit reduction of the photosynthetic electron transport chain (33). In Nostoc sp. strain PCC 7120, LexA has been previously shown to interact with a few of the genes, e.g., recA, uvrA, and ssb, involved in the SOS system in E. coli (27). The results from the bioinformatics search for additional putative LexA binding sites in the genome of Nostoc sp. strain PCC 7120 (Table 2) indicate that a number of genes connected to DNA replication, recombination, and repair may be part of the LexA regulatory network. However, many genes in the E. coli LexA regulon do not show any LexA box upstream from its coding sequence in Nostoc sp. strain PCC 7120 (27). Moreover, many identified LexA boxes correspond to intergenic regions located upstream from genes encoding either unknown or hypothetical proteins. This may indicate that LexA in Nostoc sp. strain PCC 7120 is involved in regulatory networks, so far not described, that definitely deserve further attention. Although the physiological role of the cyanobacterial bidirectional hydrogenase is unclear, the suggestions, e.g., being part of respiratory complex I, involved in fermentation, and functioning as a valve during photosynthesis, are all in agreement with an enzyme system being under redox control. Recently, LexA was suggested to be a mediator of the intracellular redox state (1, 33) in Synechocystis sp. strain PCC 6803. Nevertheless, it remains to be clarified what is the specific role of LexA on the regulation of the hox genes in Nostoc sp. strain PCC 7120 and in which signal transduction pathways LexA is directly involved.
In conclusion, the Nostoc sp. strain PCC 7120 hox genes, encoding the bidirectional hydrogenase, are separated in two operons that show similar responses to anaerobic induction. The function of the additional ORFs transcribed together with the hox genes is not apparently associated with hydrogen metabolism. Two transcriptional start sites, upstream from alr0750 and hoxU, have been identified, but it is not possible to exclude the possibility that additional transcriptional start sites are present. LexA interacts with the regulatory region of the two hox gene clusters. However, since the transcription of lexA does not seem to be clearly affected by anaerobic conditions (data not shown), it is likely that additional transcription factors are involved in the regulation of the bidirectional hydrogenase in Nostoc sp. strain PCC 7120 and maybe also in other cyanobacterial strains.
Acknowledgments
This work was supported by the Swedish Research Council, the Swedish Energy Agency, the Knut and Alice Wallenberg Foundation, the Nordic Energy Research Program (project BioHydrogen), and the EU/NEST projects SOLAR-H (contract 516510) and BioModularH2 (contract 043340).
We thank Rikard Axelsson for his initial work on this project. Fernando Lopes Pinto (Uppsala University) is gratefully acknowledged for his help with the Perl script and further analysis of the bioinformatics data.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Antal, T., P. Oliveira, and P. Lindblad. 2006. The bidirectional hydrogenase in the cyanobacterium Synechocystis sp. strain PCC 6803. Int. J. Hydrogen Energy 31:1439-1444. [Google Scholar]
- 2.Appel, J., and R. Schulz. 1996. Sequence analysis of an operon of a NAD(P)-reducing nickel hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803 gives additional evidence for direct coupling of the enzyme to NAD(P)H-dehydrogenase (complex 1). Biochim. Biophys. Acta 1298:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Appel, J., S. Phunpruch, K. Steinmüller, and R. Schulz. 2000. The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch. Microbiol. 173:333-338. [DOI] [PubMed] [Google Scholar]
- 4.Au, N., E. Kuester-Schoeck, V. Mandava, L. E. Bothwell, S. P. Canny, K. Chachu, S. A. Colavito, S. N. Fuller, E. S. Groban, L. A. Hensley, T. C. O'Brien, A. Shah, J. T. Tierney, L. L. Tomm, T. M. O'Gara, A. I. Goranov, A. D. Grossman, and C. M. Lovett. 2005. Genetic composition of the Bacillus subtilis SOS system. J. Bacteriol. 187:7655-7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axelsson, R. 2003. Cyanobacterial hydrogen metabolism—transcriptional regulation of the hydrogenases in filamentous strains. Acta Univ. Ups. 888:49. [Google Scholar]
- 6.Axelsson, R., and P. Lindblad. 2002. Transcriptional regulation of Nostoc hydrogenases: effects of oxygen, hydrogen, and nickel. Appl. Environ. Microbiol. 68:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axelsson, R., F. Oxelfelt, and P. Lindblad. 1999. Transcriptional regulation of Nostoc uptake hydrogenase. FEMS Microbiol. Lett. 170:77-81. [DOI] [PubMed] [Google Scholar]
- 8.Boison, G., H. Bothe, A. Hansel, and P. Lindblad. 1999. Evidence against a common use of the diaphorase subunits by the bidirectional hydrogenase and by the respiratory complex I in cyanobacteria. FEMS Microbiol. Lett. 174:159-165. [Google Scholar]
- 9.Boison, G., H. Bothe, and O. Schmitz. 2000. Transcriptional analysis of hydrogenase genes in the cyanobacteria Anacystis nidulans and Anabaena variabilis monitored by RT-PCR. Curr. Microbiol. 40:315-321. [DOI] [PubMed] [Google Scholar]
- 10.Boison, G., O. Schmitz, B. Schmitz, and H. Bothe. 1998. Unusual gene arrangement of the bidirectional hydrogenase and functional analysis of its diaphorase subunit HoxU in respiration of the unicellular cyanobacterium Anacystis nidulans. Curr. Microbiol. 36:253-258. [DOI] [PubMed] [Google Scholar]
- 11.Campoy, S., N. Salvador, P. Cortes, I. Erill, and J. Barbe. 2005. Expression of canonical SOS genes is not under LexA repression in Bdellovibrio bacteriovorus. J. Bacteriol. 187:5367-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9:228-237. [DOI] [PubMed] [Google Scholar]
- 13.Curtis, M. E., and J. A. Martin. 1994. Transcription in cyanobacteria, p. 613-639. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer, Dordrecht, The Netherlands.
