Abstract
The metabolite production of lactic acid bacteria (LAB) on silage was investigated. The aim was to compare the production of antifungal metabolites in silage with the production in liquid cultures previously studied in our laboratory. The following metabolites were found to be present at elevated concentrations in silos inoculated with LAB strains: 3-hydroxydecanoic acid, 2-hydroxy-4-methylpentanoic acid, benzoic acid, catechol, hydrocinnamic acid, salicylic acid, 3-phenyllactic acid, 4-hydroxybenzoic acid, (trans, trans)-3,4-dihydroxycyclohexane-1-carboxylic acid, p-hydrocoumaric acid, vanillic acid, azelaic acid, hydroferulic acid, p-coumaric acid, hydrocaffeic acid, ferulic acid, and caffeic acid. Among these metabolites, the antifungal compounds 3-phenyllactic acid and 3-hydroxydecanoic acid were previously isolated in our laboratory from liquid cultures of the same LAB strains by bioassay-guided fractionation. It was concluded that other metabolites, e.g., p-hydrocoumaric acid, hydroferulic acid, and p-coumaric acid, were released from the grass by the added LAB strains. The antifungal activities of the identified metabolites in 100 mM lactic acid were investigated. The MICs against Pichia anomala, Penicillium roqueforti, and Aspergillus fumigatus were determined, and 3-hydroxydecanoic acid showed the lowest MIC (0.1 mg ml−1 for two of the three test organisms).
Postharvest spoilage of food and animal feed by molds and yeasts is a problem worldwide. Apart from giving the food and feed an unpleasant smell, taste, or appearance, these fungi may also produce a wide array of mycotoxins, making the food and animal feed unsuitable for consumption. One traditional way of controlling pathogenic fungi in food and animal feed is to use lactic acid bacteria (LAB) to produce a variety of fermented foodstuffs, including sauerkraut and a multitude of dairy products, as well as silage for use as animal feed (27). This biological approach to control spoilage fungi is particularly interesting because of the general desire to reduce the use of chemical preservatives and fungicides in the food and feed industries, as well as in agriculture. Moreover, many LAB are generally regarded as safe, which makes these organisms particularly suitable for use in food and animal feed applications. In silage making, often the LAB naturally present in the crop are enough to initiate fermentation. However, the use of a biological silage additive will ensure that a high enough number of LAB always is present. With an additive, it is also possible to use strains with desired properties, such an antifungal activity, to further improve the final product.
The antimicrobial activity of LAB is commonly explained by the synthesis of small organic acids such as lactic, acetic, and formic acids, which may exert the biological effect either directly or by acidification of the growth medium (reviewed in reference 23). Other substances produced by LAB have been reported to have antimicrobial effects, e.g., 2,3-butadione, reuterin (3-hydroxypropionaldehyde), acetaldehyde, hydrogen peroxide, hydroxyl radical, and peptides or proteins such as the bacteriocins (10, 23). Recently, we isolated and characterized a number of antifungal substances from different strains of Lactobacillus plantarum. The strains MiLAB14 and MiLAB393 were found to produce 3-phenyllactic acid, several 3-hydroxy fatty acids, and a series of diketopiperazines (24, 25, 28). Among these compounds, the 3-hydroxy fatty acids were the most potent antifungal substances, with MICs against important spoilage yeasts and molds on the order of 10 to 100 μg ml−1, whereas 3-phenyllactic acid and the diketopiperazines were significantly less active (MICs of several milligrams per milliliter) (25, 28). These studies were performed to characterize promising biocontrol LAB strains that were isolated from various sources and have the ability to suppress the growth or spore germination of several important food and feed spoilage organisms, such as Penicillium roqueforti and Aspergillus fumigatus. In these studies, the LAB strains were cultivated in liquid media under conditions optimized for growth, and the bioactive metabolites were isolated and characterized. For future applications of LAB as biocontrol agents on, e.g., grass in airtight silos, the growth conditions will be different and probably less than optimal, including the absence of a balanced growth medium and a restricted oxygen supply. The outcome of the metabolism of a particular LAB strain may thus be significantly different under these conditions, which may have important implications for the production of, e.g., antifungal or toxic metabolites. Thus, to be able to judge the real potency of biocontrol agents, as well as to evaluate safety aspects, it is important to characterize the metabolism of the organisms also under the conditions of practical biocontrol situations, e.g., when grown in silage, or any other possible application.
Thus, the present study was performed to investigate if the antifungal metabolites produced by strains of L. plantarum in liquid media also were produced by the same strains growing on grass in airtight silos, as well as to characterize other antifungal metabolites produced in this environment. If antifungal compounds are detected at elevated concentrations in LAB-inoculated silage, this will further emphasize the use of antifungal LAB strains in biocontrol applications.
MATERIALS AND METHODS
Strains and culture conditions.
L. plantarum strains MiLAB393 (28) and MiLAB14 (16) were grown on MRS agar or in MRS broth (Oxoid Ltd., Basingstoke, England) at 30°C in anaerobic jars in a CO2-N2 atmosphere (GasPak system; BBL, Cockeysville, MD). L. plantarum MiLAB14(pLV100) and MiLAB393(pLV100) were grown on MRS agar or in MRS broth containing 15 μg ml−1 chloramphenicol (MRS-C). A. fumigatus J9 and Pichia anomala J121 were grown on malt extract agar (Oxoid Ltd.) at 30°C, and P. roqueforti J268 was grown at 25°C.
Transformation of L. plantarum MiLAB14 and MiLAB393.
