Abstract
The fate of calicivirus in oysters in a 10-day depuration was assessed. The norovirus gene was persistently detected from artificially contaminated oysters during the depuration, whereas feline calicivirus in oysters was promptly eliminated. The prolonged observation of norovirus in oysters implies the existence of a selective retention mechanism for norovirus within oysters.
Noroviruses have been one of the major causative agents of acute gastroenteritis and are often related to food poisoning from eating oysters (12, 15). Cases of food poisoning caused by norovirus-contaminated raw oysters have been increasing, and marine industries related to oysters have been significantly damaged by the contamination of cultivated oysters with noroviruses. Many researchers are devoted to developing procedures for efficient removal or inactivation of pathogenic viruses in contaminated oysters (5, 14), and viral surrogates have been employed to evaluate the removal or inactivation efficiency of noroviruses in oysters because there are no suitable cell culture systems to cultivate noroviruses (25). Feline calicivirus (FCV) is one of the most frequently used surrogates (1, 6, 11, 26). Norovirus and FCV belong to the same family, Caliciviridae, which are small, nonenveloped, icosahedral viruses with a linear, positive-sense, single-stranded RNA genome (7). Since norovirus and FCV have almost the same configuration, these two viruses are expected to have similar fates in oysters. However, the similarity or discrepancy of behaviors between norovirus and FCV in oysters has not been significantly evaluated thus far.
In this study, the persistency of norovirus genogroup II (GII) in oysters was compared with that of the FCV f4 strain. Oysters (Crassostrea gigas) were artificially contaminated with norovirus GII and FCV f4 for 72 h; the contaminated oysters were moved to a water body without viruses for a 10-day depuration. The gene concentrations of test viruses in oysters were quantified with the real-time quantitative reverse transcription-PCR (qRT-PCR) during the contamination and depuration periods. FCV strain f4 used in this study was given by Y. Tohya, University of Tokyo, and was multiplied in CRFK cells cultivated with minimum essential medium including 10% bovine serum according to Tohya et al. (23). Norovirus GII used in this study was purified from a stool specimen of a patient with acute infectious gastroenteritis in Miyagi Prefecture, Japan. About 20 g of stool was suspended in 200 ml of distilled water and centrifuged at 10,000 × g for 30 min. The supernatant, including norovirus particles, was collected and stored at −20°C. The genotype of norovirus GII in the specimen was found to be norovirus GII genotype 6 (GII.6), which was revealed by RT-PCR and sequencing of the norovirus GII gene as described previously (24).
A contamination test of oysters with norovirus GII and FCV f4 was conducted at Miyagi Prefecture Fisheries Research and Development Center. Three water baths were prepared, each containing 160 liters of sand-filtered seawater. The temperature of the water was set at 10 ± 2°C, which corresponds approximately to the average water temperature of the oyster cultivation area in November and December (oyster harvesting period) in Miyagi Prefecture. Phytoplankton (Chaetoceros calcitrans) was put into all water baths as feed for oyster cultivation. Test viruses were then added to the water baths, and the water was aerated at 4 liters of air/min for 1 h. Two of the three water baths were contaminated with norovirus GII or FCV f4, and the other one was contaminated with both norovirus GII and FCV f4. One hundred eighty oysters were prepared for the contamination test, and 50 oysters were put into each water bath. Ten oysters in each water bath (30 oysters in total) were set aside as blank. The contamination of oysters with test virus was performed for 72 h under aeration condition (4 liters of air/min), but contaminated oysters were transferred every 24 h (i.e., twice) to new water baths in which test virus and phytoplankton were freshly prepared. The quantities of input test viruses were 2.77 ± 0.59 log10 copies/liter in the norovirus GII-contaminated water bath, 2.87 ± 0.60 log10 copies/liter in the FCV f4-contaminated water bath, and 2.06 ± 0.55 log10 copies of norovirus GII and 2.86 ± 1.37 log10 copies of FCV f4 per liter in the cocontaminated water bath. Ten oysters were collected from each water bath when the oysters were transferred to the freshly prepared water bath every 24 h during the 72-h contamination. After the contamination period, oysters were transferred to new water baths (sand-filtered seawater) without test viruses and depurated for 10 days. The absence of test viruses in the prepared water bath was confirmed by real-time qRT-PCR. The flow rate of the sand-filtered seawater during the depuration was 12 liters/min. Ten oysters were collected from each water bath just after the transfer and on days 3 and 10 during the depuration.
