Abstract
Many confined-livestock farms store their wastes for several months prior to use as a fertilizer. Storing manure for extended periods could significantly bias the composition of enteric bacterial populations subsequently released into the environment. Here, we compared populations of Escherichia coli isolated from fresh feces and from the manure-holding tank (stored manure) of a commercial swine farm, each sampled monthly for 6 months. The 4,668 confirmed E. coli isolates were evaluated for resistance to amikacin, ampicillin, cephalothin, chloramphenicol, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, trimethoprim, and trimethoprim plus sulfamethoxazole. A subset of 1,687 isolates was fingerprinted by repetitive extragenic palindromic PCR (rep-PCR) with the BOXA1R primer to evaluate the diversity and the population structure of the collection. The population in the stored manure was generally more diverse than that in the fresh feces. Half of the genotypes detected in the stored manure were never detected in the fresh fecal material, and only 16% were detected only in the fresh feces. But the majority of the isolates (84%) were assigned to the 34% of genotypes shared between the two environments. The structure of the E. coli population showed important monthly variations both in the extent and distribution of the diversity of the observed genotypes. The frequency of detection of resistance to specific antibiotics was not significantly different between the two collections and varied importantly between monthly samples. Resistance to multiple antibiotics was much more temporally dynamic in the fresh feces than in the stored manure. There was no relationship between the distribution of rep-PCR fingerprints and the distribution of antibiotic resistance profiles, suggesting that specific antibiotic resistance determinants were dynamically distributed within the population.
Fecal contamination of surface water represents a threat to human and environmental health (16). This is particularly true when water resources are in proximity to land that is subject to increasing agricultural activity and burgeoning human populations, increasing the risk to adjacent waters from agricultural runoff, sewage effluent, leaking rural septic systems, and storm water discharge. Escherichia coli is a fecal indicator bacterium that has traditionally been used to evaluate the microbiological quality of surface and drinking water, using standard microbiological methods (10, 53). The presence of this organism is implicit evidence of fecal contamination and indicates a risk of contamination with viral, bacterial, or protozoan pathogens of enteric origin. Therefore, many jurisdictions mandate compliance with drinking and recreational water standards on the basis of contamination with E. coli (3, 14-16).
In most industrial countries, swine produced on commercial farms are raised confined in barns, and their waste is stored for several months as an anoxic slurry prior to being added as a fertilizer to the land when climate and crop conditions are suitable. In Canada, for example, about 85% of swine are produced on farms that use static liquid manure storage systems (49). These manure storage systems therefore represent on many farms the crucial secondary habitat that enteric bacteria must survive before they are released into the broader environment, where they could pose a threat to water quality. As well as reducing the abundance of enteric bacteria, storing waste for extended periods could significantly alter the composition of enteric bacterial populations subsequently released into the broader environment. The dynamism of bacterial populations during storage of anoxic manure slurry is somewhat unclear. The distribution of dominant bacteria in swine manure slurry is stable for at least several weeks (11, 30, 40). On the other hand, observed populations of E. coli distinguished by repetitive extragenic palindromic PCR (rep-PCR) were found to be consistently more diverse in stored manure slurry than in freshly shed feces from the corresponding swine (32). Changes in the distribution of attributes among populations of E. coli that are used to distinguish host source (e.g., antibiotic resistance and dominant host-specific genotypes detected by ribotyping, pulsed-field gel electrophoresis, or rep-PCR methods) could influence the ability of library-dependent microbial source tracking methods to correctly identify the porcine host (2, 4, 20, 26, 27, 35, 36, 38, 42, 50, 54).
In the study reported here, we examined the dynamics and characteristics of E. coli populations in fresh and in stored manure, both with respect to population structure and with respect to the frequency of multiple-antibiotic resistance. If resistance to specific antibiotic residues excreted by the animals conferred a fitness advantage to bacteria in the manure holding tank, the phenotype could be expected to be overrepresented in this habitat. Alternatively, if genes encoded resistance to antibiotics unnecessarily and imposed a fitness cost, bacteria carrying these determinants could be expected to be disadvantaged in the manure holding tank. We obtained from a single commercial farm on a monthly basis (March to August 2005) E. coli from freshly shed feces collected in the swine barn and from the farm's manure storage tank. Our specific objectives were to (i) compare the structure of E. coli populations obtained from stored and freshly shed manure by means of rep-PCR and determine how these varied with time, (ii) determine if the population from the storage lagoon differed from the population from the fresh manure with respect to the frequency and the profiles of antibiotic resistance, and (iii) determine if the distribution of antibiotic resistance profiles was associated with, or independent of, the population structure defined by rep-PCR.
MATERIALS AND METHODS
Husbandry practices and manure collection.
Characteristics of the swine farm used in this study were described by Lu et al. (32). The farm used in this study is a farrow-to-finish operation consisting of approximately 2,000 animals. The animals received a feed mix consisting of corn and soybean meal. During the course of the study described here, the animals received the following antibiotics. Nursery pigs received a growth promotion level of lincomycin and spectinomycin (Linco-Spectrin), and finishing pigs received 40 g tonne−1 (40 ppm) of tylosin phosphate (Tylan) in their feed. Penicillin G was added to the water for 2 weeks after the animals were moved from the nursery to the finishing pens. Oxytetracycline was fed to the nursing sows (330 g tonne−1; 330 ppm) in January and April and to the dry sows (550 g tonne−1; 550 ppm) in April.
The farm was sampled on a monthly basis from March to August 2005. Sampling individual animals in the barn was not possible due to the number of individuals per pen and the type of pens used in the barn. In order to obtain a sample that was as representative as possible of the entire herd, about 100 g of feces was collected on the ground of one holding pen of each room (n = 18; there were six pens per room, and the number of pigs per pen varied approximately between 10 and 30) of the barn and mixed together. Then approximately 2.5 g of fecal material of each pen was pooled and thoroughly mixed in sterile bottles with sterile sodium metaphosphate buffer (pH 6.8; 2 g per liter) to yield a composite sample. Slurry from within the barn fell through slats to an open pit below. Material from the pit was pumped from below the barn every few days to the manure holding tank, a large concrete reservoir open to the air. The holding tank was emptied during the week before the May sampling, and only a thick layer of sludge at the bottom remained until August. Samples from the manure holding tank were collected from a depth of about 0.5 m below the surface and 0.5 m from the bottom of the tank (Sludge Judge Ultra sampler; NASCO Canada, Aurora, Ontario, Canada) and when possible (April, May, and August) in three different locations around the tank and pooled in 1-liter sterile bottles (Systems Plus, Woodstock, Ontario, Canada).