L. plantarum MiLAB14 and MiLAB393 were transformed with plasmid pLV100 (26) as described by Aukrust et al. (3). The presence of the plasmid was verified by PCR with the primers CatF2 (5′-GAA AGG ATA TGA AAT TTA TCC CTC TT-3′) and CatR2 (5′-TAC CCT ATG AAT TAT TTG AAA TTC A-3′) and Ready-To-Go PCR beads (Amersham Biosciences, Uppsala, Sweden) at an annealing temperature of 47°C. A small amount of colony material was used as the template.
The stability of the plasmid in the absence of selection was verified by repeated inoculation into MRS broth. After 50 generations, all colonies were resistant to chloramphenicol and carried the plasmid, as verified by PCR (data not shown).
Growth of L. plantarum MiLAB14(pLV100) and MiLAB393(pLV100) in silage.
A grass-dominated crop sample that had been stored at −20°C was inoculated by spraying with a suspension of L. plantarum MiLAB14(pLV100) or MiLAB393(pLV100) in peptone water (4 × 107 CFU ml−1) to give a final inoculum of 1 × 106 CFU g−1 of crop sample. The crop sample used for control silos was treated with peptone water only. The material was tightly packed into 500-ml glass jars (approximately 200 g per silo), and the jars were sealed with metal lids fitted with a water-filled plastic fermentation trap. Silos were incubated at 20°C. The total number of viable LAB before inoculation was determined by plating on MRS agar plates, and the number of inoculated bacteria was determined after spraying onto MRS-C agar plates.
Three MiLAB393(pLV100)-inoculated silos were opened at days 5, 13, and 26, and the total number of viable LAB was determined on MRS agar, and the number of viable MiLAB393(pLV100) bacteria was determined on MRS-C agar. For MiLAB14(pLV100), one single time point, 13 days, was investigated. The silage was also processed for the identification of substances produced by the bacteria as described below.
Extraction of silage.
The material from each silo was divided into four portions. After the addition of 300 ml of peptone water (0.2% Bacto peptone with 0.01% Tween 80) to the first portion in a plastic bag, it was treated for 2 min in a stomacher. The supernatant was then transferred to the next portion, which was processed in the same way and transferred to the third portion and so on. This process was repeated twice; i.e., a total of 900 ml of peptone water was used to process each sample. Any remaining solid material was removed by centrifugation at 22,000 × g for 30 min twice, after which each supernatant was passed through three filter papers with decreasing pore sizes (Munktell 3, 1F, and 00H; Munktell Filter AB, Grycksbo, Sweden), a glass fiber prefilter (Nalgene, NY), and finally 0.8-μm and 0.45-μm PES filters (Nalgene, NY, or PALL Corporation, NY). Thereafter, each supernatant was fractionated on a solid-phase extraction (SPE) column (Isolute, C18, EC, 10 g; International Sorbent Technology Ltd., Hengoed, United Kingdom). The column was activated with 30 ml of 95% acetonitrile (MeCN) and equilibrated with 50 ml of 5% aqueous MeCN. After sample loading, the column was washed with 30 ml of 5% aqueous MeCN and the retained compounds were eluted with 2 × 20 ml of 95% MeCN. Subsequently, the internal standard 3-hydroxyundecanoic acid was added to the combined MeCN fractions {1.0 μg g−1 [fresh weight (FW)] of grass}.
Metabolite identification and quantification.
Samples (500 μl) of the MeCN fractions obtained by SPE were taken in triplicate and dried in a vacuum centrifuge in 1.5-ml plastic tubes. Each dried sample was treated with 40 μl of pyridine and 100 μl of N,O-bis-trimethylsilyltrifluoroacetamide-trimethylsilylchloride (99:1) at 80°C for 40 min. Following cooling to room temperature, the samples were diluted with 1 ml of ethyl acetate and transferred to gas chromatography (GC) vials. Analysis was performed on an HP5890/5970 GC-mass spectrometry (MS) apparatus (Hewlett-Packard, Palo Alto, CA) equipped with a fused silica capillary column (0.25 mm by 30 m, 25 μm, HP5-MS; Agilent Technologies, Palo Alto, CA). Samples (1 μl) were injected in splitless mode, and helium was used as the carrier gas at 1.0 ml min−1. The temperature gradient was 80°C for 3 min, followed by 80 to 240°C at 5°C min−1 and 240°C for 5 min. The injector was kept at 240°C, the transfer line to the MS was kept at 260°C, and data were collected in full scan mode. Compounds were identified by using a mass spectral database (Wiley), and the identities were subsequently verified by comparison with commercial standards or substances synthesized (as described below).
