Abstract
Listeria monocytogenes is a facultative intracellular pathogen thought to be widely distributed in the environment. We investigated the prevalence and characteristics of L. monocytogenes isolates from surface waters derived from catchments within the South Nation River watershed (Ontario, Canada). This watershed is dominated by urban and rural development, livestock and crop production, and wildlife habitats. From June to November 2005, a total of 314 surface water samples were collected biweekly from 22 discrete sampling sites characterized by various upstream land uses. Presumptive Listeria spp. were isolated using a selective enrichment and isolation procedure, and 75 L. monocytogenes isolates were identified based on colony morphology, hemolytic activity, and amplification of three pathogenicity genes: iap, inlA, and hlyA. Thirty-two of 314 (10%) surface water samples were positive for the presence of L. monocytogenes, but detection ranged between 0 and 27% depending on the sampling date. Isolates belonging to serovar group 1/2a, 3a (50%) and group 4b, 4d, 4e (32%) were dominant. L. monocytogenes populations were resolved into 13 EcoRI ribotypes and 21 ApaI and 21 AscI pulsotypes. These had Simpson indexes of discrimination of up to 0.885. Lineage I-related isolates were dominant (61%) during the summer, whereas lineage II isolates were dominant (77%) in the fall. Isolates were, on average, resistant to 6.1 ± 2.1 antibiotics out of 17 tested. Half of the L. monocytogenes isolates exhibited potential virulence linked to the production of a functional internalin A, and some isolates were found to be moderately to highly virulent by in vitro Caco-2 plaque formation assay (up to 28% of entry). There was a statistically significant link between the occurrence of L. monocytogenes and proximity to an upstream dairy farm and degree of cropped land. Our data indicate that L. monocytogenes is widespread in the studied catchments, where it could represent a public health issue related to agricultural land use.
Listeria monocytogenes is a gram-positive facultative intracellular pathogen responsible for severe food-borne infections in humans and causes 20 to 50% mortality in susceptible populations, such as newborn children, the elderly, and immunocompromised persons (22, 56). This bacterium is thought to be a saprophytic organism living naturally in the plant-soil environment, where it can survive for up to several months, being able to multiply in decaying vegetation but unlikely to multiply in soil (16). In the natural environment, biotic and abiotic factors shown to reduce L. monocytogenes survival are predation, high temperature, UV exposure, and low moisture (12, 16, 20). Soil texture also influences L. monocytogenes survival, but the availability of inorganic nutrients (nitrogen and phosphorus) does not influence survival (12).
A possible agricultural route of human exposure is through the ingestion of uncooked food crops grown in soil irrigated with contaminated water and/or fertilized with Listeria-contaminated manure or biosolids (5, 53) or through the consumption of fish or shellfish grown in contaminated fresh- or seawater (20, 30). Listeria spp. are carried in asymptomatic livestock and poultry and excreted in feces (31); therefore, humans can also be exposed to this pathogen through normal farming operations where human-livestock interaction can readily occur. A recent survey of 1,549 agricultural waste samples in the United Kingdom found 15.4% (stored poultry samples) to 44.4% (stored sheep samples) of fecal samples to be positive for pathogenic L. monocytogenes and Listeria ivanovii (24). Although L. monocytogenes was detected at very low frequencies (0.12%) in fresh human feces in the United States, a survey of the organism in sewage treatment discharges in France found 49 of 77 wastewater treatment plant effluent and 56 of 83 raw sludge samples to be L. monocytogenes positive (38, 48).
Wildlife represents another environmental reservoir for Listeria (5, 14), and there is little information on the prevalence and characteristics of L. monocytogenes in natural surface waters, particularly in areas that are characterized by a mix of anthropogenic (e.g., farming, urban) and nonanthropogenic (e.g., natural forests, wetlands) land uses. For instance, 5 of 128 (3.9%) river or lake water samples collected from a Northern Greece study site were positive for Listeria spp. (1). L. monocytogenes was also isolated in 62% of freshwater samples from a California coast estuary (9) as well as in 6.4% of surface and groundwater samples from a mountainous region in Switzerland (50). The widespread distribution of this bacterium in watersheds that contain mixed agricultural and human recreational activity could be significant with respect to the transmission and epidemiology of zoonotic infections in livestock and wildlife (16). Furthermore, ruminants in particular can shed significant numbers of L. monocytogenes (up to 5 × 102 CFU g−1 of feces) (15), increasing their density in surrounding soil and drainage waters (31). Thus, the presence of Listeria spp. could serve as a potentially robust indicator of fecal contamination originating from beef and dairy operations (49).