Regional climate data during the experiment were obtained from Environment Canada (http://www.climate.weatheroffice.ec.gc.ca/climateData/canada_e.html).
E. coli enumeration, isolation, and identification.
Isolation of E. coli was performed as previously described (32). Briefly, samples were serially diluted in sterile sodium metaphosphate buffer and spread plated on mFC-BCIG agar (8), made with mFC basal agar (Difco, Fisher Scientific, Ottawa, Ontario, Canada) and 100 μg 5-bromo-4-chloro-3-indolyl β-d-glucuronide cyclohexyl ammonium salt (Medox Diagnostics, Ottawa, Ontario, Canada) per liter and then restreaked twice on Luria-Bertani (LB) agar (Difco, Fisher Scientific) (8). Isolates were considered E. coli if they grew at 44.5°C, had a positive reaction for β-glucuronidase (blue color on mFC-BCIG agar), fermented lactose, and produced indole. Isolates confirmed to be E. coli were inoculated into sterile 96-well microtiter plates containing 100 μl well−1 of LB broth and incubated overnight at 37°C. Sterile glycerol (Sigma-Aldrich Canada Ltd., Mississauga, Ontario, Canada) was then added to each well at a final concentration of 15% (vol/vol), and the plates were stored at −70°C. Approximately 400 isolates were picked from each sample, when populations allowed it (see Table 2).
TABLE 2.
Antibiotic resistance of E. coli isolates from fresh feces and stored manure
| Antibiotic | % Resistant isolates (fresh feces/stored)a
|
||||||
|---|---|---|---|---|---|---|---|
| March (396/1001) | April (383/352) | May (383/476) | June (378/172) | July (341/142) | August (312/350) | Total (2,193/2,475) | |
| Am | 73.0/86.2* | 95.3/74.7* | 100.0/88.2* | 37.8/72.1* | 52.5/91.1* | 78.8/43.4* | 73.2/78.2* |
| Ce | 0.8/1.0 | 7.0/11.4* | 6.5/8.2 | 0.3/1.2 | 0.0/5.6* | 0.0/0.3 | 2.6/4.0* |
| Sm | 42.2/13.4* | 79.6/42.9* | 84.3/45.4* | 36.8/48.8* | 26.7/46.0* | 47.1/21.1* | 53.4/28.9* |
| Te | 98.5/66.9* | 99.7/96.0* | 100.0/92.9* | 100.0/100.0 | 99.7/100.0 | 98.7/96.6 | 99.5/84.2* |
| Cl | 37.9/4.7* | 74.7/12.2* | 90.6/10.3* | 7.4/55.8* | 9.4/56.5* | 16.7/6.3* | 40.8/13.2* |
| Na | 0.0/0.0 | 0.0/0.3 | 0.5/1.5 | 0.3/0.0 | 0.0/3.2* | 0.0/0.0 | 0.1/0.5 |
| Ak | 0.0/0.0 | 0.0/0.0 | 0.0/0.2 | 0.0/0.0 | 0.0/3.2* | 0.0/0.0 | 0.0/0.2 |
| Ka | 26.3/5.7* | 11.7/18.2* | 43.7/3.8* | 3.5/9.3* | 3.5/29.8* | 29.7/7.7* | 19.8/8.9* |
| Tm | 59.6/41.9* | 36.9/43.8 | 39.4/56.5* | 52.0/46.5 | 27.1/51.6* | 17.1/37.4* | 39.7/45.1* |
| Su | 98.5/47.9* | 76.8/50.0* | 95.8/57.3* | 48.8/64.5* | 40.7/79.0* | 42.3/44.6 | 68.7/52.2* |
| Ct | 51.3/25.1* | 5.5/30.7* | 13.9/41.1* | 3.7/26.7* | 7.1/8.1 | 1.6/11.7* | 14.7/26.3* |
Total numbers of isolates are in parentheses. *, values for fresh feces and the stored manure populations are significantly different.
Determination of antibiotic resistance.
Microtiter plates containing 100 μl of Mueller-Hinton broth (Difco, Fisher Scientific) were inoculated with bacteria from frozen glycerol stock and were grown statically for 16 to 24 h at 37°C. An aliquot of 4 μl was then transferred with a floating pin replicator (VP Scientific, San Diego, CA) into microtiter plates containing 200 μl of a sterile 0.02% solution of Tween 20 to improve the wetting of the replicator in sterile Milli-Q water. Using the 96-pin replicator, the cell suspension (experimentally adjusted to yield about 104 CFU per spot) was spotted onto a series of Mueller-Hinton agar plates containing one of the following antibiotics at the indicated breakpoint concentrations (μg ml−1): amikacin (Ak), 64; ampicillin (Am), 32; cephalothin (Ce), 32; chloramphenicol (Cl), 32; kanamycin (Ka), 64; nalidixic acid (Na), 32; streptomycin (Sm), 64; sulfamethoxazole (Su), 512; trimethoprim (Tm), 16; trimethoprim-sulfamethoxazole (Ct), 4 to 76; or tetracycline (Te), 16. The breakpoint were specified by the National Antimicrobial Resistance Monitoring System (NARMS) (9) or the Société Française de Microbiologie Comité de l'antibiogramme (for trimethoprim) (48). The plates were incubated at 37°C for 20 to 24 h, and growth was scored by eye. Isolates were considered resistant to each antibiotic when growth at that antibiotic's breakpoint concentration was not limited to visibly isolated colonies. Isolates resistant to 4 or more antibiotics were considered multiresistant. The repeatability and validity of the method was evaluated using E. coli strain ATCC 25922.
Serotyping.
Representative isolates were sent to the Laboratory for Food-borne Zoonoses, Public Health Agency of Canada (Guelph, Ontario, Canada), for serotyping by standard protocols (39).
BOX PCR fingerprinting.