Each oyster was assayed for norovirus GII and FCV. Viruses in oysters were extracted by the spallation method, in which an aseptically separated digestive diverticulum from an oyster was mashed up by a spallation apparatus to efficiently extract viruses (24). Viruses in water were concentrated by polyethylene precipitation as previously described (24). Noroviral RNA was extracted from specimens with the QIAmp viral RNA mini kit and quantified by real-time qRT-PCR according to Kageyama et al. (10). The quantitative detection limit of 10 copies per reaction was employed in the real-time qRT-PCR, which has been admissible in other investigations (9, 10). FCV RNA was extracted as well and quantified by real-time qRT-PCR using the set of primers and probe indicated in Table 1. The sensitivity and specificity of the primers were ensured by the end-point RT-PCR and sequencing of the RT-PCR product (data not shown). The PCR product amplified with the primers in Table 1 was used to make a standard curve, and a quantitative detection limit of 10 copies/reaction was also employed.
TABLE 1.
Oligonucleotide primer and probe sequences used for quantification of FCV by real-time qRT-PCR
| Primer or probea | Sequence (5′→3′) | Location |
|---|---|---|
| FCVf4fwd | TCGATTCCTTCGGACCTGATC | 6319-6339 |
| FCVf4rev | AAGTCGAAATGACGGTTTGCTT | 6422-6440 |
| FCVf4pro | TAATCGCTACTGGACTGAC | 6365-6383 |
fwd, forward primer; rev, reverse primer; pro, probe.
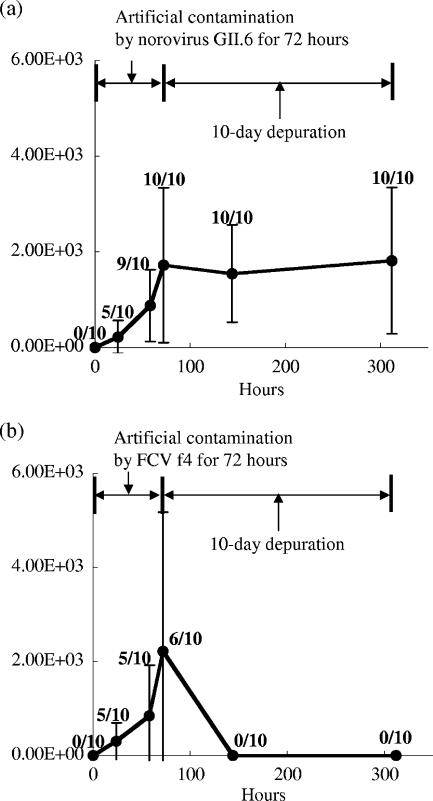
Figure 1a shows the average value of norovirus GII gene concentration in digestive diverticulum of oysters during artificial contamination by norovirus GII.6 (0 to 72 h) and with a 10-day depuration process (72 to 312 h). The average value of norovirus GII gene in oysters was increasing during the artificial contamination and reached 1.7 × 103 copies/g of the digestive diverticulum of oyster (n = 10; standard deviation [SD] = 1.6 × 103) after the 72-h contamination. The value of SD is large because each oyster has a bioaccumulation rate. The average concentrations of the norovirus GII gene in oysters during the depuration process were not significantly decreased and were 1.5 × 103 and 1.8 × 103 copies/g after the 3-day and 10-day depurations, respectively. It was difficult to reduce the concentration of noroviruses in contaminated oysters with the 10-day depuration. On the other hand, the concentration of the FCV gene in artificially-contaminated oysters was changed in a totally different manner. The average concentration of FCV reached 2.2 × 103 copies/g (n = 10; SD = 3.0 × 103) after the 72-h contamination, but all oysters collected during the depuration were negative by real-time qRT-PCR (Fig. 1b). These results mean that a large amount of FCV in contaminated oysters was promptly eliminated from the digestive diverticulum of oyster in the depuration.
FIG. 1.