Quantification was achieved by comparing the peak area of the internal standard and the areas of relevant peaks by using the indicated extracted ion chromatograms {internal standard, m/z 233 (25.08 min); 2-hydroxy-4-methylpentanoic acid, m/z 159 (11.97 min); benzoic acid, m/z 179 (12.11 min); catechol (1,2-dihydroxybenzene), m/z 254 (14.19 min); hydrocinnamic acid (3-phenylpropanoic acid), m/z 222 (16.83 min); salicylic acid (2-hydroxybenzoic acid), m/z 267 (19.43 min); 3-phenyllactic acid, m/z 193 (21.34 min); 4-hydroxybenzoic acid, m/z 282 (22.25 min); 3,4-dihydroxycyclohexane-1-carboxylic acid, m/z 286 (23.74 min); p-hydrocoumaric acid [3-(4-hydroxyphenyl)propanoic acid], m/z 310 (25.36 min); vanillic acid (4-hydroxy-3-methoxybenzoic acid), m/z 312 (25.50 min); azelaic acid (1,9-nonadioic acid), m/z 317 (26.26 min); hydroferulic acid [3-(4-hydroxy-3-methoxyphenyl)propanoic acid], m/z 340 (28.37 min); p-coumaric acid [3-(4-hydroxyphenyl)propenoic acid], m/z 308 (29.19 min); hydrocaffeic acid [3-(3,4-dihydroxyphenyl)propanoic acid], m/z 398 (29.46 min); ferulic acid [3-(4-hydroxy-3-methoxyphenyl)propenoic acid], m/z 338 (32.29 min); caffeic acid [3-(3,4-dihydroxyphenyl)propenoic acid], m/z 396 (33.18 min) } and by correcting for differences in response factors (obtained by analyzing samples containing known amounts of the internal standard and the respective compounds). For analysis of 3-hydroxy fatty acids, the oven temperature settings were 120°C for 3 min, followed by 120 to 240°C at 5°C min−1 and 240°C for 5 min, and data were collected in selected-ion monitoring mode by using the ion at m/z 233 (all 3-hydroxy fatty acids) for quantification together with the ions at m/z 275 and 317 (3-hydroxydecanoic acid), m/z 289 and 331 (3-hydroxyundecanoic acid), m/z 303 and 345 (3-hydroxydodecanoic acid), and m/z 331 and 373 (3-hydroxytetradecanoic acid) for identification.
Determination of MICs.
The antifungal activities of the substances identified were determined in triplicate in microtiter plates by using A. fumigatus J9, P. roqueforti J268, and P. anomala J121 as target organisms. The compounds were dissolved in 200 mM lactic acid (pH 4.1) at the highest possible concentration in the interval from 0.02 to 20 mg ml−1 and diluted in steps of 10 in 200 mM lactic acid. Spores or cells were suspended in 4% malt extract broth (Oxoid), at pH 4.1, to a final concentration of 104 spores or cells per ml. Fifty microliters of the suspension was added to 50 μl of the respective substance solution. A. fumigatus J9 and P. anomala J121 were incubated at 30°C and P. roqueforti J268 was incubated at 25°C for 48 h, after which the degree of inhibition was determined with the naked eye and an inverted microscope. The MICs were determined as the lowest concentrations at which no growth was observed.
HPLC fractionation of silage extracts.
A peptone water extract of silage (200 g of grass) inoculated with MiLAB393, harvested after 13 days, was subjected to SPE as described above. The MeCN extract was dried under reduced pressure, dissolved in 1 ml of aqueous 50% MeCN, and injected onto a reversed-phase high-performance liquid chromatography (HPLC) column (20 by 100 mm, 20-by-30-mm guard column, 5 μm; A. Maisch High-Performance LC GmbH, Germany) eluted with 10 to 100% MeCN in 10 min, followed by 100% MeCN for 10 min, at 10 ml min−1, with UV detection at 210 nm. Fractions were collected in 2.2-ml deep-well plates (12 s per fraction).
For analysis of antifungal activity, aliquots (300 μl) from all fractions were transferred to microtiter plates and air dried. Subsequently, the antifungal activity of each fraction was evaluated as described before (15), by observing the inhibition of A. fumigatus J9 spore germination. Subsequently, antifungal fractions were pooled and analyzed by nuclear magnetic resonance (NMR) analysis (Bruker DRX600, 5-mm QXI probe head, 2H2O, 30°C).
For isolation of one unidentified component detected during GC-MS analysis, a small portion from each HPLC fraction was dried in a vacuum centrifuge, trimethylsilylated, and subjected to GC-MS analysis (as described above). HPLC fractions containing the unidentified component, as guided by GC-MS data, were pooled and dried in a vacuum centrifuge. The material was dissolved in aqueous 6% MeCN and fractionated with the same column as above, with aqueous 6% MeCN as the solvent, at 10 ml min−1. Fractions containing the unidentified substance, as indicated by GC-MS (as described above), were dried and subjected to NMR analysis as described below.
Identification of unknown components in HPLC fractions.
NMR analysis (Bruker DRX600, 2.5-mm SEI probe head, 2H2O, 30°C) of the unidentified compound yielded data consistent with (trans, trans)-3,4-dihydroxycyclohexane-1-carboxylic acid. Subsequently, this compound was synthesized by a method previously published (13). 3-Cyclohexene-1-carboxylic acid (99.5 mg, 0.79 mmol) was dissolved in 100 ml of 20 mM KOH and cooled to 0°C. To this solution, 100 mg of KMnO4 (0.63 mmol) in 20 ml of H2O was added dropwise at 0°C, and the solution was stirred for 1 h at 0°C. Subsequently, 4 ml of 1 M HCl was added, the precipitated MnO2 was removed by filtration, and the resulting solution was freeze-dried. The resulting material was dissolved in 6 ml of aqueous 7.5% MeCN (0.1% trifluoroacetic acid) and fractionated (12 × 0.5 ml injected) on a graphitized carbon HPLC column (21.2 by 100 mm, 5 μm; ThermoQuest, Runcorn, Cheshire, United Kingdom) eluted with aqueous 7.5% MeCN (0.1% trifluoroacetic acid) at 10 ml min−1, with UV detection at 210 nm. Fractions were pooled and subjected to NMR analysis, resulting in 27 mg of (trans, trans)-3,4-dihydroxycyclohexane-1-carboxylic acid, which was the main product.
HPLC-electrospray ionization (ESI)-tandem MS (MS/MS) analysis of silage extracts.