Cell suspensions of E. coli were prepared by inoculating 100 μl of fresh LB broth per well in a sterile 96-well microtiter plate with frozen stock cultures. Cells were grown statically at 37°C overnight and centrifuged at 710 × g for 25 min (Centra CL3 microplate centrifuge; Thermo IEC, Needham Heights, MA). The cells were resuspended in 100 μl of sterile Milli-Q H2O and agitated at 1,000 rpm with a microplate shaker (Sarstedt, Montréal, QC, Canada) for 5 min. The resuspended cells were used directly as a template for the PCR or frozen at −20°C until required. Rep-PCR fingerprinting was done with the BOXA1R primer as described by Versalovic et al. (55). The final reaction mix (25 μl) consisted of 1× PCR buffer (Promega, Madison, WI), 1.5 mM MgCl2, 1% dimethyl sulfoxide, 200 μM of each deoxynucleoside triphosphate (Invitrogen, Burlington, Ontario, Canada), 2 μM of the primer BOXA1R, 1 U of Taq polymerase (Promega), and 2 μl of suspended E. coli cells as the template. Amplification was performed with a Thermo MBS Satellite 0.2 Thermocycler instrument (VWR International, Mississauga, Ontario, Canada) as follows: after an initial denaturation at 94°C for 10 min, 34 cycles of denaturation (94°C, 3 s), (92°C, 30 s), annealing (50°C, 1 min), and extension (65°C, 8 min) were performed, followed by a final extension (65°C, 8 min). Six microliters of loading dye was added to 25 μl of PCR product, and 7 μl of this mixture was loaded into wells prepared with an 8-mm by 1-mm comb tooth size. Every eighth well received the MassRuler DNA ladder (Fermentas, Burlington, Ontario, Canada). PCR products were resolved by horizontal gel electrophoresis (2.5 V/cm for 16 h) in 1× Tris-borate-EDTA buffer. The gel was stained with 1 μg ml−1 ethidium bromide solution for 10 min and destained in Milli-Q water for 10 min. Gel images were captured as 16-bit TIFF images, using Alphaease FC software and an Alpha Innotech digital gel documentation system (Fisher Scientific, Ottawa, Ontario, Canada).
Computer-assisted image and data analysis.
Normalization of gel images and assignment of fingerprints to isolates were done with the Bionumerics software package (version 4.5; Applied Maths, Kortrijk, Belgium) as published earlier (32). Filtering and background subtraction were optimized for each image independently according to methods available at http://www.ecolirep.umn.edu/addinggelimages.shtml. Positions of fingerprints on gels were normalized using the MassRuler DNA ladder as the external standard in the range of 400 bp to 4,000 bp. The assignment of strains to different clusters was performed by calculating the similarity coefficients with the curve-based Pearson similarity coefficient. Similarity trees were generated using the unweighted-pair group method using average linkage. Repeated experiments where the same isolate was amplified with BOX primers and run on different gels under similar conditions consistently showed an average similarity of 80% in our laboratory. Hence, clusters were initially assigned using the software on the basis of 80% similarity, and the final assignments were determined on the basis of careful visual inspection.
All data were grouped in an Excel database and used to perform basic statistical analyses. The chi-square test was used for the analysis of the distribution of antibiotic resistances in the different subsets of the collection. Associations were considered significant when P was <0.05. The diversity captured in the E. coli collections was estimated by rarefaction analysis using the analytical approximation algorithm of Hurlbert (23) and 95% confidence intervals estimated as described by Heck et al. (21). The calculations were carried out on a random subsample (n = 84) from each monthly sample to prevent sensitivity of the calculation to the size of the sample. The isolates were individually assigned a pseudorandom number between 1 and 10000 using Excel, and the 84 isolates with the lowest values were used for the calculation. Calculations were performed with the freeware program Analytical Rarefaction 1.3, available at http://www.uga.edu/strata/software/. Curves were plotted using SigmaPlot (version 9.1; SPSS Inc., Chicago, IL). The asymptotes of the rarefaction curves were estimated using the Michaelis-Menten equation, which is available in SigmaPlot as the one-site saturation ligand model (22). The asymptote is a measure of richness at sampling saturation and was used to estimate the fraction of total community diversity captured within the E. coli collections. The SigmaPlot curve fitter uses the Marquardt-Levenberg algorithm to find the coefficients that give the best fit between the equation and the data (33).
The Shannon-Wiener and Simpson diversity indices for populations of E. coli obtained from each manure sample were evaluated using randomly picked isolates (n = 84) from each monthly sample. The isolates were individually assigned a pseudorandom number between 1 and 1000 using Excel, and the 84 isolates with the lowest values were used for the calculation. The number of individuals sampled was normalized to match the smallest sample to account for the sensitivity to the sample size of both diversity indices. Diversity indices were determined with the software calculator available at http://www.changbioscience.com/genetics/shannon.html. Confidence intervals were calculated according to Grundmann et al. (19).
Significance of differences between distribution of genotypes in the aggregated populations were determined by the method described by Kropf et al. (29) using the abundance of all the genotypes in each sample as the unit of comparison.
RESULTS
In this study, the average population size of viable E. coli in fresh feces was 1.4 × 107 ± 1.0 × 107 cells g (wet weight)−1 (n = 4, May to August). The average population size in the stored manure slurry was 1.2 × 104 ± 0.2 × 104 ml−1 in March, April, and May, which had average monthly air temperatures of −2.3°C, 7.8°C, and 11.8°C, respectively. The average population size in the stored manure slurry declined to 1.5 × 103 ± 1.8 × 103 ml−1 in June through August, when the average monthly air temperature was 21.8 ± 0.5°C.
E. coli isolates obtained from fresh feces and within the farm's manure holding tank (stored manure) were fingerprinted by means of rep-PCR, and rarefaction curves were used to estimate the abundance of genotypes within the collections (Table 1). The rarefaction data were fitted with the Michaelis-Menten equation and used to estimate the asymptote (saturation of richness) and the number of isolates required to capture half of the diversity. The Michaelis-Menten fit with the experimental data was excellent (r2 > 0.91), and the estimated saturation of richness indicated that between 48% and 79% of the diversity of the collections were captured. Nine (June) to 35 (April) distinct genotypes were detected in the fresh feces, and 17 (August) to 63 (April) were detected in the stored manure. Diversity (expressed as the Shannon-Wiener or Simpson indices or by predicted number of genotypes) was consistently greater in the stored manure than in the fresh feces in March through June (differences were significant in March, May, and June). This was not the case in July and August, when the diversity in the stored manure declined dramatically to be significantly smaller in August. The diversity of E. coli in the fresh feces was much lower in June than in any other month.
TABLE 1.
