Abstract
A loop-mediated isothermal amplification (LAMP) procedure for the detection of Cryptosporidium in environmental and fecal samples was developed and evaluated. This is the first demonstration of LAMP applied to detection of Cryptosporidium. Due to its specificity and simplicity, the method could become a useful diagnostic tool for epidemiologic studies of Cryptosporidium presence.
Cryptosporidiosis is a disease of major public health concern caused by several genotypically and phenotypically diverse Cryptosporidium species. The role of water and food in the epidemiology of this disease is now well recognized (3, 5). Cryptosporidium hosts excrete large numbers of infective, transmissive stages (oocysts) in feces. Apart from the difficulties of isolation, detection of Cryptosporidium from environmental samples by currently available methods remains difficult and costly, restricting their use by diagnostic laboratories. Loop-mediated isothermal amplification (LAMP), developed originally by Notomi et al. (10), is a novel method that amplifies DNA with high specificity, efficiency, and rapidity under isothermal conditions. It is based on autocycling strand displacement DNA synthesis facilitated by a Bst DNA polymerase (10). The primer architecture as applied with appropriate DNA polymerase generates, as described originally by Notomi et al., “stem-loop DNAs with several inverted repeats of the target and cauliflower-like structures with multiple loops” (10). These are double-stranded DNA fragments in multiples of a given length producing characteristic ladder banding patterns when electrophoresed. The LAMP method can amplify a few copies of DNA to 109 copies in less than an hour under isothermal conditions. In LAMP, the use of four primers that recognize six sequences of the target gene at the initial stage and four during next stages eliminates nonspecific binding, thereby ensuring the specificity of LAMP. The successful development of LAMP procedures has been reported for many clinical applications, including viral and bacterial infections, and for diagnoses of protozoan diseases, including trypanosomiasis (8, 11) and both canine and equine piroplasmosis (1, 4).
The critical element in developing LAMP for a previously untested organism is primer target selection and design, which is facilitated by computer programs available online (http://primerexplorer.jp). A non-DNA-containing (e.g., distilled water) negative control is typically used to ensure no amplification from the reaction mixture alone. Here, we report on the development and preliminary evaluation of LAMP for detection of Cryptosporidium presence and address its possible usefulness in epidemiologic studies.
The LAMP primer set used here was designed from the 60-kDa glycoprotein (gp60) gene of Cryptosporidium parvum (GenBank accession no. AB237136), which amplifies a 189-bp product, using primer explorer software (http://primerexplorer.jp). The LAMP primer sequences are as follows: F3, 5′-TCGCAC CAG CAA ATA AGG C-3′; B3, 5′-GCCGCA TTC TTC TTT TGG AG-3′; FIP, 5′-ACCCTG GCT ACC AGA AGC TTC AGA ACT GGA GAC GCA GAA-3′; BIP, 5′-GGCCAA ACT AGT GCT GCT TCC CGT TTC GGT AGT TGC GCC TT-3′. The LAMP reaction was conducted as described previously (10). Briefly, each reaction mixture (total volume, 25 μl) contained 12.5 μl reaction buffer [40 mM Tris-HCl (pH 8.8), 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 0.2% Tween 20, 1.6 M Betaine, 2.8 mM each deoxynucleoside triphosphate], 1 μl (8 units) Bst DNA polymerase (Eiken Chemicals Co., Japan), 0.9 μl primer mixture (20 pmol each of the FIP and BIP primers, 5 pmol each of the F3 and B3 primers), 2 μl DNA, and 8.6 μl distilled water). Samples were incubated at 63°C for 60 min and then heated at 80°C for 3 min for termination of the reaction.
The PCR mixture for the F3 and B3 LAMP primers, for PCR yielding a product of 189 bp, consisted of 5 μl of 10× PCR buffer (500 mM KCl, 100 mM Tris-HCl, pH 8.3, 15 mM MgCl2, and 0.01% [wt/vol] gelatin), 200 μM of each deoxynucleoside triphosphate, 200 nM of each primer, 2.5 U of AmpliTaq Gold polymerase, and 2 μl of DNA in a total volume of 50 μl. The reaction mixture was incubated at 94°C for 10 min of denaturation and then subjected to 30 cycles at 94°C for 45 s, annealing at 60°C for 1 min, and extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min. Both LAMP and PCR products were electrophoresed in a 1.5% Tris-acetic acid-EDTA agarose gel and stained with ethidium bromide for visualization under UV light.