The objectives of this study were (i) to conduct a systematic survey of the prevalence and seasonal distribution of L. monocytogenes in surface waters of the South Nation River watershed, eastern Ontario, Canada, (ii) to characterize L. monocytogenes isolates phenotypically, genotypically, and with respect to attributes that influence pathogenicity, and (iii) to evaluate the spatial distribution of L. monocytogenes with respect to land use practices in the watershed study area.
MATERIALS AND METHODS
Sample sites and surface water sampling.
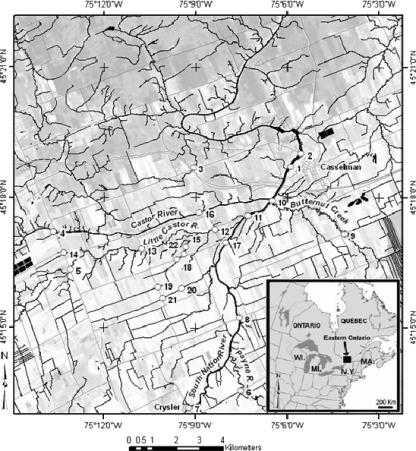
The surface water sampling sites were located within the South Nation River watershed in eastern Ontario, Canada. The total area of the watershed is 3,900 km2. Topography in the region is generally flat, with subsurface tile drainage and groundwater being the primary flow contributors. Roughly 60% of the land use in the South Nation watershed is farming, consisting primarily of dairy operations and cash/livestock cropping systems (Statistics Canada's 1996 agriculture census data are available at http://ceps.statcan.ca/english/profil/PlaceSearchForm1.cfm). Surface waters are frozen in the winter months, and manure from livestock operations is typically applied in the spring and in the fall. At Plantagenet Springs, mean monthly discharge peaks in April, at 197 m3 s−1, and is lowest on average in August, at 6 m3 s−1 (archived hydrometric data for station 02LB005 from Environment Canada's 2004 water survey; http://www.wsc.ec.gc.ca/). For this study, water sampling took place within an area of approximately 200 km2, with sampling sites located on the South Nation River proper and on selected water courses that feed the South Nation River (Fig. 1).
FIG. 1.
Location of water sampling sites on subwatersheds located within the broader South Nation River watershed, Ontario, Canada.
Land use, hydrological, and ancillary water quality information.
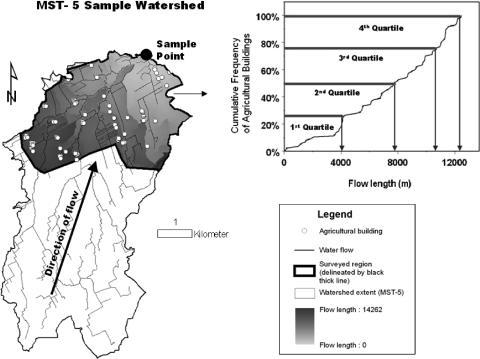
Primary land use information was collected via an intensive roadside survey in the study region (47). In addition, land surface attributes were classified in a spatially exhaustive manner for each catchment on the basis of SPOT 5 panchromatic imagery (2.5-m lateral resolution), and a K-means unsupervised classification (40) using LANDSAT-5 Thematic Mapper imagery from 2 June 2005 (30-m resolution covering bands 1 to 5 and 7). Catchment limits were derived in geographic information systems using a digital elevation model with integration of a vector stream layer. Three general classes of land use variables based on survey as well as remote sensing information were generated (Fig. 2), namely, (i) within the roadside survey section of a given sample site catchment area, the closest distance upstream of the sample site to critical land use activity or emplacement (e.g., dairy operation, pasture lands, etc.), (ii) from the cumulative frequency of a specified land use attribute with distance upstream of the sample site (within roadside survey section of given sample site catchment area), the distance upstream at which the first, second, third, and fourth frequency quartiles are achieved, and (iii) the percentage of total watershed coverage for selected land surface attributes (e.g., via LANDSAT-5 image processing) upstream of the sample site. Distance upstream calculations are flow length distances of the path of water from land use activity to a particular sample location. Land use attributes considered in this study include pasture land, land under commercial and noncommercial development, dairy operations (e.g., barns), forage land, shrub land (natural lands), forests, pasture lands, and crop lands. Specific land use variables are given in Table 1.
FIG. 2.
Example of how nearest upstream distance and first-quartile upstream distance variables were derived for analysis. The land use attribute example given here is agricultural buildings.
TABLE 1.