Estimates of genotypic diversity and total richness in E. coli populations
| Origin | Month (no. of isolates) | Shannon-Wiener indexa | Simpson index (1/D)a | r2a,b | Genotypes
|
No. of isolates to capture 50% of predicted genotypes (mean ± SD)a,d | ||
|---|---|---|---|---|---|---|---|---|
| Predicted no. (mean ± SD)a,c | Detected
|
|||||||
| No.a | % of predicted value | |||||||
| Fresh feces | March (188) | 1.93 | 4.34 | 0.97 | 30 ± 2 | 18 | 60 | 64 ± 8 |
| April (187) | 2.84 | 11.06 | 0.97 | 50 ± 2 | 29 | 58 | 65 ± 4 | |
| May (168) | 2.71 | 10.50 | 0.95 | 36 ± 1 | 24 | 67 | 43 ± 2 | |
| June (94) | 1.11 | 1.94 | 0.92 | 11 ± 0.5 | 9 | 79 | 29 ± 3 | |
| July (93) | 2.92 | 13.36 | 0.95 | 41 ± 0.4 | 27 | 66 | 44 ± 1 | |
| August (84) | 2.64 | 10.86 | 0.92 | 26 ± 0.2 | 20 | 76 | 27 ± 0.5 | |
| Stored manure | March (188) | 2.70 | 9.82 | 0.97 | 42 ± 1 | 26 | 62 | 57 ± 4 |
| April (237) | 3.28 | 18.57 | 0.98 | 80 ± 3 | 39 | 49 | 94 ± 5 | |
| May (127) | 3.40 | 24.16 | 0.97 | 69 ± 0.5 | 38 | 55 | 69 ± 1 | |
| June (139) | 3.18 | 17.04 | 0.97 | 59 ± 1 | 34 | 57 | 65 ± 2 | |
| July (91) | 2.46 | 8.66 | 0.91 | 23 ± 0.3 | 18 | 78 | 26 ± 1 | |
| August (91) | 2.05 | 5.02 | 0.95 | 24 ± 1 | 17 | 70 | 43 ± 4 | |
Calculated on a randomly selected subsample of equal size (n = 84) from each collection.
Coefficient of determination of goodness of fit to the Michaelis-Menten equation.
The Vmax parameter of the Michaelis-Menten equation.
The KD parameter of the Michaelis-Menten equation.
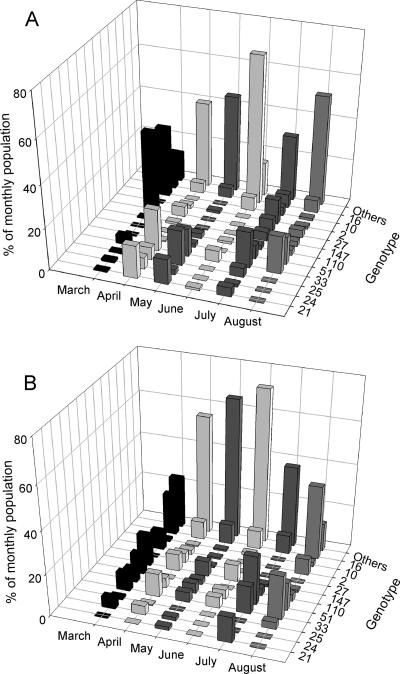
One hundred fifty distinct genotypes were identified in the collection, with 51 (34%) detected in both fresh feces and stored manure, 24 (16%) detected in fresh feces only, and 75 (50%) detected in stored manure only. When individual isolates were considered, the genotypes detected both within the fresh feces and the stored manure comprised 84% (n = 1,411), the genotypes unique to the fresh feces comprised only 4% (n = 69), and the genotypes unique to the stored manure comprised 12% (n = 208) of the total E. coli collection. More than half of the isolates (59.8% of the total collection) shared one of 11 dominant genotypes, while 54 genotypes were detected in only one isolate, and 36 were shared by only two isolates. The distribution of the genotypes between the two aggregated samples (fresh feces versus stored manure) showed that there were no significant differences between the structures of the two populations (chi-square, underrepresented genotypes aggregated; P = 0.08). The greater diversity in the stored manure than in the fresh feces during March through to June is reflected in the relative abundance of underrepresented genotypes (defined as those detected in fewer than 2% of the total collection; aggregated in Fig. 1 as “others”). The transient profound decline in diversity in E. coli obtained from fresh feces in June was associated with the dominance of a single genotype (designated genotype 16). The precipitous decline in diversity in stored manure following the June sampling was associated with a decline in underrepresented genotypes. Genotypes (e.g., 16 and 33) that were consistently detected in fresh feces were also in the stored manure. Genotypes (e.g., 2 and 24) that were detected only sporadically in fresh feces were likewise sporadically detected in stored manure.
FIG. 1.
Monthly variation in the distribution of BOX genotypes of E. coli obtained from fresh feces (A) (n = 873) and stored manure (B) (n = 730). “Others” denotes an aggregate of all fingerprints that were detected in less than 2% of all isolates in the collection.
Four representative isolates of each of the 11 dominant genotypes were serotyped. These consisted of two isolates from the fresh feces and two isolates from the stored manure each chosen from a different month to reduce the likelihood of clonality. All four isolates from genotype 27 had the same serotype (O139:NM), whereas all the other genotypes exhibited little or no homogeneity in the expressed antigens. Other serotypes detected within the collection included O116:H30, O101:NM, O41:H32, O178:H32, OR:H48, O86:H51, O154:H48, O8:H49, O98:H39, OR:H12, O88:H12, O2:NM, O139:NM, O101:NM, O153:NM, O140:H32, O154:H25, O21:H25, O51:NM, O99:NM, O51:NM, and O8:H9. The serotypes of 11 isolates could not be determined.
Over the entire experiment, the frequencies of resistance to specific antibiotics in the fresh feces collection (n = 2,193) were not significantly different from those in the stored manure collection (n = 2,475), due to the very significant monthly (n = 6) variations. These frequencies (fresh feces and holding tank [mean ± standard deviation]) were as follows: Te, 99% ± 1% and 84% ± 12%; Su, 69% ± 32% and 52% ± 13%; Am, 73% ± 24% and 78% ± 18%; Sm, 53% ± 24% and 29% ± 15%; Tm, 40% ± 38% and 45% ± 7%; Cl, 41% ± 36% and 13% ± 25%; Ka, 20% ± 11% and 9% ± 10%; Ct, 15% ± 7% and 26% ± 12%; Na, 0.1% ± 0.2% and 0.5% ± 1.3%; Ce, 3% ± 3% and 4% ± 5%; and Ak, 0% and 0.2% ± 1.3%. However, when considered on a monthly basis, the resistance to a number of antibiotics varied widely and dynamically (Table 2). Perhaps most striking were the trends for Am, Sm, and Cl resistance frequency in the fresh feces collection. The frequencies increased from March through May, when almost all of the isolates were resistant to these antibiotics. Resistance decreased abruptly in June and thereafter increased through August. The June collection also had lower frequencies of resistance to Ka and Su, but trends for these antibiotics were less coherent during the experiment. The lowest frequencies of resistance to the antibiotics in the holding tank populations were detected in March and August. Resistance to Ce, Na, and Ak remained low throughout the experiment, and Te resistance remained uniformly high. About half the isolates were resistant to Tm through the experiment. There were no consistent differences with respect to frequency of resistance in populations from the fresh feces and the holding tank. In the March-to-May period, in 17 (74%) of the 23 instances where there was a significant difference with respect to frequency of resistance to an antibiotic, it was higher in the fresh feces collection. In contrast, in the June-to-August period, in only 4 (19%) of the 21 instances of a significant difference was it higher in the fresh-feces collection.