Double-distilled water (DDW) samples were also used as a negative control for all sensitivity test reactions. In these tests, cryptosporidial DNA was extracted by freeze-thaw cycling. Briefly, 500 μl genome extraction buffer (0.2 M NaCl, 10 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% sodium dodecyl sulfate) was added to oocyst suspensions and then vortexed, followed by 10 freeze-thaw cycles of 2 min in liquid nitrogen and 2 min at 100°C. Afterwards, 10 μl of proteinase K (final concentration, 100 μg/ml) was added and samples were incubated at 55°C overnight. The following day, DNA was extracted by phenol-chloroform-isoamyl (25:24:1) and precipitated with 1 ml of 100% ethanol and 50 μl of 1 M sodium acetate. Finally, precipitated DNA was dissolved in 30 μl of DDW. Serial DNA dilutions were prepared as follows: extracted DNA was quantified using a spectrophotometer (Ultrospec 2100) and then serially diluted 1:10 seven times from 4 μg/μl (initial concentration) down to 400 fg/μl (1:107).
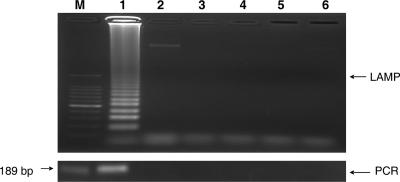
The genomic DNA used to test the LAMP primer set specificity was from C. parvum of a bovine origin that has been identified as C. parvum genotype II (referred to hereinafter as C. parvum) (data not shown). For negative controls, distilled water and the DNA of various parasites, including Trypanosoma brucei (GUTat 1.3), Babesia bovis (USDA strain), Theileria parva (Muguga strain), and Toxoplasma gondii (RH strain), as well as uninfected bovine blood DNA were used. Enteric pathogens more common in fecal or water samples but more distantly related to the target organism and thus unlikely to generate false positives are being included in continuing work. The LAMP outer primers (F3 and B3) were used for PCR with a product size of 189 bp for comparison of the two methods. Clearly, both LAMP and PCR specifically amplified Cryptosporidium DNA whereas negative-control DNA was not amplified (Fig. 1).
FIG. 1.
LAMP and PCR specificity for protozoan DNA. Lane M, 100-bp DNA ladder; lane 1, Cryptosporidium parvum of a bovine origin; lane 2, Trypanosoma brucei; lane 3, Babesia bovis; lane 4, Theileria parva; lane 5, Toxoplasma gondii; lane 6, uninfected bovine blood. Upper panel, LAMP; lower panel, PCR.
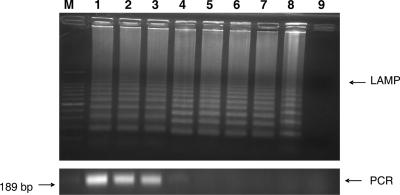
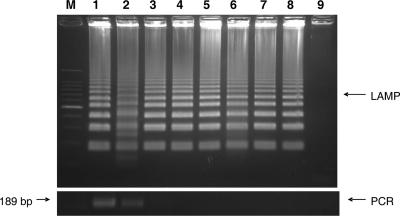
The sensitivity of this LAMP application was assessed in two ways. For the first sensitivity test, DNA was extracted from ca. 106 C. parvum oocysts purified from a bovine fecal sample. The DNA was quantified by a spectrophotometer and then serially diluted sevenfold to a minimum DNA concentration equivalent to that of 0.1 oocysts. The LAMP procedure amplified the serially diluted C. parvum DNA for each dilution from the highest sample DNA concentration of 4 μg/μl to as little as 400 fg/μl (dilution of 1:107) (Fig. 2, upper). In comparison, PCR amplified the diluted DNA only to a 1:103 dilution (Fig. 2, lower). For the second sensitivity test, 1 × 106 oocysts from the stock isolate were diluted 1:10 serially sevenfold to a minimum concentration equivalent to that of a single oocyst. Then, DNA was extracted from each oocyst dilution. The LAMP procedure amplified the DNA of C. parvum oocysts diluted to a minimum concentration equivalent to that of a single oocyst (Fig. 3, upper). In comparison, PCR amplified the same DNA only to a minimum concentration equivalent to that of 1 × 105 oocysts (Fig. 3, lower).
FIG. 2.
LAMP and PCR sensitivity for serially diluted C. parvum DNA. Lane M, 100-bp DNA ladder; lane 1, 4 μg/μl of DNA (initial); lane 2, 1:10 dilution; lane 3, 1:102 dilution; lane 4, 1:103 dilution; lane 5, 1:104 dilution; lane 6, 1:105 dilution; lane 7, 1:106 dilution; lane 8, 1:107 dilution; lane 9, DDW. Upper panel, LAMP; lower panel, PCR.
FIG. 3.
LAMP and PCR sensitivity for DNA extracted from serially diluted C. parvum oocysts. Lane M, 100-bp DNA ladder; lane 1, 106 oocysts; lane 2, 105 oocysts; lane 3, 104 oocysts; lane 4, 103 oocysts; lane 5, 102 oocysts; lane 6, 101 oocysts; lane 7,100 oocysts; lane 8,10−1 oocysts; and lane 9, DDW. Upper panel, LAMP; lower panel, PCR.