Land use variables used in this studya
| Variable name | Description |
|---|---|
| NU_PAS | Nearest upstream distance to pasture (km) |
| NU_DEVHI | Nearest upstream distance to commercial and nonresidential land (km) |
| NU_DEVL | Nearest upstream distance to a residential house (km) |
| NU_DAI | Nearest upstream distance to a dairy farm (km) |
| NU_FOREST | Nearest upstream distance to forest observation (km) |
| Q1_PAS | First quartile upstream distance to pasture (km) |
| Q1_DAI | First quartile upstream distance to a dairy farm (km) |
| L5_CROP | Percent crop land coverage in subwatershed (% of land) |
| L5_DEVb | Percent commercial and residential building coverage in watershed (% of land) |
| L5_FOR | Percent forage coverage in watershed (% of land) |
| L5_PAS | Percent pasture coverage in watershed (% of land) |
| L5_SHR | Percent shrub land coverage in watershed (% of land) |
All variables are referenced spatially with respect to all sampling sites within the study area (Fig. 1).
Excluding agricultural buildings.
Water samples (n = 314) were collected from 29 June 2005 to 30 November 2005, at 22 sample sites, on a biweekly basis (Table 2). Some smaller tributaries did not flow during the whole sampling period and yielded fewer samples. Some sampling was undertaken to capture rain events, and 74 samples were obtained within 24 h of a major precipitation event. The mean monthly air temperature during the experiment ranged from 3°C to 22°C. The catchment area contributing surface waters to the sample locations ranged between 2,371.1 km2 (South Nation River proper) and 1.8 km2 (rural drainage ditch) (Table 2). Detailed site descriptions of sites MST 1 to MST 15 are given by Ruecker et al. (47). Sites MST 16 and MST 17 are river-based sampling locales and represent significant upstream catchment areas. Sites MST 18 to MST 22 are rural drainage ditches fed by tile drainage waters.
TABLE 2.
Land use and surface water catchment area characteristics for the suite of catchments within the South Nation River watershed
| Site | Catchment size (km2) | % Catchment area surveyed in 2005 | Upstream distance from sampling site to upper margin of surveyed area (km) | % Land surveyedd
|
|||
|---|---|---|---|---|---|---|---|
| Dairy or cattle pasture | Cropa | Urbanb | Naturalc | ||||
| MST 1 | 2,370.6 | 16 | 53.0 | 4 | 38 | 1 | 15 |
| MST 2 | 2,371.1 | 16 | 53.5 | 4 | 38 | 1 | 15 |
| MST 3 | 3.0 | 72 | 3.4 | 0 | 17 | 2 | 27 |
| MST 4 | 723.5 | 2 | 12.9 | 2 | 41 | 8 | 13 |
| MST 5 | 80.9 | 38 | 24.0 | 2 | 46 | 0 | 11 |
| MST 6 | 176.2 | 76 | 41.3 | 5 | 32 | 1 | 23 |
| MST 7 | 1,215.9 | 4 | 26.5 | 5 | 48 | 0 | 10 |
| MST 8 | 1,412.8 | 14 | 44.5 | 4 | 37 | 1 | 18 |
| MST 9 | 54.3 | 70 | 15.8 | 4 | 36 | 0 | 19 |
| MST 10 | 67.5 | 75 | 21.0 | 4 | 38 | 1 | 15 |
| MST 11 | 1,548.0 | 18 | 49.6 | 4 | 39 | 1 | 15 |
| MST 12 | 95.6 | 48 | 32.9 | 3 | 46 | 0 | 11 |
| MST 13 | 88.0 | 43 | 28.3 | 3 | 44 | 0 | 11 |
| MST 14 | 2.2 | 100 | 3.4 | 9 | 36 | 1 | 5 |
| MST 15 | 2.9 | 100 | 6.9 | 3 | 54 | 0 | 0 |
| MST 16 | 738.5 | 3 | 19.7 | 2 | 28 | 3 | 19 |
| MST 17 | 1,450.3 | 16 | 47.9 | 4 | 38 | 1 | 16 |
| MST 18 | 2.8 | 100 | 6.3 | 0 | 56 | 0 | 0 |
| MST 19 | 1.8 | 100 | 4.4 | 1 | 70 | 0 | 0 |
| MST 20 | 4.3 | 100 | 6.0 | 2 | 53 | 0 | 13 |
| MST 21 | 1.8 | 100 | 4.5 | 3 | 56 | 0 | 14 |
| MST 22 | 2.8 | 100 | 6.4 | 0 | 55 | 0 | 0 |
Crop land is land under corn, soybean, wheat, or other production, excluding farmland devoted to forages (e.g., alfalfa, grass, clover).
Urban land excludes housing associated with a farming operation.
Natural land includes wetland, exposed rock, shrub land, or forest.
Rounded to the nearest percent.