One hundred eighty-eight resistance phenotypes representing combinations of resistance to up to nine antibiotics were found in the collection (Table 3). Eighty-seven distinct phenotypes (47% of the 188 resistance phenotypes) were detected in both the fresh feces and the stored manure. Only 22 phenotypes (12%) were found only in the fresh feces, and these represented only 35 isolates (0.75% of the collection). There were 79 (42%) profiles specific for the stored manure, representing 244 isolates (5.2% of all isolates). Fourteen (7.5%) phenotypes, each representing at least 2% of the total collection, accounted for 57% of the total collection. Very few isolates were resistant to no antibiotics or to more than eight antibiotics. There was no relationship between rep-PCR genotype and antibiotic resistance pattern: isolates from any one genotype had a wide variety of resistance phenotypes (data not shown). Trends in temporal variation were again highlighted by the June transition in the fresh feces population. Notably, in May, 28.7% of the population had the resistance phenotype AmClSmSuTe, and 11% were AmClSmSuTeTm. In June and thereafter, those phenotypes never represented more than 5.1% of the isolates. Phenotypes that were previously infrequently observed or undetected in the fresh feces population were obtained in June, namely, AmSuTe (12.4%), TeSu (19%), and SmTeTm (22.2%). In June, July, and August, the previously rare Te and AmTe phenotypes were prominent. There were no obvious trends or significant transitions in the holding tank population. The most consistently detected phenotypes in the holding tank were AmTe and AmTeTm.
TABLE 3.
Monthly variation in the frequency of antibiotic resistance phenotypes in E. colia
| Resistance pattern | % of population (fresh feces/stored manure) with pattern in:
|
|||||
|---|---|---|---|---|---|---|
| March | April | May | June | July | August | |
| Am | 0.0/14.0 | 0.0/0.0 | 0.0/1.9 | 0.0/0.0 | 0.0/0.0 | 0.3/0.0 |
| Te | 0.0/2.6 | 0.0/4.8 | 0.0/3.2 | 6.9/7.6 | 18.8/0.0 | 6.4/19.4 |
| AmTe | 0.3/13.1 | 2.1/7.7 | 0.5/10.9 | 4.0/1.7 | 14.1/3.2 | 15.7/7.4 |
| TeSu | 18.7/1.6 | 0.3/1.1 | 0.0/0.4 | 19.0/1.7 | 8.8/0.8 | 1.6/6.9 |
| AmSmTe | 0.0/1.4 | 1.6/9.7 | 0.0/5.9 | 0.3/1.2 | 3.5/0.0 | 11.2/1.1 |
| AmSuTe | 9.8/6.3 | 1.8/2.3 | 1.0/3.4 | 12.4/7.0 | 7.0/2.4 | 2.9/7.1 |
| AmTeTm | 0.3/7.8 | 1.8/2.8 | 0.0/4.4 | 4.0/1.7 | 8.2/5.6 | 5.1/7.7 |
| SmTeTm | 0.0/0.2 | 0.0/0.3 | 0.0/0.2 | 22.2/0.0 | 0.6/0.0 | 0.0/1.1 |
| AmSmSuTe | 0.5/2.3 | 6.0/2.6 | 2.1/3.2 | 1.6/4.1 | 3.5/0.8 | 3.5/4.3 |
| AmClSmSuTe | 1.3/1.2 | 32.9/0.6 | 28.7/1.7 | 0.5/5.2 | 4.4/8.1 | 5.1/1.1 |
| AmCtSuTeTm | 5.3/9.3 | 0.0/4.5 | 0.0/9.5 | 0.5/2.9 | 1.2/0.8 | 0.0/2.3 |
| AmClSmSuTeTm | 0.5/0.3 | 10.4/0.3 | 11.0/0.4 | 1.3/2.3 | 0.9/10.5 | 0.0/0.3 |
| AmCtSmSuTeTm | 5.8/1.7 | 0.0/5.4 | 0.0/18.5 | 0.0/2.3 | 2.9/0.0 | 0.3/2.0 |
| AmClCtSmSuTeTm | 15.4/0.2 | 3.1/1.1 | 2.1/2.1 | 0.3/8.1 | 0.6/0.8 | 0.0/0.3 |
| Othersb | 42.2/38.1 | 39.9/56.8 | 54.6/34.5 | 27.0/54.1 | 25.5/66.9 | 47.8/38.9 |
Sample sizes are identical to those used for Table 2.
Aggregate of all isolates with phenotypes that were each detected in less than 2% of the total collection.
DISCUSSION
It has been suggested that conditions outside the digestive tract can alter the genetic composition of E. coli populations once shed by the host, and there is some evidence to suggest that some E. coli strains may become adapted for survival in secondary habitats such as some soils and water (2, 7, 17, 24, 25, 51, 57). Were this to be the case in large-scale livestock manure slurry holding tanks, the population structure could be expected to be significantly skewed compared to the structures of populations shed by the animals, as less fit individuals perish and fitter genotypes become increasingly well represented. Swine manure slurry typically is at least 95% water, representing an approximately 50-fold dilution of freshly shed material. Recognizing that the holding tank is continuously inoculated with fresh material, and that the water content will vary with precipitation and with evaporation, there was significant attrition in the stored E. coli population, particularly in the warmer months. Throughout the experiment, the genotypes that dominated the E. coli population in freshly shed feces also dominated the community in the stored manure (Fig. 1). Fully 84% of all the isolates obtained from both the fresh feces and the holding tank shared common genotypes, and only 12% of the holding tank isolates had genotypes that were not detected in the fresh feces. Genotypes that were consistently detected in fresh feces (e.g., 16 and 33) were also similarly consistently present in the stored manure. Likewise, the same genotypes (e.g., 21 and 10) were sporadically and periodically detected in fresh feces and in the stored manure. The observed holding tank population was more diverse than the freshly shed population, except in the hot summer months of July and August (Table 1). The generally higher diversity in the holding tank is consistent with previous observations on this same farm, which had been investigated in the winter of 2003-2004 (32). Genotypes that were detected only in the manure (75 of them) could be characteristic of isolates that have superior fitness in this secondary habitat, and likewise, genotypes (24 of them) detected only in the fresh feces could be characteristic of isolates with superior fitness in the primary habitat. These genotypes (all 99) were detected in only 16% of the total collection. Furthermore, none of the 99 genotypes that were detected in only one habitat or the other represented 2% or more of the overall collections. Taken together, these results indicate that the dominant genotypes were well represented in both habitats, suggesting that these did not have a particular fitness advantage in either.