The MeCN fractions obtained by SPE were analyzed by HPLC-ESI-MS and HPLC-ESI-MS/MS (HP1100 [Hewlett-Packard, Palo Alto, CA], Bruker Esquire [Bruker Daltonik GmbH, Bremen, Germany]), with a Hypercarb graphitized carbon HPLC column (2.1 by 100 mm) eluted with a gradient of MeOH in H2O (35 to 100% in 8 min, followed by 12 min at 100%) containing 0.1% formic acid at 0.2 ml min−1. Diketopiperazines were identified by comparison of retention times and fragmentation patterns with those of authentic standards.
RESULTS AND DISCUSSION
After thawing, the level of viable LAB in the frozen, grass-dominated crop samples turned out to be unexpectedly low, only 700 CFU g−1. This makes a comparison between the quality of the silages obtained without inoculation and after inoculation with either L. plantarum MiLAB393 or MiLAB14 unfeasible. However, it provides a good opportunity to relate the detection of compounds to the presence of the L. plantarum strains. Therefore, this study focused on the metabolites detected rather than on the effect on silage quality.
After inoculation to 6 × 109 CFU g−1, L. plantarum MiLAB393(pLV100) grew to 7 × 105 CFU g−1 at day 5 and 2 × 109 CFU g−1 at day 13. For L. plantarum MiLAB14(pLV100), the initial inoculation level was 6 × 105 CFU g−1 and 2 × 109 CFU g−1 were isolated at day 13. The total number of LAB at day 13 in the uninoculated control was 1 × 108 CFU g−1, suggesting that the added strains dominated the fermentation in the inoculated silos.
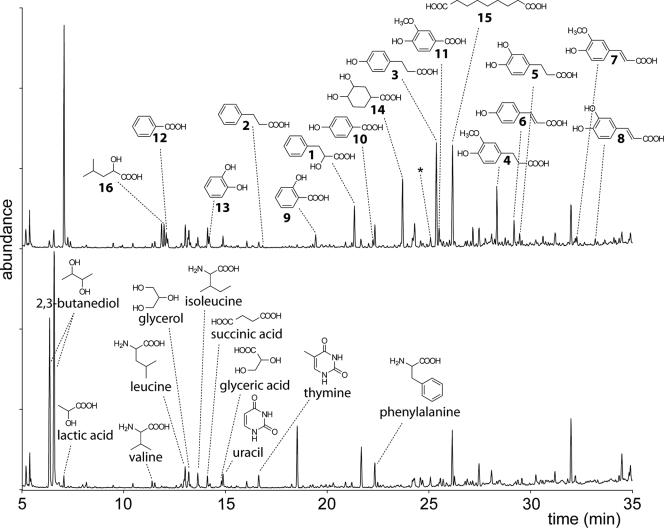
The metabolite profiles of the silage extracts were found to be highly dependent on whether the grass was inoculated with a LAB strain or not, as well as the time of harvesting (Fig. 1 and Table 1). One major difference between the LAB-inoculated samples and the control samples was the peak eluting after 7.10 min (Fig. 1), which was found to consist of the trimethylsilyl derivative of lactic acid. As expected, lactic acid was present at higher concentrations in the silos inoculated with a LAB strain than in control silos. Other major differences were the peaks eluting after 6.41 and 6.63 min (Fig. 1), corresponding to the trimethylsilyl derivatives of the diastereomers of 2,3-butanediol, which were much more pronounced in the samples from control silos than in samples from LAB-inoculated silos. Although 2,3-butanediol has been shown to be formed by LAB from pyruvate via diacetyl and/or acetoin (20), it is possible that it is produced by enterobacteria, if they are present in the starting material. For example, several species of the genera Klebsiella, Bacillus, Serratia, and Pseudomonas have shown the ability to produce 2,3-butanediol (11).
FIG. 1.
Reconstructed total ion mass chromatograms from GC-MS analysis of water-extracted silage harvested after 13 days. The top silage sample was inoculated with MiLAB393 at day 0, whereas the bottom sample was an uninoculated control silage. Samples were analyzed by GC-MS in full scan mode after trimethylsilylation, and the denoted compounds were identified by using a mass spectral database. The compounds indicated in the top chromatogram were quantified relative the internal standard 3-hydroxyundecanoic acid (*) by using selected ions as described in Materials and Methods. The numbering refers to compounds quantified and presented in Table 1.
TABLE 1.