Environmental fecal and water samples previously (6) proven positive by an immunofluorescence test (Cellabs Pty. Ltd., Australia) or by PCR (data not shown) used in this application were from a variety of both geographic and host origins (1 cattle and 1 snake, Germany; 1 human and 1 cattle, Japan; 1 leopard, China; 11 cattle, Mongolia; 1 cattle, Spain; 1 cattle, Hungary; 7 river water samples, Bulgaria). LAMP detected Cryptosporidium DNA in all of the above-mentioned samples. The seven river water samples have previously been reported to be positive for Cryptosporidium presence by an immunofluorescence test (6). LAMP amplified Cryptosporidium DNA from all of the water samples tested, while PCR did not detect DNA in any of them (unpublished data). The negative PCR results could have been due to the presence of DNA polymerase inhibitors likely present in water samples (7). Such inhibitors have no effect on LAMP (2). A positive LAMP reaction can be easily detected by the white turbidity visible to the naked eye in the reaction tube (9) and/or by addition of fluorescent dyes (Fig. 4).
FIG. 4.
LAMP negative and positive controls using fluorescent detection reagent.
While further evaluation and validation of this method for detection of the presence of Cryptosporidium spp. continue in our laboratory, due to its specificity, simplicity, and cost-effectiveness, we suggest LAMP as a useful diagnostic tool for examination of Cryptosporidium spp. in samples for clinical laboratories as well as for water industries.
Acknowledgments
This study was supported by a Grant-in-Aid for Young Scientists and a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and from the 21st Century COE Program (A-1), Ministry of Education, Sports, Science, and Technology of Japan.
We thank the Department of Parasitology, University of Leipzig, Germany, for providing the Cryptosporidium parvum isolate that we used for evaluation studies.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Alhassan, A., O. M. M. Thekisoe, N. Yokoyama, N. Inoue, M. Y. Motloang, P. A. Mbati, H. Yin, Y. Katayama, T. Anzai, C. Sugimoto, and I. Igarashi. 2007. Development of loop-mediated isothermal amplification (LAMP) method for diagnosis of equine piroplasmosis. Vet. Parasitol. 143:155-160. [DOI] [PubMed] [Google Scholar]
- 2.Grab, D. J., J. Lonsdale-Eccles, and N. Inoue. 2005. LAMP for tadpoles. Nat. Methods 2:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter, P. R., and R. C. A. Thompson. 2005. The zoonotic transmission of Giardia and Cryptosporidium. Int. J. Parasitol. 35:1181-1190. [DOI] [PubMed] [Google Scholar]
- 4.Ikadai, H., H. Tanaka, N. Shibahara, A. Matsuu, M. Uechi, N. Itoh, S. Oshiro, N. Kudo, I. Igarashi, and T. Oyamada. 2004. Molecular evidence of infections with Babesia gibsoni parasites in Japan and evaluation of the diagnostic potential of a loop-mediated isothermal amplification method. J. Clin. Microbiol. 42:2465-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karanis, P., C. Kourenti, and H. Smith. 2006. Water borne transmission of protozoan parasites: a review of world-wide outbreaks and lessons we learnt. J. Water Health 5:1-38. [DOI] [PubMed] [Google Scholar]
- 6.Karanis, P., I. Sotiriadou, V. Kartashev, C. Kourenti, N. Tsvetkova, and K. Stojanova. 2006. Investigations on Giardia and Cryptosporidium in drinking water supplies of Rostov region (Southern Russia) and Sofia (Bulgaria). Environ. Res. 102:475-481. [DOI] [PubMed] [Google Scholar]
- 7.Kourenti, C., and P. Karanis. 2006. Evaluation and applicability of a purification method coupled with nested PCR for the detection of Toxoplasma gondii in water. Lett. Appl. Microbiol. 43:475-481. [DOI] [PubMed] [Google Scholar]
- 8.Kuboki, N., N. Inoue, T. Sakurai, F. Di Cello, D. J. Grab, H. Suzuki, C. Sugimoto, and I. Igarashi. 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 4:5517-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 10.Notomi, T., O. Hiroto, M. Harumi, Y. Toshihiro, K. Watanabe, A. Nobuyuki, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thekisoe, O. M. M., N. Inoue, N. Kuboki, D. Tuntasuvan, W. Bunnoy, S. Borisutsuwan, I. Igarashi, and C. Sugimoto. 2005. Evaluation of loop-mediated isothermal amplification (LAMP), PCR and parasitological tests for detection of Trypanosoma evansi in experimentally infected pigs. Vet. Parasitol. 130:327-330. [DOI] [PubMed] [Google Scholar]