Average gene concentration (copies/g of digestive diverticulum) of norovirus GII (a) and FCV (b) in digestive diverticulum of oyster during artificial contamination by norovirus GII.6 (a) or FCV f4 (b) (0 to 72 h) and the depuration process (72 to 312 h). Ten oysters were sampled at each time point, and each oyster was assayed by real-time qRT-PCR. The error bar indicates the SD from 10 oysters in a single test of the contamination and depuration. The ratio of the number of positive samples to the number of analyzed samples is indicated above each dot.
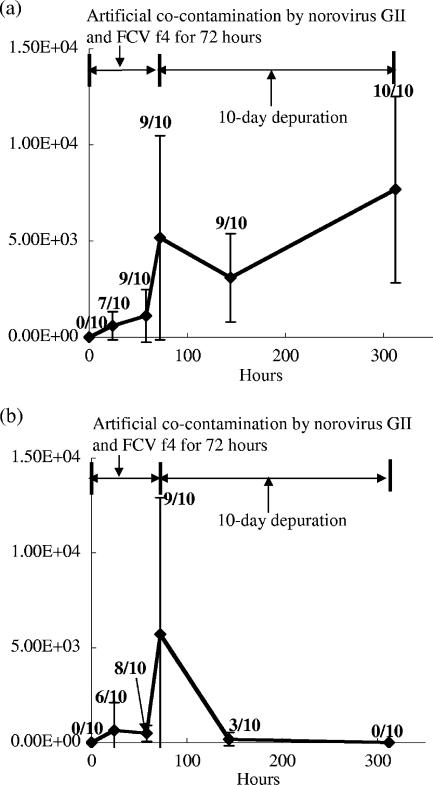
Similar results with the former experiments using norovirus GII.6 or FCV f4 as a contaminant were observed in the cocontamination experiment with norovirus and FCV. Figure 2a and b show the average concentrations of the norovirus GII and FCV genes in oysters cocontaminated with norovirus GII.6 and FCV f4. The concentration of the norovirus GII gene in oysters reached 5.2 × 103 copies/g after the 72-h contamination, and the concentrations in the depuration process were 3.1 × 103 and 7.7 × 103 copies/g after the 3-day and 10-day depurations, respectively. These results indicate that norovirus GII had accumulated in the oyster's digestive tissue despite the coexistence of FCV. On the other hand, FCV also accumulated during the contamination process but the accumulated FCV in the digestive diverticulum of oyster disappeared after the 10-day depuration (Fig. 2b).
FIG. 2.
Average gene concentration (copies/g of digestive diverticulum) of norovirus GII (a) and FCV (b) in digestive diverticulum of oysters during artificial cocontamination by norovirus GII.6 and FCV f4 (0 to 72 h) and the depuration process (72 to 312 h). Ten oysters were sampled at each time point, and each oyster was assayed by real-time qRT-PCR. The error bar indicates the SD from 10 oysters in a single test of the cocontamination and depuration. The ratio of the number of positive samples to the number of analyzed samples is indicated above each dot.
Burkhardt and Calci showed that virus (F-specific coliphage) was selectively accumulated up to 99-fold in oysters (Crossostrea virginica), but the accumulation of the other indicator microorganisms (fecal coliforms, Escherichia coli, and Clostridium perfringens) was not observed (4). As shown in Fig. 1 and 2, not only norovirus but also FCV accumulated in oysters when the surrounding water was contaminated with these viruses, which means that the occurrence of viral contamination of oysters does not depend on virus species. However, the results in this study clearly indicate that there is an apparent difference between the behaviors of norovirus GII and FCV in the oyster body during the depuration process. The persistency of norovirus GII in oysters could be attributed to the specific binding of noroviral virion on carbohydrate expressed on the tissue of the oyster. Le Guyader et al. showed that there is a carbohydrate on the surface of the digestive tissue of the oyster (Crassostrea gigas) that can specifically bind with the norovirus particle (16). If FCV cannot participate in the selective bindings with oyster tissues, noroviruses could be trapped in the oyster body for a longer period in the depuration process. Although FCV has been used as a surrogate of norovirus in several studies (1, 6, 11, 26), it is important to pay attention to the difference in the behaviors of norovirus and FCV in oyster digestive tissue during the depuration when FCV is employed as a surrogate for norovirus.