GC-MS quantification of compounds identified in silage inoculated or not inoculated with LABa
| Compound (structure no.) | Mean concn (μg g−1 [FW] of grass) ± SD
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 days, negb | 5 days, neg | 5 days, MiLAB393c | 13 days, neg | 13 days, MiLAB393 | 13 days, MiLAB14d | 26 days, neg | 26 days, MiLAB393 | |
| 3-Phenyllactic acid (1) | 0.03 ± 0.01 | 0.057 ± 0.006 | 3.5 ± 0.2 | 0.072 | 5 ± 1 | 5 ± 1 | 0.095 ± 0.009 | 2.6 ± 0.2 |
| 3-Hydroxydecanoic acid | 0.019 ± 0.003 | 0.036 ± 0.005 | 0.15 ± 0.02 | 0.080 | 0.15 ± 0.02 | 0.16 ± 0.03 | 0.05 ± 0.03 | 0.21 ± 0.03 |
| Hydrocinnamic acid (2) | NDe | ND | 0.25 ± 0.04 | ND | 0.26 ± 0.03 | 0.30 ± 0.08 | ND | 0.34 ± 0.03 |
| p-Hydrocoumaric acid (3) | 0.05 ± 0.01 | 0.11 ± 0.01 | 8.1 ± 0.1 | 0.11 | 12.8 ± 0.5 | 13 ± 2 | 0.11 ± 0.02 | 5 ± 1 |
| Hydroferulic acid (4) | 0.09 ± 0.03 | 0.18 ± 0.02 | 7.1 ± 0.2 | 0.18 | 9.3 ± 0.6 | 9.8 ± 0.2 | 0.18 ± 0.03 | 5.0 ± 0.8 |
| Hydrocaffeic acid (5) | ND | ND | 0.60 ± 0.07 | ND | 1.16 ± 0.01 | 1.4 ± 0.7 | ND | 0.3 ± 0.2 |
| p-Coumaric acid (6) | 0.8 ± 0.2 | 0.82 ± 0.04 | 5.1 ± 0.8 | 0.30 | 5.6 ± 0.4 | 4 ± 1 | 0.19 ± 0.09 | 3 ± 1 |
| Ferulic acid (7) | ND | 0.9 ± 0.1 | 2.9 ± 0.4 | 0.58 | 3.7 ± 0.4 | 3.5 ± 0.9 | 0.6 ± 0.2 | 5 ± 1 |
| Caffeic acid (8) | ND | ND | 0.77 ± 0.01 | ND | 1.17 ± 0.03 | 1.0 ± 0.3 | ND | 0.6 ± 0.3 |
| Salicylic acid (9) | 0.11 ± 0.02 | 0.25 ± 0.01 | 3.8 ± 0.7 | 0.38 | 4 ± 1 | 5.3 ± 0.6 | 0.44 ± 0.04 | 3.1 ± 0.6 |
| 4-Hydroxybenzoic acid (10) | ND | ND | 0.57 ± 0.04 | ND | 0.79 ± 0.02 | 0.8 ± 0.1 | 0.08 ± 0.04 | 0.41 ± 0.07 |
| Vanillic acid (11) | 0.09 ± 0.02 | 0.14 ± 0.02 | 1.7 ± 0.1 | 0.15 | 2.2 ± 0.1 | 2.1 ± 0.2 | 0.18 ± 0.02 | 1.4 ± 0.1 |
| Benzoic acid (12) | 0.037 ± 0.007 | 0.09 ± 0.01 | 0.90 ± 0.08 | 0.087 | 0.9 ± 0.1 | 1.0 ± 0.1 | 0.096 ± 0.004 | 0.7 ± 0.2 |
| Catechol (13) | ND | ND | 0.8 ± 0.2 | ND | 0.97 ± 0.07 | 0.26 ± 0.03 | 0.3 ± 0.1 | 0.8 ± 0.2 |
| 3,4-Dihydroxycyclo-hexane-1-carboxylic acid (14) | ND | ND | 2.6 ± 0.3 | ND | 3.7 ± 0.1 | 1.6 ± 0.4 | ND | 2.5 ± 0.7 |
| Azelaic acid (15) | 0.35 ± 0.04 | 0.5 ± 0.1 | 12.2 ± 0.7 | 0.45 | 15 ± 2 | 14 ± 2 | 0.6 ± 0.1 | 11 ± 3 |
| 2-Hydroxy-4-methyl-pentanoic acid (16) | ND | ND | 1.1 ± 0.1 | ND | 1.4 ± 0.2 | 1.5 ± 0.1 | ND | 0.94 ± 0.09 |
Quantification is relative to the internal standard 3-hydroxyundecanoic acid. Substance numbering refers to structures in Fig. 1.
neg, negative control.
Strain MiLAB393.
Strain MiLAB14.
ND, not detected.
The compound 3-phenyllactic acid, previously isolated from liquid cultures of MiLAB393 and MiLAB14, was found to be produced also on grass in airtight silos. The concentration was the highest (5 μg g−1 [FW] of grass) in silos inoculated with MiLAB14(pLV100) or MiLAB393(pLV100) and harvested after 13 days, whereas the concentrations after 5 and 26 days were slightly lower (3.5 and 2.6 μg g−1, respectively). The 3-phenyllactic acid concentrations were approximately 25- to 100-fold lower in silos not inoculated with LAB and additionally 2- to 3-fold lower in the starting grass.
3-Hydroxy fatty acids have previously been detected in liquid cultures of L. plantarum (25), and of these compounds, 3-hydroxydecanoic acid was detected on grass incubated in airtight silos. The differences in the concentrations of this compound between inoculated and uninoculated grass were less pronounced than for 3-phenyllactic acid. The concentrations were between 0.15 and 0.21 μg g−1 (FW) of grass for samples inoculated with MiLAB14(pLV100) or MiLAB393(pLV100) and between 0.036 and 0.080 μg g−1 for uninoculated grass. The concentration in the starting grass was 0.019 μg g−1 (FW). The observation of only 3-hydroxydecanoic acid of the 3-hydroxy fatty acids in these samples is in accord with data recorded on MiLAB14 in liquid cultures, which indicated the concentration of this 3-hydroxyfatty acid to be the highest among these compounds (25). The relatively small difference between the inoculated silos and the control silos (two- to fivefold higher concentrations in the inoculated silos) may depend on the production of 3-hydroxydecanoic acid by other bacteria in the silage, or the production of 3-hydroxydecanoic acid by the added LAB strains may be regulated in order to avoid reaching concentrations detrimental to the LAB.