For bacterial analysis, 1 liter of surface water was collected from within a 0.5-m depth of the surface directly into sterile containers (Systems Plus, Woodstock, Ontario, Canada) by using a collection pole at each sampling site. The samples (packed with cold packs) were shipped by overnight courier to Agriculture and Agri-Food Canada laboratories in London, Ontario, Canada.
Enrichment and isolation of Listeria spp. from water samples.
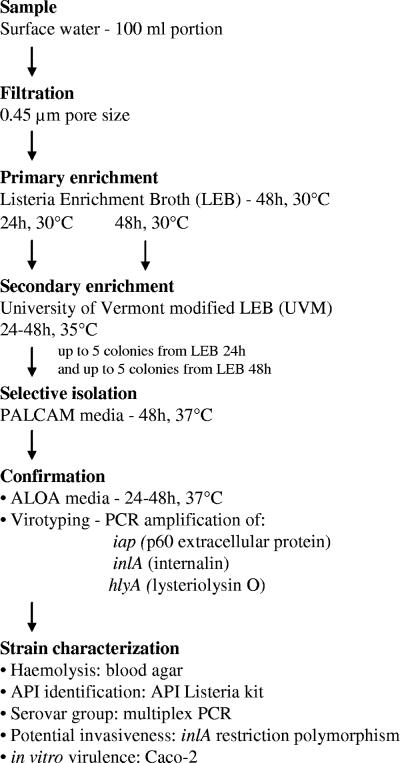
Listeria sp. enrichments as described below were initiated within 24 h of sampling. Water samples were processed according to the MFHPB-30 method (36) with some modifications (Fig. 3). Primary enrichment was conducted with Listeria enrichment broth (LEB; Difco, Toronto, Ontario, Canada), followed by a secondary enrichment in UVM modified Listeria enrichment broth (UVM; Difco). One hundred-milliliter portions of water were filtered through sterile, 0.45-μm-pore-size, 47-mm cellulose acetate filters (Pall Gelman GN-6; VWR International, Mississauga, Ontario, Canada), and the filters were aseptically transferred into 9 ml of LEB. Inoculated LEB was incubated for 48 h at 30°C. At 24 and 48 h, 100 μl of primary enrichment broth was transferred into 10 ml of UVM and incubated for a further 48 h at 35°C. The UVM secondary enrichment (50 μl), inoculated from the 24- and 48-h LEB, was streaked onto polymyxin-acriflavin-lithium chloride-ceftazidime-aesculin-mannitol (PALCAM) agar (Difco) and incubated for 24 to 48 h at 37°C. For each sample and from each PALCAM plate, five (or all, when fewer were present) well-isolated Listeria presumptive colonies that were olive green in color and surrounded by a dark halo were picked, inoculated into 100 μl of brain heart infusion broth (Fisher Scientific, Ottawa, Ontario, Canada), and grown overnight at 37°C. Cultures were then stored at 4°C for a maximum of 2 weeks until used in confirmatory tests. Two samples spiked with L. monocytogenes ATCC19112 were used as positive controls.
FIG. 3.
Flowchart of the procedures for enrichment, isolation, confirmation, and characterization of L. monocytogenes strains.
Confirmation of L. monocytogenes.
PALCAM isolates (presumptive L. monocytogenes; n = 2,826) were confirmed as L. monocytogenes on the basis of colony morphology on agar Listeria Ottaviani & Agosti (ALOA; AES Laboratories, Combourg, France) and the presence of at least two of three virulence genes. Isolates were plated on ALOA and incubated at 37°C for 24 to 48 h. L. monocytogenes on this medium yields blue-green colonies surrounded by an opaque halo. L. monocytogenes can also be distinguished from other species of Listeria on the basis of the presence of three virulence genes that can be revealed by PCR: iap (encoding the p60 extracellular protein) (21), inlA (encoding internalin) (46), and hlyA (encoding lysteriolysin O) (34). DNA template was prepared from overnight brain heart infusion broth cultures by using a DNeasy extraction kit (QIAGEN, Mississauga, Ontario, Canada) according to the manufacturer's “purification of genomic DNA from gram-positive bacteria” protocol. A 300-bp iap fragment was amplified using primer LIM2 (21) and modified reverse primer LIM2REV (5′-AACGTGAGAAATTCCGCTACC-3′) (modified from reference 21), according to the protocols of these authors. A 733-bp inlA fragment, encoding a region between A10 and part of repeat B1 of InlA, was amplified using primers seq01 and seq02 as described previously (46). An 858-bp hlyA fragment was amplified using primers α-1 and β-1 as described elsewhere (34). Five L. monocytogenes strains (ATCC7644, ATCC15313, ATCC19112, ATCC19114, ATCC19115) and a strain from each of the other Listeria spp. (L. grayi ATCC25401, L. innocua ATCC33090, L. ivanovii ATCC19119, L. seeligeri ATCC35967, and L. welshimeri ATCC35897) were included as positive and negative controls. These strains were also included as positive and negative controls in the assays described below, unless mentioned otherwise.