In this study, the fate of norovirus in the oyster's digestive tissue during the depuration process was compared to that of FCV. FCV accumulated in oysters was promptly eliminated from the digestive diverticulum of artificially contaminated oysters, whereas noroviruses were persistently observed from the oyster body even though the oyster was processed for the 10-day depuration. The dependency of the elimination rate on virus species during the depuration of bivalve shellfishes has been also reported by the other researchers with regards to an oyster species (Crassostrea virginica) (13), mussels (Mytilus species) (2, 19), and a soft-shelled clam (Mya arenaris) (18). These results suggest that the employment of appropriate surrogates is a crucial issue in the evaluation of shellfish depuration. One alternative surrogate of human norovirus could be murine norovirus 1, which is cultivable and more persistent than FCV under several conditions such as a low-pH environment (<3) and a wet environment with a fecal matrix at room temperature (5a). It is of great interest to evaluate the applicability of murine norovirus 1 as a surrogate for human norovirus in the study of oyster depuration.
We used one of the norovirus GII strains (norovirus GII.6) in this study because this genogroup of norovirus is an important etiological agent all over the world (3). Of further interest is the fate of the other strains of noroviruses, especially norovirus GI strains, in the oyster body. It was reported that Norwalk virus, a prototype of norovirus GI, is poorly depurated from oysters compared to E. coli (22), but the high variability of the noroviral capsid gene implies that each genogroup of norovirus has its own capsid surface characteristics and exhibits distinctive behavior in the oyster body. Furthermore, the water temperature in the contamination and depuration test of oysters is also an important parameter that was not evaluated here. The water temperature was set at 10°C ± 2°C in this study, which is approximately the average water temperature of the oyster farming area during the harvesting period in the Miyagi Prefecture, Japan. Since norovirus contamination in the water environment is found throughout the year (8, 21), the fate of noroviruses in oysters under several conditions of water temperature is worth elucidating in a further study.
Since the removal or inactivation of noroviruses in contaminated oysters is not practical now, it is desirable to cultivate oysters in coastal areas not influenced by domestic wastewater or treated wastewater, major sources of contamination of noroviruses in water environments (17, 20, 21), in order to produce safe oysters with regards to noroviruses.
Acknowledgments
We are deeply grateful to Y. Tohya, University of Tokyo, for the kind distribution of the FCV f4 strain and CRFK cells.
This study was supported by the Fishing Port and Fishing Area Development Division of Miyagi Prefecture with Grants-in-Aid from the Raw Oyster Safety and Reassurance Measures Program, Miyagi Prefecture, Japan.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Bidawid, S., N. Malik, O. Adegbunrin, S. A. Sattar, and J. M. Farber. 2003. A feline kidney cell line-based plaque assay for feline calicivirus, a surrogate for Norwalk virus. J. Virol. Methods 107:163-167. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, A., R. M. Pinto, and F. X. Abad. 1995. Differential accumulation and depuration of human enteric viruses by mussels. Water Sci. Technol. 31:447-451. [Google Scholar]
- 3.Bull, R. A., E. T. V. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkhardt, W., III, and K. R. Calci. 2000. Selective accumulation may account for shellfish-associated viral illness. Appl. Environ. Microbiol. 66:1375-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calci, K. R., G. K. Meade, R. C. Tezloff, and D. H. Kingsley. 2005. High-pressure inactivation of hepatitis A virus within oysters. Appl. Environ. Microbiol. 71:339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Cannon, J. L., E. Papafragkov, G. W. Park, J. Osborne, L.-A. Jaykus, and J. Vinje. 2006. Surrogates for the study of norovirus stability and inactivation in the environment: a comparison of murine norovirus and feline calicivirus. J. Food Prot. 69:2761-2765. [DOI] [PubMed] [Google Scholar]
- 6.D'Souza, D. H., A. Sair, K. Williams, E. Papafragkou, J. Jean, C. Moore, and L. Jaykus. 2006. Persistence of caliciviruses on environmental surfaces and their transfer to food. Int. J. Food Microbiol. 108:84-91. [DOI] [PubMed] [Google Scholar]