The finding of increased concentrations of 3-phenyllactic acid and 3-hydroxydecanoic acid on silage inoculated with LAB is in accordance with the hypothesis that these compounds contribute to the antimicrobial activity of the added LAB, as suggested by previous studies of MiLAB14 and MiLAB393 in liquid cultures (24, 25, 28). In this study, MICs were determined at pH 4.1 and in the presence of 100 mM lactic acid. This was done to mimic the conditions in silage, as a previous study using fresh grass and L. plantarum MiLAB14 as an additive resulted in silage with a pH of 4.1 at day 90 (unpublished results). This concentration of lactic acid had no effect on the growth of any of the fungi included in the assay. The MIC of 3-phenyllactic acid against P. roqueforti, A. fumigatus, and P. anomala was 10 mg ml−1 (Table 2). The corresponding MIC of 3-hydroxydecanoic acid was 0.1 mg ml−1, except for P. roqueforti, which was unaffected at this concentration (the highest tested because of the limited solubility of the compound). It is difficult to compare the MICs with the concentrations measured in silage because of the problem of establishing the total volume of the water solution present on the surface of the grass constituting the crop sample. However, it is possible that locally, on the surface of the grass, the concentrations of the metabolites studied reach values close to or even above the MICs. If this is the case, these substances can be important to the antifungal effects of LAB.
TABLE 2.
MICs against food and feed spoilage fungi of compounds identified in grass silage inoculated with L. plantarum strains
| Compound | MIC (mg ml−1)
|
||
|---|---|---|---|
| P. anomala | P. roqueforti | A. fumigatus | |
| 3-Phenyllactic acid | 10 | 10 | 10 |
| 3-Hydroxydecanoic acid | 0.1 | >0.1 | 0.1 |
| Hydrocinnamic acid | 1 | 1 | 0.1 |
| p-Hydrocoumaric acid | 10 | 10 | 10 |
| Hydroferulic acid | >1 | >1 | >1 |
| Hydrocaffeic acid | >10 | >10 | >10 |
| p-Coumaric acid | >0.1 | >0.1 | >0.1 |
| Ferulic acid | >0.1 | >0.1 | >0.1 |
| Salicylic acid | 1 | 1 | 0.1 |
| 4-Hydroxybenzoic acid | >1 | >1 | >1 |
| Vanillic acid | >0.1 | >0.1 | >0.1 |
| Benzoic acid | 1 | 1 | 0.1 |
| Catechol | 10 | 1 | 1 |
| 3,4-Dihydroxycyclohexane-1-carboxylic acid | >10 | >10 | >10 |
| Azelaic acid | >0.1 | >0.1 | >0.1 |
| 2-Hydroxy-4-methylpentanoic acid | 10 | 10 | 10 |
In addition to the compounds mentioned above, i.e., lactic acid, 2,3-butanediol, 3-phenyllactic acid, and 3-hydroxydecanoic acid, the GC-MS data demonstrated the presence of many additional compounds in the silage and grass extracts, and many of these compounds were found to be present at increased concentrations when the grass was inoculated with LAB strains. A compound eluting after 26.26 min was found to be azelaic acid, and the concentration of this compound was highly dependent on the presence of LAB. In silos inoculated with MiLAB14(pLV100) or MiLAB393(pLV100), the concentrations were between 11 and 15 μg g−1 (FW) of grass, whereas in silos without LAB inoculation, the concentrations were 20- to 30-fold lower (Table 1). Azelaic acid has been shown to inhibit the growth of both gram-positive and gram-negative bacteria and to do so more efficiently at low pH than at neutral pH (reviewed in reference 5). Inhibitory activity has also been shown against dermatophytic fungi at concentrations between 0.1% for Scopulariopsis brevicaulis and 2% for Candida albicans at low pH (6). The azelaic acid detected in the LAB-inoculated silage may be synthesized by the LAB strains, which would be a novel finding. Alternatively, the compound could be released from the grass by the action of the added LAB strains, as azelaic acid previously has been found in extracts of the grasses wheat (9) and rice (21), or produced by oxidative cleavage of the double bond of oleic acid (18).
The peak at 23.74 min (Fig. 1) was only present in samples inoculated with LAB, but this substance could not be identified by using the mass spectral database. Instead, this compound was isolated from an SPE product by HPLC and identified as (trans, trans)-3,4-dihydroxycyclohexane-1-carboxylic acid by NMR analysis and GC-MS, as well as by the synthesis of a reference sample. This compound has previously been isolated from L. plantarum and has been described as a metabolite formed from quinic acid and shikimic acid (29, 30). The role of this metabolite is, however, unknown.