Both the fresh feces and the holding tank collections exhibited important monthly variation in the population structure (Fig. 1). This was particularly evident with respect to the importance of genotypes that were poorly represented in the collection. For example, in the holding tank, the proportion of genotypes that were detected in less than 2% of the overall collection (aggregated as “other”) steadily increased from March through to May and then gradually declined through to August. In the fresh feces, the “other” genotypes increased in frequency from March to May, were much less frequently observed in June, and then steadily increased through August. In contrast, the dynamics of the fresh feces population was the transient dominance of genotype 16 during the month of June, causing most of the underrepresented groups to drop below the detection level. The genetic composition of E. coli varies between individual animals, changes during the lifetime of the animal, and is influenced by feed composition (28, 43). We are unable to explain the May-to-June transition on the basis of any variation in husbandry (e.g., change in antibiotic regimen or feed composition), herd health (there were no clinical problems), herd composition (the proportions of animals of different ages and reproductive statuses were uniform throughout the study), or in-barn sanitation practices (e.g., an unusual use of disinfectant before sampling).
The manure holding tank was emptied during the week prior to the May sampling. The March and April samples represented waste that had accumulated since the previous autumn when the tank was last emptied, whereas in May and thereafter the material was much fresher. On this basis, we reasoned that the holding tank E. coli population in May and thereafter would more closely resemble the fresh feces population than in the previous months. This was not the case, with respect to either the population structure or the distribution of antibiotic resistance profiles. In fact, the tank is never completely emptied, and the sludge left at the bottom was undoubtedly carrying over an important preexisting population as the tank was subsequently refilled.
When the entire collection (fresh feces, n = 2,193, and holding tank, n = 2,475) was considered, there were no significant differences in the frequency of resistance to any antibiotic. However, when the collection was considered on the basis of specific rep-PCR-defined genotypes, in some cases there were significant differences in the frequency of antibiotic resistance. In 26 (21.5%) of 121 comparative observations (fresh feces versus holding tank; 11 antibiotics and 11 genotypes), the two populations differed in the frequency of resistance to an antibiotic. Of the 26 observations that showed significant differences, 21 (80.9%) revealed a lower frequency of resistance in the holding tank isolates than in the fresh-feces isolates. For example, the frequency of resistance to Cl and Sm was lower in the holding tank isolates of genotypes 16, 24, and 33 than in the fresh-feces isolates of these same genotypes (data not shown). However, the relative abundances of these three genotypes in the fresh feces and in the holding tank populations were similar (Fig. 1). Taken together, these results suggest that in some cases antibiotic resistance genes were lost in the holding tank, but this did not confer a selective advantage. Fitness in the holding tank was neutral with respect to resistance to any of the antibiotics; these attributes conferred neither a detectable advantage or disadvantage in this habitat.
Most of the antibiotics we evaluated for resistance were not used on this farm. The exceptions were oxytetracycline, which was briefly used during the experiment, and penicillin G, which was constantly administered to a portion of the herd and which could promote resistance to ampicillin. The frequency of ampicillin resistance measured on a monthly basis varied from 38% to 100% of the isolates. The very high frequency of Te resistance is consistent with what has been observed on other Ontario farms (6, 52). Overall, these results illustrate that there are factors beyond short-term on-farm antibiotic use that influence the frequency of antibiotic resistance and patterns of multiple antibiotic resistance in bacteria shed by livestock. In some cases, antibiotic resistance genes may be mobilized into environmental bacteria in soils receiving manure (18, 47). The role of environmental contamination from livestock wastes in promoting antibiotic resistance is difficult to evaluate against the background of the high frequency of resistance to antibiotics found in soil bacteria (12, 44, 45). Nevertheless, prudent use of antibiotics, particularly with respect to the chronic provision of growth-promoting agents and the use of antibiotics that are important for human and animal health, is advised (34).
The temporal flux of multiple antibiotic resistance phenotypes, particularly within the fresh feces collection, was striking, both in its tempo and in its temporal coherence (Table 3). There was no apparent relationship between resistance phenotype and rep-PCR-defined genotype. For example, during the course of the experiment, the AmSuTe phenotype was detected in organisms of 28 genotypes, the SmTeTm in 5, and the AmClSmSuTe in 28. Likewise, 65 distinct antibiotic resistance phenotypes were detected in genotype 16, 55 in genotype 33, 19 in genotype 27, and 44 in genotype 51. Clearly, variation in the frequency of various resistance phenotypes was not entirely due to the proliferation of distinct clonal populations that carried a specific complement of resistance genes. Rather, the results suggest that multiply resistant phenotypes varied in their nature and frequency of detection according to the accrual or the loss of resistance determinants. Conditions in the mammalian gastrointestinal tract are conducive to conjugal transfer of plasmid-borne antibiotic resistance genes (reviewed in reference 31). In our study, the frequencies of resistance to Cl, Su, Sm, Ka, and Am but not Te in the fresh feces collection all declined as of the June sampling. Linkage of these markers is consistent with what has previously been observed in swine isolates of E. coli, with chloramphenicol resistance being conferred by plasmid-borne cmlA (5). The cmlA gene was linked to sul3 or sul1 and to aadA1 and aadA2 in various configurations of class 1 integrons (5). Conjugative transfer of cmlA was accompanied by acquisition of resistance to Su, Te, and Ka. The AmClSmSuTe phenotype has also been identified in Salmonella and been shown to be able to be transferred between serotypes (13, 41). Overall, the genetic elements underlying our observations remain to be determined, but the mechanisms underlying instability and horizontal transfer of antibiotic resistance genes in the porcine model are well established.