Biochemical identification.
Hemolysis was assessed by streaking isolated colonies on 5% sheep Columbia blood agar (Oxoid, Nepean, Ontario, Canada). After 24 h of incubation at 35°C, plates were examined for the presence of a zone of hemolysis. Some isolates were also identified using an API Listeria kit (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions.
Antibiotic resistance analysis.
Amoxicillin, ampicillin, cephalosporin C, ciprofloxacin, chloramphenicol, erythromycin, florfenicol, gentamicin, kanamycin, lincomycin, nalidixic acid, penicillin G, rifampin, streptomycin, tetracycline, trimethoprim, and vancomycin were purchased from Sigma-Aldrich (Oakville, Ontario, Canada). Antibiotics were dissolved in appropriate solvents to prepare stock solutions and were added to Mueller-Hinton agar (Difco, Toronto, Ontario, Canada) after autoclaving. Media were prepared without antibiotics, or with each antibiotic at the breakpoint concentration, as specified previously (52). Breakpoints of 1, 8, and 8 μg ml−1 were used for ciprofloxacin, lincomycin, and nalidixic acid, respectively (based on the January 2006 release of Communiqué 2006 from the Comité de l'Antibiogramme de la Société Française de Microbiologie, Société Française de Microbiologie, Paris, France; http://www.sfm.asso.fr/). L. monocytogenes isolates (n = 75) from frozen glycerol stock were inoculated into 100 μl of Mueller-Hinton broth in 96-well microtiter plates and grown overnight at 37°C. Cultures were then diluted in sterile water containing 0.02% Tween 20 to ensure that cells were well dispersed to an optical density suitable to transfer 104 CFU by using a floating pin replicator (V&P Scientific, Inc., San Diego, CA) to inoculate the Mueller-Hinton plates. The plates were incubated for 36 h at 37°C and scored visually for growth. If the culture grew in the presence of a particular antibiotic, it was considered resistant, whereas if the culture did not grow, it was considered susceptible.
Serovar group characterization.
The serovar group of 75 L. monocytogenes strains was determined using a multiplex PCR assay that detects the presence of five genes whose distribution is a reliable indicator of serovar: lmo0737 (unknown protein), lmo1118 (unknown protein), the open reading frame (ORF) 2819 gene (putative transcriptional regulator), the ORF 2110 gene (putative secreted protein), and prs (putative phosphoribosyl pyrophosphate synthetase) (10, 11). Amplification was carried out as described elsewhere (10), and PCR products were resolved by 2% agarose gel electrophoresis.
EcoRI ribotyping.
Ribotyping was performed with the 75 L. monocytogenes strains by using the restriction enzyme EcoRI and a RiboPrinter microbial characterization system (Qualicon, Inc., Wilmington, DE) according to the manufacturer's instructions and as previously described (7). Automated ribotyping data allowed for the classification of the isolates into the lineages defined previously (58). Ribotyping patterns were named according to the DuPont database of existing patterns. When a pattern exhibited a one-band difference from the DuPont database designation, the symbol “<” precedes its designation. EcoRI patterns were compared against the Pathogen Tracker database (http://www.pathogentracker.net).
PFGE.
L. monocytogenes isolates (n = 75) were subtyped by using PFGE with AscI and ApaI restriction endonucleases as previously described (19). Pulsed-field gel electrophoresis (PFGE) and pattern analyses were performed by the Listeriosis Reference Service Laboratory, a member of PulseNet Canada, the National Molecular Subtyping Network for Food-Borne Disease Surveillance. The PFGE patterns were analyzed using the BioNumerics software package (version 4.5; Applied Maths, Kortrijk, Belgium). Pattern clustering was performed using the unweighted-pair group method using arithmetic averages and the Dice correlation coefficient with a position tolerance of 1.0%. The PFGE patterns were compared against data in the PulseNet Canada National Listeria Database.
inlA PCR-RFLP.
Potentially noninvasive L. monocytogenes strains (n = 75) were screened using a PCR-restriction fragment length polymorphism (RFLP) method based on inlA polymorphism as described previously (46). PCR-amplified (as described above) inlA fragments were digested using the restriction endonuclease AluI (Qbiogene, Illkirch, France), and PCR-RFLP fragments were resolved by electrophoresis in 3.5% agarose gel (MetaPhor agarose; TEBU, Le Perray-en-Yvelines, France). The position of restriction fragments was normalized using an internal DNA size marker (Marker VIII; Roche, Meylan, France).