The data also pointed to the presence of a series of 3-phenylpropenoic and 3-phenylpropanoic acids, i.e., substituted cinnamic acids and hydrocinnamic acids, respectively. The compounds were p-coumaric acid, caffeic acid, ferulic acid, hydrocinnamic acid, p-hydrocoumaric acid, hydrocaffeic acid, and hydroferulic acid. The identities of these compounds were subsequently verified by comparison with authentic reference samples. The concentrations of the different hydrocinnamic acids were found to be substantially higher in silos inoculated with LAB strains than in control silos without added LAB, and the highest concentration found was that of p-hydrocoumaric acid (13 μg g−1 of grass) in silos inoculated with MiLAB14(pLV100) or MiLAB393(pLV100) harvested after 13 days (Table 1). In the control silos, the concentrations ranged from 0.18 μg g−1 of grass (hydroferulic acid, 5 days) down to below detection (hydrocinnamic and hydrocaffeic acids, all control silos). The concentrations of the substituted cinnamic acids were also significantly higher in LAB-enriched silage than in control silos. The highest concentration found was that of p-coumaric acid (5.6 μg g−1 of grass) in silos inoculated with MiLAB393(pLV100) and harvested after 13 days, whereas those in the control silos ranged from 0.9 μg g−1 of grass (ferulic acid, 5 days) down to below detection (caffeic acid, all control silos). The increase in the concentrations of the substituted hydrocinnamic and cinnamic acids when treating the grass in airtight silos inoculated with LAB indicates that microbial processes either are involved in the synthesis of these compounds or enhance the release of the compounds from the grass. Substituted cinnamic acids are well known to occur in all plants, since they are precursors of, e.g., coniferyl alcohol and sinapyl alcohol, which are polymerized into lignin (reviewed in reference 4). Hydrocinnamic acids are less frequently found in plants, but some have been detected in the grass Vulpia myuros (1). The collected data indicate that the added LAB strains have the enzymatic ability to reduce the C2-C3 double bond of the cinnamic acids to form the corresponding hydrocinnamic acids. Similar enzymatic activity has previously been reported for the microflora of rat intestine (22) and cow rumen (19), for anaerobic methanogenic consortia (12), for several strains of Pseudomonas (2), for the bacterium Nocardia autotrophica (17), and for Clostridium glycolicum and C. bifermentans (7, 8). In addition to the substituted hydrocinnamic acids produced in the presence of the LAB strains, the GC-MS analysis also pointed to increased concentrations of a series of related substances, e.g., catechol and vanillic acid (Fig. 1 and Table 1). These and related compounds may be produced during the microbial degradation of the corresponding substituted cinnamic acids, possibly via the hydrocinnamic acids (17). Several of these compounds show moderate antifungal activity in vitro (Table 2), but their importance during ensiling remains to be investigated.
When MeCN fractions obtained by SPE were analyzed by HPLC-ESI-MS and HPLC-ESI-MS/MS, three of the previously identified diketopiperazines were detected, i.e., cyclo(Phe-Pro), cyclo(Phe-OHPro), and cyclo(Leu/Ile-OHPro) (24, 28). For cyclo(Phe-OHPro) and cyclo(Leu/Ile-OHPro), one major peak was observed, together with two and one, respectively, smaller peaks (∼10% of the major corresponding peak) probably caused by epimerization during sample handling and analysis. No significant differences in diketopiperazine concentrations were observed between samples from silage samples obtained with or without the presence of LAB strains or samples from original grass samples. This leaves the origin of the diketopiperazines as an open question; they may either be produced by the microorganisms present on the grass or be present in the grass itself. However, diketopiperazines have been reported to be involved in quorum sensing in some gram-negative bacteria (14). So far, there are no reports of gram-positive organisms using diketopiperazines in cell-to-cell communication, but the possibility cannot be excluded that diketopiperazines produced by gram-positive organisms may interfere with the quorum-sensing systems in gram-negative bacteria or even influence eukaryotic organisms in the environment.
The growth of LAB strains in grass silage may hypothetically promote the production of antifungal substances other than the compounds isolated from liquid cultures. To investigate this, the MeCN fractions obtained by SPE were fractionated by preparative reversed-phase HPLC and the fractions were investigated by a spore germination-based bioassay. For all of the samples tested, this resulted in one single antifungal fraction which, according to NMR analysis, mainly contained lactic acid. On the other hand, the GC-MS analyses showed the presence of several other antifungal metabolites (Table 1 and Fig. 1). The concentrations in the extracts were too low to be detected by the bioassay but may locally, in the silage, be high enough to affect the growth of fungi. The procedure used to extract and wash the silage with peptone water in several rounds may bias the results toward hydrophilic metabolites. This method was chosen to mainly extract metabolites present in aqueous solution on the surface of the grass and to avoid the extraction of lipophilic constituents from the grass.
This study shows that antifungal compounds found in liquid cultures of L. plantarum strains also can be detected in silage inoculated with the same strains. In addition, other compounds differing in concentration between inoculated and uninoculated silos were identified. The importance of these compounds in silage preservation remains to be investigated.
Acknowledgments
The present study is part of the MASE program, financed by MISTRA (Swedish Foundation for Strategic Environmental Research).