- 14.Domain, F., L. Houot, F. Chauvat, and C. Cassier-Chauvat. 2004. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 53:65-80. [DOI] [PubMed] [Google Scholar]
- 15.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 16.Gutekunst, K., S. Phunpruch, C. Schwarz, S. Schuchardt, R. Schulz-Friedrich, and J. Appel. 2005. LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 58:810-823. [DOI] [PubMed] [Google Scholar]
- 17.Happe, T., K. Schutz, and H. Bohme. 2000. Transcriptional and mutational analysis of the uptake hydrogenase of the filamentous cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 182:1624-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houchins, J. P. 1984. The physiology and biochemistry of hydrogen metabolism in cyanobacteria. Biochim. Biophys. Acta 768:227-255. [Google Scholar]
- 19.Houchins, J. P., and R. H. Burris. 1981. Comparative characterization of two distinct hydrogenases from Anabaena sp. strain 7120. J. Bacteriol. 146:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houchins, J. P., and R. H. Burris. 1981. Occurrence and localization of two distinct hydrogenases in the heterocystous cyanobacterium Anabaena sp. strain 7120. J. Bacteriol. 146:209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howitt, C. A., and W. E. J. Vermaas. 1999. Subunits of the NAD(P)-reducing nickel containing hydrogenase do not act as part of the type-1 NAD(P)H-dehydrogenase in the cyanobacterium Synechocystis sp. PCC 6803, p. 595-601. In G. A. Perschek and W. Löffelhardt (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, NY.
- 22.Kentemich, T., M. Casper, and H. Bothe. 1991. The reversible hydrogenase in Anacystis nidulans is a component of the cytoplasmic membrane. Naturwissenschaften 78:559-560. [Google Scholar]
- 23.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 24.Kovács, Á. T., G. Rákhely, D. F. Browning, A. Fülöp, G. Maróti, S. J. W. Busby, and K. L. Kovács. 2005. An FNR-type regulator controls the anaerobic expression of Hyn hydrogenase in Thiocapsa roseopersicina. J. Bacteriol. 187:2618-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvint, K., L. Nachin, A. Diez, and T. Nyström. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140-145. [DOI] [PubMed] [Google Scholar]
- 26.Masukawa, H., M. Mochimaru, and H. Sakurai. 2002. Disruption of the uptake hydrogenase gene, but not of the bidirectional hydrogenase gene, leads to enhanced photobiological hydrogen production by the nitrogen-fixing cyanobacterium Anabaena sp. PCC 7120. Appl. Microbiol. Biotechnol. 58:618-624. [DOI] [PubMed] [Google Scholar]
- 27.Mazón, G., J. M. Lucena, S. Campoy, A. R. Fernández de Henestrosa, P. Candau, and J. Barbé. 2004. LexA-binding sequences in gram-positive and cyanobacteria are closely related. Mol. Genet. Genomics 271:40-49. [DOI] [PubMed] [Google Scholar]
- 28.Montesinos, M. L., A. M. Muro-Pastor, A. Herrero, and E. Flores. 1998. Ammonium/methylammonium permeases of a cyanobacterium: identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J. Biol. Chem. 273:31463-31470. [DOI] [PubMed] [Google Scholar]
- 29.Nachin, L., U. Nannmark, and T. Nystrom. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira, P., and P. Lindblad. 2005. LexA, a transcription regulator binding in the promoter region of the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 251:59-66. [DOI] [PubMed] [Google Scholar]
- 31.Oliveira, P., E. Leitão, P. Tamagnini, P Moradas-Ferreira, and F. Oxelfelt. 2004. Characterization and transcriptional analysis of hupSLW in Gloeothece sp. ATCC 27152: an uptake hydrogenase from a unicellular cyanobacterium. Microbiology 150:3647-3655. [DOI] [PubMed] [Google Scholar]
- 32.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson-Fortin, L. M., K. R. Colvin, and G. W. Owttrim. 2006. A LexA-related protein regulates redox-sensitive expression of the cyanobacterial RNA helicase, crhR. Nucleic Acids Res. 34:3446-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pohlmeyer, K., B. K. Paap, J. Soll, and N. Wedel. 1996. CP12: a small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol. Biol. 32:969-978. [DOI] [PubMed] [Google Scholar]