In vitro virulence test for L. monocytogenes isolates.
The capacity of L. monocytogenes to propagate in Caco-2 cells was evaluated with a plaque formation assay as described previously (46), using human carcinoma cell line Caco-2, obtained from the European Collection of Cell Cultures (ECACC 86010202) and used between passages 43 and 46. The initial entry was determined by the ratio (expressed as a percentage) of the number of lysis plaques observed to the initial number of bacteria added. Strains Scott A and EGD-e (obtained from the Institut Pasteur Collection, Paris, France) were used as virulent reference strains for comparative analysis.
CART analysis.
The 314 water samples that were tested were categorized by the presence (positive samples) or absence of L. monocytogenes. This information was used as the dependent variable in a “decision tree”-based classification and regression tree analysis (CART) (6, 54) to uncover affinities between L. monocytogenes and land use attributes (Table 1). The nonparametric, binary, recursive, and computer-driven CART method is a well-established data mining tool that has been used, for instance, to uncover spatial relationships between Escherichia coli populations in manure holding tanks (28). Because the L. monocytogenes data are categorical, a classification tree methodology was employed with CART using the Gini splitting criteria (54). For this heuristic study, default CART parameters were used exclusively. In addition to evaluating which variables, in statistical terms, “optimally” classified the data in terms of independent variable criteria, grouping conditions that closely mimicked, on a case by case basis, the “optimal” classification were examined. These surrogate analyses helped reveal the structure of intercorrelations between the predictor variables as well as the most robust predictors in the classification tree.
Statistical treatments.
Summer and fall distributions of L. monocytogenes isolates into lineages were compared using the chi-square test. Caco-2 entry percentages reported in relation to inlA polymorphism types and antibiotic resistance levels for summer and fall populations were compared using the Mann-Whitney test. Nonparametric tests were chosen due to the nonnormality of our data as revealed using the Kolmogorov-Smirnov test. The SPSS 11.0 for Windows program (SPSS, Inc.) was used for all statistical analyses; all significance tests and correlations were considered significant statistically at a P value of ≤0.05.
RESULTS
Prevalence of L. monocytogenes in surface water.
Three hundred fourteen water samples, collected from 22 sites, were analyzed in this study over a 5-month period (Table 3). After enrichment, 200 water samples (64%) exhibited presumptive Listeria colonies on PALCAM (olive green colonies surrounded by a dark halo). After confirmation tests were applied, 32 water samples (10%) were found to be positive for the presence of at least one L. monocytogenes isolate (Table 3). Depending on the sampling date, between 0 and 27% (average, 9%) of the collected water samples were positive for L. monocytogenes isolation. Out of the 2,826 presumptive isolates collected in the present work (up to 10 per tested sample), 75 were confirmed to be L. monocytogenes isolates. All L. monocytogenes isolates exhibited typical colony morphology on ALOA. All but eight isolates showed PCR amplification for the three virulence genes tested: one and seven isolates did not exhibit amplification for the iap gene and the hlyA gene, respectively. A subset of 60 confirmed isolates was tested for β-hemolysis on sheep blood agar, and two did not exhibit any hemolytic activity. An attempt to confirm identification by using an API Listeria kit was carried out with 10 confirmed L. monocytogenes isolates. Four isolates were correctly identified as L. monocytogenes, and six isolates yielded ambiguous codes which did not allow for their identification by this method. The method thus proved to be unsatisfactory with environmental isolates and was abandoned.
TABLE 3.