- 7.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 8.Haramoto, E., H. Katayama, K. Oguma, and S. Ohgaki. 2005. Application of cation-coated filter method to detection of noroviruses, enteroviruses, adenoviruses, and torque teno viruses in the Tamagawa River in Japan. Appl. Environ. Microbiol. 71:2403-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jothikumar, N., J. A. Lowther, K. Henshilwood, D. N. Lees, V. R. Hill, and J. Vinjé. 2005. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl. Environ. Microbiol. 71:1870-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kageyama, H., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kampf, G., D. Grotheer, and J. Steinmann. 2005. Efficacy of three ethanol-based hand rubs against feline calicivirus, a surrogate virus for norovirus. J. Hosp. Infect. 60:144-149. [DOI] [PubMed] [Google Scholar]
- 12.Kingsley, D. H., G. K. Meade, and G. O. Richards. 2002. Detection of both hepatitis A virus and Norwalk-like virus in imported clams associated with food-borne illness. Appl. Environ. Microbiol. 68:3914-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kingsley, D. H., and G. P. Richards. 2003. Persistence of hepatitis A virus within oysters. J. Food Prot. 66:331-334. [DOI] [PubMed] [Google Scholar]
- 14.Lees, D. 2000. Viruses and bivalve shellfish. Int. J. Food Microbiol. 59:81-116. [DOI] [PubMed] [Google Scholar]
- 15.Le Guyader, F. S., F. Bon, D. DeMedici, S. Parnaudeau, A. Bertone, S. Crudeli, A. Doyle, M. Zidane, E. Suffredini, E. Kohli, F. Maddalo, M. Monini, A. Gallay, M. Pommepuy, P. Pothier, and F. M. Ruggeri. 2006. Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. J. Clin. Microbiol. 44:3878-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Guyader, F. S., F. Loisy, R. L. Atmar, A. M. Hutson, M. K. Estes, N. Ruvoen-Clouet, M. Pommepuy, and J. Le Pendu. 2006. Norwalk virus-specific binding to oyster digestive tissues. Emerg. Infect. Dis. 12:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodder, W. J., and A. M. de Roda Husman. 2005. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl. Environ. Microbiol. 71:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metcalf, T. G., B. M. Mullin, D. Eckerson, E. Moulton, and E. P. Larkin. 1979. Bioaccumulation and depuration of enteroviruses by the soft-shelled clam, Mya arenaria. Appl. Environ. Microbiol. 38:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Power, U. F., and J. K. Collins. 1989. Differential depuration of poliovirus, Escherichia coli, and a coliphage by the common mussel, Mytilus edulis. Appl. Environ. Microbiol. 55:1386-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutjes, S. A., R. Italiaander, H. H. J. L. van den Berg, W. J. Lodder, and A. M. de Roda Husman. 2005. Isolation and detection of enterovirus RNA from large-volume water samples by using the NucliSens miniMAG system and real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 71:3734-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sano, D., Y. Ueki, T. Watanabe, and T. Omura. 2006. Genetic variation in the conservative gene region of Norovirus genogroup II strains in environmental and stool samples. Environ. Sci. Technol. 40:7423-7427. [DOI] [PubMed] [Google Scholar]
- 22.Schwab, K. J., F. H. Neill, M. K. Estes, T. G. Metcalf, and R. L. Atmar. 1998. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by using RT-PCR. J. Food. Prot. 61:1674-1680. [DOI] [PubMed] [Google Scholar]
- 23.Tohya, Y., K. Masuoka, E. Takahashi, and T. Mikami. 1991. Neutralizing epitopes of feline calicivirus. Arch. Virol. 117:173-181. [DOI] [PubMed] [Google Scholar]
- 24.Ueki, Y., D. Sano, T. Watanabe, K. Akiyama, and T. Omura. 2005. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res. 39:4271-4280. [DOI] [PubMed] [Google Scholar]
- 25.Wobus, C. E., S. M. Karst, L. B. Thackray, K. O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackenzie, K. Y. Green, and H. W. Virgin IV. 2004. Replication of Norovirus in cell culture reveals a tropism for dendritic cells macrophages. PLoS Biol. 2:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wollants, E., P. Maes, I. Thoelen, F. Vanneste, M. Rahman, and M. van Ranst. 2004. Evaluation of a norovirus sampling method using sodium dodecl sulfate/EDTA-pretreated chromatography paper strips. J. Virol. Methods 122:45-48. [DOI] [PubMed] [Google Scholar]