- 35.Ramasubramanian, T. S., T. F. Wei, A. K. Oldham, and J. W. Golden. 1996. Transcription of the Anabaena sp. strain PCC 7120 ntcA gene: multiple transcripts and NtcA binding. J. Bacteriol. 178:922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reents, H., I. Gruner, U. Harmening, L. H. Bottger, G. Layer, P. Heathcote, A. X. Trautwein, D. Jahn, and E. Hartig. 2006. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol. Microbiol. 60:1432-1445. [DOI] [PubMed] [Google Scholar]
- 37.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stainer. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Schmitz, O., and H. Bothe. 1996. The diaphorase subunit HoxU of the bidirectional hydrogenase as electron transferring protein in cyanobacterial respiration? Naturwissenschaften 83:525-527. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz, O., G. Boison, and H. Bothe. 2001. Quantitative analysis of expression of two circadian clock-controlled gene clusters coding for the bidirectional hydrogenase in the cyanobacterium Synechococcus sp. PCC7942. Mol. Microbiol. 41:1409-1417. [DOI] [PubMed] [Google Scholar]
- 41.Schmitz, O., G. Boison, H. Salzmann, H. Bothe, K. Schutz, S.-H. Wang, and T. Happe. 2002. HoxE—a subunit specific for the pentameric bidirectional hydrogenase complex (HoxEFUYH) of cyanobacteria. Biochim. Biophys. Acta 1554:66-74. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz, O., G. Boison, R. Hilscher, B. Hundeshagen, W. Zimmer, F. Lottspeich, and H. Bothe. 1995. Molecular biological analysis of a bidirectional hydrogenase from cyanobacteria. Eur. J. Biochem. 233:266-276. [DOI] [PubMed] [Google Scholar]
- 43.Scott, C., J. D. Partridge, J. R. Stephenson, and J. Green. 2003. DNA target sequence and FNR-dependent gene expression. FEBS Lett. 541:97-101. [DOI] [PubMed] [Google Scholar]
- 44.Serebryakova, L., N. A. Zorin, and P. Lindblad. 1994. Reversible hydrogenase in Anabaena variabilis ATCC 29413: presence and localization in non-N2-fixing cells. Arch. Microbiol. 161:140-144. [Google Scholar]
- 45.Serebryakova, L. T., M. E. Sheremetieva, and P. Lindblad. 2000. H2-uptake and evolution in the unicellular cyanobacterium Chroococcidiopsis thermalis CALU 758. Plant Physiol. Biochem. 38:525-530. [Google Scholar]
- 46.Sheremetieva, M. E., O. Y. Troshina, L. T. Serebryakova, and P. Lindblad. 2002. Identification of hox genes and analysis of their transcription in the unicellular cyanobacterium Gloeocapsa alpicola CALU 743 growing under nitrate-limiting conditions. FEMS Microbiol. Lett. 214:229-233. [DOI] [PubMed] [Google Scholar]
- 47.Stal, L. J., and R. Moezelaar. 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21:179-211. [Google Scholar]
- 48.Tamagnini, P., O. Troshina, F. Oxelfelt, R. Salema, and P. Lindblad. 1997. Hydrogenases in Nostoc sp. strain PCC 73102, a strain lacking a bidirectional enzyme. Appl. Environ. Microbiol. 63:1801-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamagnini, P., R. Axelsson, P. Lindberg, F. Oxelfelt, R. Wunschiers, and P. Lindblad. 2002. Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol. Mol. Biol. Rev. 66:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamoi, M., T. Miyazaki, T. Fukamizo, and S. Shigeoka. 2005. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 42:504-513. [DOI] [PubMed] [Google Scholar]
- 51.Troshina, O., L. Serebryakova, M. Sheremetieva, and P. Lindblad. 2002. Production of H2 by the unicellular cyanobacterium Gloeocapsa alpicola CALU 743 during fermentation. Int. J. Hydrogen Energy 27:1283-1289. [Google Scholar]
- 52.Vioque, A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20:6331-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner, R. 2000. Transcription regulation in prokaryotes. Oxford University Press, New York, NY.
- 54.Wedel, N., and J. Soll. 1998. Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. Proc. Natl. Acad. Sci. USA 95:9699-9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winterling, K. W., D. Chafin, J. J. Hayes, J. Sun, A. S. Levine, R. E. Yasbin, and R. Woodgate. 1998. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 180:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wünschiers, R., M. Batur, and P. Lindblad. 2003. Presence and expression of hydrogenase specific C-terminal endopeptidases in cyanobacteria. BMC Microbiol. 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]