Distribution of L. monocytogenes-positive water samples per sample localea
| Sampling site | No. of samples | No. of presumptive Listeria-positive enrichments (%) | No. of L. monocytogenes-positive samples (%) | No. of L. monocytogenes isolates |
|---|---|---|---|---|
| MST 1 | 16 | 3 (19) | 0 (0) | 0 |
| MST 2 | 15 | 6 (40) | 1 (7) | 5 |
| MST 3 | 14 | 12 (86) | 3 (21) | 4 |
| MST 4 | 14 | 5 (36) | 1 (7) | 1 |
| MST 5 | 16 | 15 (94) | 2 (13) | 3 |
| MST 6 | 15 | 10 (67) | 1 (7) | 10 |
| MST 7 | 16 | 5 (31) | 1 (6) | 1 |
| MST 8 | 16 | 7 (44) | 0 (0) | 0 |
| MST 9 | 15 | 14 (93) | 4 (27) | 8 |
| MST 10 | 14 | 4 (29) | 1 (7) | 10 |
| MST 11 | 14 | 5 (36) | 1 (7) | 1 |
| MST 12 | 17 | 11 (65) | 1 (6) | 2 |
| MST 13 | 16 | 12 (75) | 3 (19) | 6 |
| MST 14 | 11 | 8 (73) | 1 (9) | 1 |
| MST 15 | 14 | 12 (86) | 1 (7) | 1 |
| MST 16 | 14 | 5 (36) | 1 (7) | 1 |
| MST 17 | 14 | 7 (50) | 0 (0) | 0 |
| MST 18 | 15 | 13 (87) | 4 (27) | 8 |
| MST 19 | 12 | 12 (100) | 1 (8) | 2 |
| MST 20 | 13 | 12 (92) | 2 (15) | 2 |
| MST 21 | 11 | 11 (100) | 2 (18) | 8 |
| MST 22 | 12 | 11 (92) | 1 (8) | 1 |
| Total | 314 | 200 (64) | 32 (10) | 75 |
The numbers and percentages of presumptive Listeria enrichment-positive samples were assessed on the basis of PALCAM plating results.
Antibiotic resistance analysis.
The 75 L. monocytogenes surface water isolates were tested for antibiotic resistance (17 antibiotics at breakpoint concentrations). All isolates were resistant to ampicillin, cephalosporin C, and nalidixic acid, and most isolates were resistant to chloramphenicol (n = 74) and lincomycin (n = 61). Some isolates were resistant to ciprofloxacin (n = 17), erythromycin (n = 4), gentamicin (n = 20), kanamycin (n = 15), penicillin G (n = 3), rifampin (n = 14), nalidixic acid (n = 27), and tetracycline (n = 5). All isolates were susceptible to amoxicillin, florfenicol, trimethoprim, and vancomycin. Water isolates were resistant to an average of 6.2 ± 2.1 (standard deviation) antibiotics. No significant difference was observed between isolates obtained in summer and in fall (Mann-Whitney test, P = 0.777), with an average resistance to 6.2 ± 2.1 and 6.1 ± 2.1 antibiotics, respectively.
L. monocytogenes serological, ribotype, and pulsotype characterization.
The 75 L. monocytogenes isolates were characterized by serovar group, EcoRI ribotyping, and PFGE with ApaI and AscI restriction endonucleases (Table 4). Isolates were distributed in four of the five possible serovar groups described using the multiplex PCR used in the present work. No isolate was related to serovar group 1/2c, 3c. Dominant serovar groups were 1/2a, 3a (49%) and 4b, 4d, 4e (32%), followed by serovar groups 1/2b, 3b, 7 (11%) and Listeria sp. 4a, 4c (8%).
TABLE 4.
Distribution of L. monocytogenes lineages, serovar groups, EcoRI ribotypes, and ApaI and AscI pulsotypes among surface water isolatesa
| Lineage | Serovar group(s) | EcoRI ribotypeb | Pulsotypec
|
No. of isolates | |
|---|---|---|---|---|---|
| ApaI | AscI | ||||
| I | 1/2b, 3b, 7 | <DUP-1052* | 2 (a) | Fa (C, F) | 1 |
| E (C) | E (C, F) | 3 | |||
| F (C) | F (C, F) | 1 | |||
| X (C, F) | X (a) | 1 | |||
| DUP-19175* | D (a) | D (C) | 2 | ||
| 4b, 4d, 4e | DUP-19175* | Z (a) | Da (C) | 1 | |
| DUP-1038* | J (C, F) | J (C, E) | 1 | ||
| DUP-1044* | H (a) | H (a) | 2 | ||
| DUP-18611 | B (C, F) | B (C, F) | 20 | ||
| Total | 32 | ||||
| II | 1/2a, 3a | <DUP-1062* | L (a) | L (C, F) | 2 |
| <DUP-19177 | La (a) | L (C, F) | 1 | ||
| DUP-1030* | S (C, F) | S (C) | 1 | ||
| DUP-1039* | N (a) | N (a) | 6 | ||
| P (a) | P (C) | 1 | |||
| U (C) | U (C, F) | 9 | |||
| Ua (C) | Ua (a) | 11 | |||
| DUP-19157 | T (a) | T (a) | 2 | ||
| Ta (C) | Ta (C) | 1 | |||
| DUP-19171* | V (a) | V (a) | 1 | ||
| DUP-19188 | I (a) | I (C) | 2 | ||
| Total | 37 | ||||
| III | Listeria spp. | <DUP-1059* | A (C) | A (C) | 6 |
| Total | 6 | ||||
Lineages are defined according to reference 58, based on serovar group and ribotyping.