Both rep-PCR and antibiotic resistance profiling have been used to ascribe host source (e.g., human, livestock, wildlife) to environmental isolates of E. coli (20, 27, 35, 36, 38, 42, 50, 54). Typically, the host source is inferred on the basis of comparative analysis of the environmental isolates with a reference collection of E. coli strains obtained from the various potential fecal sources in the study area. One factor that will influence the accuracy of source identification is the temporal fidelity of the library with respect to the attributes being evaluated and compared. Results from this study suggest that rep-PCR fingerprints generated from a library constructed from a commercial swine manure storage facility would remain representative of the population structure over a period of at least several months. However, the frequency of resistance to specific antibiotics varied widely on a monthly basis, supporting previous findings that this temporal variability must be captured in the library construction (2, 4, 26, 58). These findings are consistent with the chromosomal location and apparent stability of repeated sequences detected by PCR with the BOXA1R primer and with the frequent association of antibiotic resistance determinants with potentially unstable plasmids, integrons, and transposons (1, 17, 37, 46, 56).
Acknowledgments
This research was funded by the AAFC GAPS research program. P. Duriez was funded through the NSERC Visiting Fellowship in Government Laboratories program.
We sincerely thank C. and M. Bontje for access to their farm and P. Morris for veterinary advice. L. Coates provided excellent technical assistance. We thank the Laboratory for Food-borne Zoonoses (Guelph, Ontario, Canada) for serotyping isolates. We thank several anonymous reviewers for comments that significantly improved the manuscript.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Agerso, Y., and D. Sandvang. 2005. Class 1 integrons and tetracycline resistance genes in Alcaligenes, Arthrobacter, and Pseudomonas spp. isolated from pigsties and manured soil. Appl. Environ. Microbiol. 71:7941-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. A., J. E. Whitlock, and V. J. Harwood. 2006. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 72:6914-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2003. Assessing microbial safety of drinking water: improving approaches and methods. OECD, WHO, Paris, France.
- 4.Aslam, M., F. Nattress, G. Greer, C. Yost, C. Gill, and L. McMullen. 2003. Origin of contamination and genetic diversity of Escherichia coli in beef cattle. Appl. Environ. Microbiol. 69:2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff, K. M., D. G. White, M. E. Hume, T. L. Poole, and D. J. Nisbet. 2005. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 243:285-291. [DOI] [PubMed] [Google Scholar]
- 6.Boerlin, P., R. Travis, C. L. Gyles, R. Reid-Smith, N. J. H. Lim, V. Nicholson, S. A. McEwen, R. M. Friendship, and M. Archambault. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71:6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 8.Ciebin, B. W., M. H. Brodsky, R. Eddington, G. Horsnell, A. Choney, G. Palmateer, A. Ley, R. Joshi, and G. Shears. 1995. Comparative evaluation of modified m-FC and m-TEC media for membrane filter enumeration of Escherichia coli in water. Appl. Environ. Microbiol. 61:3940-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CIPARS. 2005. Canadian Integrated Program for Antimicrobial Resistance Surveillance annual report (2003). Health Canada, Ottawa, Canada.
- 10.Committee on Indicators for Waterborne Pathogens, NRC. 2004. Indicators for waterborne pathogens. National Academy of Sciences, Washington, DC.
- 11.Cote, C., A. Villeneuve, L. Lessard, and S. Quessy. 2006. Fate of pathogenic and nonpathogenic microorganisms during storage of liquid hog manure in Quebec: biosecurity of livestock effluents. Livestock Sci. 102:204-210. [Google Scholar]
- 12.D'Costa, V. M., K. M. McGrann, D. W. Hughes, and G. D. Wright. 2006. Sampling the antibiotic resistome. Science 311:374-377. [DOI] [PubMed] [Google Scholar]
- 13.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 14.Edberg, S. C., E. W. Rice, R. J. Karlin, and M. J. Allen. 2000. Escherichia coli: the best biological drinking water indicator for public health protection. J. Appl. Microbiol. 88:106S-116S. [DOI] [PubMed] [Google Scholar]
- 15.Federal-Provincial-Territorial Committee on Drinking Water. March 2007, posting date. Guidelines for Canadian drinking water quality. Health Canada. http://www.hc-sc.gc.ca/ewh-semt/alt_formats/hecs-sesc/pdf/pubs/water-eau/doc-sup-appui/sum_guide-res_recom/summary-sommaire_e.pdf.
- 16.Fewtrell, L., and J. Bartram. 2001. Water quality—guidelines, standards and health: assessment of risk and risk management for water-related infectious disease. World Health Organization, London, United Kingdom.