Cecilia Berglund is gratefully acknowledged for technical assistance.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.An, M., T. Haig, and J. E. Pratley. 2000. Phytotoxicity of Vulpia residues. II. Separation, identification, and quantitation of allelochemicals from Vulpia myuros. J. Chem. Ecol. 26:1465-1476. [Google Scholar]
- 2.Andreoni, V., and G. Bestetti. 1986. Comparative analysis of different Pseudomonas strains that degrade cinnamic acid. Appl. Environ. Microbiol. 52:930-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aukrust, T. W., M. B. Brurberg, and I. F. Nes. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201-208. [DOI] [PubMed] [Google Scholar]
- 4.Boerjan, W., J. Ralph, and M. Baucher. 2003. Lignin biosynthesis. Annu. Rev. Plant Biol. 54:519-546. [DOI] [PubMed] [Google Scholar]
- 5.Bojar, R. A., and K. T. Holland. 1993. Azelaic acid: a review of its antimicrobial properties. Rev. Contemp. Pharmacother. 4:403-414. [Google Scholar]
- 6.Brasch, J., and E. Christophers. 1993. Azelaic acid has antimycotic properties in vitro. Dermatology 186:55-58. [DOI] [PubMed] [Google Scholar]
- 7.Chamkha, M., M. Labat, B. K. C. Patel, and J.-L. Garcia. 2001. Isolation of a cinnamic acid-metabolizing Clostridium glycolicum strain from oil mill wastewaters and emendation of the species description. Int. J. Syst. Evol. Microbiol. 51:2049-2054. [DOI] [PubMed] [Google Scholar]
- 8.Chamkha, M., B. K. C. Patel, J.-L. Garcia, and M. Labat. 2001. Isolation of Clostridium bifermentans from oil mill wastewaters converting cinnamic acid to 3-phenylpropionic acid and emendation of the species. Anaerobe 7:189-197. [Google Scholar]
- 9.Chaves das Neves, H. J., and E. M. M. Gaspar. 1990. Identification of active compounds in wheat straw extractives with allelopathic activity by HRGC-MS and HRGC-FTIR. J. High Resolut. Chromatogr. 13:550-554. [Google Scholar]
- 10.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 11.Garg, S. K., and A. Jain. 1995. Fermentative production of 2,3-butanediol: a review. Bioresour. Technol. 51:103-109. [Google Scholar]
- 12.Grbić-Galić, D., and L. Y. Young. 1985. Methane fermentation of ferulate and benzoate: anaerobic degradation pathways. Appl. Environ. Microbiol. 50:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grewe, R., A. Heinke, and C. Sommer. 1956. Preparation and structure of 3,4-disubstituted cyclohexanecarboxylic acids. Chem. Ber. 89:1978-1988. [Google Scholar]
- 14.Holden, M. T. G., S. R. Chhabra, R. de Nys, P. Stead, N. J. Bainton, P. J. Hill, M. Manefield, N. Kumar, M. Labatte, D. England, S. Rice, M. Givskov, G. P. C. Salmond, G. S. A. B. Stewart, B. W. Bycroft, S. Kjelleberg, and P. Williams. 1999. Quorum-sensing cross talk: isolation and chemical characterisation of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33:1254-1266. [DOI] [PubMed] [Google Scholar]
- 15.Magnusson, J., and J. Schnürer. 2001. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 67:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnusson, J., K. Ström, S. Roos, J. Sjögren, and J. Schnürer. 2003. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 219:129-135. [DOI] [PubMed] [Google Scholar]
- 17.Malarczyk, R., J. Rogalski, and A. Leonowicz. 1994. Transformation of ferulic acid by soil bacteria Nocardia provides various valuable phenolic compounds. Acta Biotechnol. 14:235-241. [Google Scholar]
- 18.Narukawa, M., K. Kawamura, N. Takeuchi, and T. Nakajima. 1999. Distribution of dicarboxylic acids and carbon isotope compositions in aerosols from 1997 Indonesian forest fires. Geophys. Res. Lett. 26:3101-3104. [Google Scholar]
- 19.Ohmiya, K., M. Takeuchi, W. Chen, S. Shimizu, and H. Kawakami. 1986. Anaerobic reduction of ferulic acid to dihydroferulic acid by Wolinella succinogenes from cow rumen. Appl. Microbiol. Biotechnol. 23:274-279. [Google Scholar]
- 20.Pasteris, S. E., and A. M. Strasser de Saad. 2005. Aerobic glycerol catabolism by Pediococcus pentosaceus isolated from wine. Food Microbiol. 22:399-407. [DOI] [PubMed] [Google Scholar]
- 21.Rimando, A. M., M. Olofsdotter, R. E. Dayan, and S. O. Duke. 2001. Searching for rice allelochemicals: an example of bioassay-guided isolation. Agron. J. 93:16-20. [Google Scholar]
- 22.Scheline, R. R. 1968. Metabolism of phenolic acids by the rat intestinal microflora. Acta Pharmacol. Toxicol. 26:189-205. [DOI] [PubMed] [Google Scholar]
- 23.Schnürer, J., and J. Magnusson. 2005. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 16:70-78. [Google Scholar]
- 24.Sjögren, J. 2005. Bioassay-guided isolation and characterisation of antifungal metabolites. Studies of lactic acid bacteria and propionic acid bacteria. Ph.D. thesis. Swedish University of Agricultural Sciences, Uppsala, Sweden. (http://diss-epsilon.slu.se/archive/00000798/01/AvhandlingJS.pdf).
- 25.Sjögren, J., K. Magnusson, A. Broberg, J. Schnürer, and L. Kenne. 2003. Antifungal 3-hydroxy fatty acids from Lactobacillus plantarum MiLAB 14. Appl. Environ. Microbiol. 69:7554-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sørvig, E., M. Skaugen, K. Naterstad, V. G. Eijsink, and L. Axelsson. 2005. Plasmid p256 from Lactobacillus plantarum represents a new type of replicon in lactic acid bacteria, and contains a toxin-antitoxin-like plasmid maintenance system. Microbiology 151:421-431. [DOI] [PubMed] [Google Scholar]
- 27.Stiles, M. E. 1996. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek 70:331-345. [DOI] [PubMed] [Google Scholar]
- 28.Ström, K., J. Sjögren, A. Broberg, and J. Schnürer. 2002. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(l-Phe-l-Pro) and cyclo(l-Phe-trans-4-OH-l-Pro) and 3-phenycllactic acid. Appl. Environ. Microbiol. 68:4322-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiting, G. C., and R. A. Coggins. 1971. The role of quinate and shikimate in the metabolism of lactobacilli. Antonie Leeuwenhoek 37:33-49. [DOI] [PubMed] [Google Scholar]
- 30.Whiting, G. C., and R. A. Coggins. 1973. (−)-3t,4t-Dihydroxycyclohexane-1c-carboxylate, a new quinate metabolite of Lactobacillus plantarum. J. Sci. Food Agric. 24:897-904. [DOI] [PubMed] [Google Scholar]