- 17.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 18.Götz, A., and K. Smalla. 1997. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl. Environ. Microbiol. 63:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood, V. J., B. Wiggins, C. Hagedorn, R. D. Ellender, J. Gooch, J. Kern, M. Samadpour, A. C. H. Chapman, B. J. Robinson, and B. C. Thompson. 2003. Phenotypic library-based microbial source tracking methods: efficacy in the California collaborative study. J. Water Health 1:153-166. [PubMed] [Google Scholar]
- 21.Heck, K. L., Jr., G. van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 22.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 24.Ishii, S., D. L. Hansen, R. E. Hicks, and M. J. Sadowsky. 2007. Beach sand and sediments are temporal sinks and sources of Escherichia coli in Lake Superior. Environ. Sci. Technol. 41:2203-2209. [DOI] [PubMed] [Google Scholar]
- 25.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins, M. B., P. G. Hartel, T. J. Olexa, and J. A. Stuedemann. 2003. Putative temporal variability of Escherichia coli ribotypes from yearling steers. J. Environ. Qual. 32:305-309. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katouli, M., A. Lund, P. Wallgren, I. Kuhn, O. Soderlind, and R. Mollby. 1995. Phenotypic characterization of intestinal Escherichia coli of pigs during suckling, postweaning, and fattening periods. Appl. Environ. Microbiol. 61:778-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kropf, S., H. Heuer, M. Gruning, and K. Smalla. 2004. Significance test for comparing complex microbial community fingerprints using pairwise similarity measures. J. Microbiol. Methods 57:187-195. [DOI] [PubMed] [Google Scholar]
- 30.Leung, K. Y., and E. Topp. 2001. Bacterial community dynamics in liquid swine manure during storage: molecular analysis using DGGE/PCR of 16S rDNA. FEMS Microbiol. Ecol. 38:169-177. [Google Scholar]
- 31.Licht, T. R., and A. Wilcks. 2005. Conjugative gene transfer in the gastrointestinal environment. Adv. Appl. Microbiol. 58C:77-95. [DOI] [PubMed] [Google Scholar]
- 32.Lu, Z., D. Lapen, A. Scott, A. Dang, and E. Topp. 2005. Identifying host sources of fecal pollution: diversity of Escherichia coli in confined dairy and swine production systems. Appl. Environ. Microbiol. 71:5992-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marquardt, D. W. 1963. An algorithm for least squares estimation of parameters. J. Soc. Ind. Appl. Math. 11:431-441. [Google Scholar]
- 34.McEwen, S. A. 2006. Antibiotic use in animal agriculture: what have we learned and where are we going? Anim. Biotechnol. 17:239-250. [DOI] [PubMed] [Google Scholar]
- 35.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murinda, S. E., P. D. Ebner, L. T. Nguyen, A. G. Mathew, and S. P. Oliver. 2005. Antimicrobial resistance and class 1 integrons in pathogenic Escherichia coli from dairy farms. Foodborne Pathog. Dis. 2:348-352. [DOI] [PubMed] [Google Scholar]
- 38.Myoda, S. P., C. A. Carson, J. J. Fuhrmann, B. K. Hahm, P. G. Hartel, I. H. Yampara, L. Johnson, R. L. Kuntz, C. H. Nakatsu, M. J. Sadowsky, and M. Samadpour. 2003. Comparison of genotypic-based microbial source tracking methods requiring a host origin database. J. Water Health 1:167-180. [PubMed] [Google Scholar]
- 39.Ørskov, F., and I. Ørskov. 1984. Serotyping of E. coli. Methods Microbiol. 14:43-112. [Google Scholar]
- 40.Peu, P., H. Brugere, A.-M. Pourcher, M. Kerouredan, J.-J. Godon, J.-P. Delgenes, and P. Dabert. 2006. Dynamics of a pig slurry microbial community during anaerobic storage and management. Appl. Environ. Microbiol. 72:3578-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poppe, C., N. Smart, R. Khakhria, W. Johnson, J. Spika, and J. Prescott. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559-565. [PMC free article] [PubMed] [Google Scholar]
- 42.Ritter, K. J., E. Carruthers, C. A. Carson, R. D. Ellender, V. J. Harwood, K. Kingsley, C. Nakatsu, M. J. Sadowsky, B. Shear, B. West, J. E. Whitlock, B. A. Wiggins, and J. D. Wilbur. 2003. Assessment of statistical methods used in library-based approaches to microbial source tracking. J. Water Health 1:209-223. [PubMed] [Google Scholar]
- 43.Russell, J. B., F. Diez-Gonzalez, and G. N. Jarvis. 2000. Effects of diet shifts on Escherichia coli in cattle. J. Dairy Sci. 83:863-873. [DOI] [PubMed] [Google Scholar]
- 44.Schmitt, H., K. Stoob, G. Hamscher, E. Smit, and W. Seinen. 2006. Tetracyclines and tetracycline resistance in agricultural soils: microcosm and field studies. Microb. Ecol. 51:267-276. [DOI] [PubMed] [Google Scholar]
- 45.Sengelov, G., Y. Agerso, B. Hallingsorensen, S. B. Baloda, J. S. Andersen, and L. B. Jensen. 2003. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 28:587-595. [DOI] [PubMed] [Google Scholar]
- 46.Seurinck, S., W. Verstraete, and S. D. Siciliano. 2003. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl. Environ. Microbiol. 69:4942-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smalla, K., H. Heuer, A. Gotz, D. Niemeyer, E. Krogerrecklenfort, and E. Tietze. 2000. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 66:4854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Société Française de Microbiologie, Comité de l'antibiogramme. 2004. Recommandations du CA-SFM Jan 2004. Société Française de Microbiologie, Paris, France.
- 49.Statistics Canada. 2001. Farm environmental management survey. Collection registration: STC/AGR—450-75054. Statistics Canada, Ottawa, Canada.
- 50.Stoeckel, D. M., M. V. Mathes, K. E. Hyer, C. Hagedorn, H. Kator, J. Lukasik, T. L. O'Brien, T. W. Fenger, M. Samadpour, K. M. Strickler, and B. A. Wiggins. 2004. Comparison of seven protocols to identify fecal contamination sources using Escherichia coli. Environ. Sci. Technol. 38:6109-6117. [DOI] [PubMed] [Google Scholar]
- 51.Topp, E., M. Welsh, Y.-C. Tien, A. Dang, G. Lazarovits, K. Conn, and H. Zhu. 2003. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiol. Ecol. 44:303-308. [DOI] [PubMed] [Google Scholar]
- 52.Travis, R. M., C. L. Gyles, R. Reid-Smith, C. Poppe, S. A. McEwen, R. Friendship, N. Janecko, and P. Boerlin. 2006. Chloramphenicol and kanamycin resistance among porcine Escherichia coli in Ontario. J. Antimicrob. Chemother. 58:173-177. [DOI] [PubMed] [Google Scholar]
- 53.U.S. Environmental Protection Agency. 2000. Improved enumeration methods for the recreational water quality indicator: enterococci and Escherichia coli. Document EPA/821/R-97/004. Office of Science and Technology, Washington, DC.
- 54.U.S. Environmental Protection Agency. 2005. Microbial source tracking guide. Document EPA-600/R-05/064. Office of Research and Development, Washington, DC.
- 55.Versalovic, J., M. Schneider, F. J. De Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 56.Versalovic, J., T. Koeuth, and R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitman, R. L., M. B. Nevers, and M. N. Byappanahalli. 2006. Examination of the watershed-wide distribution of Escherichia coli along southern Lake Michigan: an integrated approach. Appl. Environ. Microbiol. 72:7301-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiggins, B. A., P. W. Cash, W. S. Creamer, S. E. Dart, P. P. Garcia, T. M. Gerecke, J. Han, B. L. Henry, K. B. Hoover, E. L. Johnson, K. C. Jones, J. G. McCarthy, J. A. McDonough, S. A. Mercer, M. J. Noto, H. Park, M. S. Phillips, S. M. Purner, B. M. Smith, E. N. Stevens, and A. K. Varner. 2003. Use of antibiotic resistance analysis for representativeness testing of multiwatershed libraries. Appl. Environ. Microbiol. 69:3399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]