EcoRI ribotype names preceded by a less than sign (<) correspond to patterns exhibiting a one-band difference from the DuPont database designation; those followed by an asterisk correspond to ribotypes associated with human sporadic or epidemic strains, according to the Pathogen Tracker database.
For each PFGE pattern, matches against the PulseNet Canada National Listeria Database are indicated in parentheses. C, clinical; F, food; E, environment; a, no match.
Thirteen unique ribotypes were identified. Ribotypes DUP-1039 (n = 27), DUP-18611 (n = 20), <DUP-1052 (n = 6), and <DUP-1059 (n = 6) were the most commonly observed in this study and grouped 79% of the isolates. Ribotype diversity, as determined by the Simpson index of discrimination (D), was 0.791. Nine of the 13 ribotypes described here were found in the Pathogen Tracker database and have been associated with sporadic and epidemic listeriosis in humans (Table 4).
Both ApaI and AscI PFGE revealed 21 pulsotypes (Table 4). Nine ribotypes, representing 48% of the isolates, were not further discriminated using PFGE, but four ribotypes (DUP-19175, DUP-19157, DUP-1039, and <DUP-1052), representing 52% of our isolates, were further discriminated into 12 pulsotypes. The Simpson index of discrimination calculated for AscI and ApaI pulsotypes was 0.885. Of 75 isolates tested against the PulseNet Canada National Listeria Database, 64 had patterns identical to clinical isolates, 39 were identical to isolates obtained from food, and 1 was of environmental origin (Table 4). Fifteen isolates had patterns that did not match any of the isolates present in the national database.
Serovar group and ribotype data were used to define isolate lineages (58). A significant change was observed between summer and fall L. monocytogenes-positive samples (chi-square test, P < 0.05): summer samples (mid-June to mid-September) were dominated by lineage I isolates (61% of the samples), whereas during the fall (mid-September to the end of November), 77% of the positive samples carried isolates related to lineage II. Samples exhibiting lineage III isolates were found more consistently throughout the sampling period (11 and 8% for summer and fall, respectively).
Potential virulence characterization of L. monocytogenes isolates.
Sequence polymorphism of the inlA gene encoding internalin A was assessed for all isolates by using PCR-RFLP with the AluI restriction enzyme. Four polymorphisms were detected for the L. monocytogenes isolates (corresponding to four of the five profiles previously described [46]; profile 3 was not detected in the present work). Isolates belonged mainly to polymorphism profile 2 (polymorphism 2) (52%) and polymorphism 1 (42%), followed by polymorphism 4 (5%) and polymorphism 5 (1%). Thus, the polymorphism profiles associated with the production of a truncated InlA and a deficient ability to invade Caco-2 (polymorphisms 1 and 4) represented 47% of the tested water isolates. A strong relationship was also observed between inlA polymorphism and serovar groups; polymorphism 2 was recovered only for isolates belonging to lineages I and III (serovar groups 1/2b, 3b, 7; 4b, 4d, 4e; and 4a, 4c), whereas isolates belonging to lineage II (serovar group 1/2a, 3a) were characterized by inlA polymorphism types 1, 4, and 5, which were not found within isolates belonging to lineages I and III.
The abilities of 14 L. monocytogenes isolates to invade epithelial cells were evaluated using the in vitro human enterocyte-like Caco-2 cell model. These 14 strains were chosen to be representative of the dominant inlA polymorphisms (1 and 2). No differences were observed (Mann-Whitney test, P = 0.982) between entry percentages when reported in relation to inlA polymorphism. Entry percentages ranged from less than 1% to 17.2% for polymorphism 1 (mean = 6.7%, n = 6) and from less than 1% to 28.1% for polymorphism 2 (mean = 6.6%, n = 8).
L. monocytogenes and land use relationships.
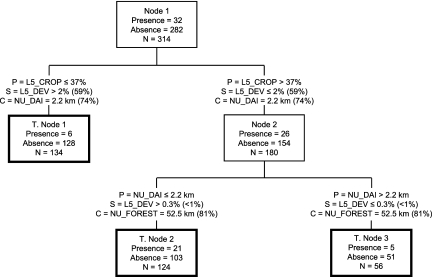
A three-terminal node (i.e., a terminal node [T. node], a terminal classification group produced in CART) classification tree was selected for analysis (Fig. 4) using the 12 independent land use variables given in Table 1. The cross-validated relative cost (i.e., error rate of the tree, relative to node 1, using the learn-test cross validation method [54]) was the second lowest, relative to a tree with two terminal nodes stemming from node 1; a two-terminal-node model was deemed too simplistic for the heuristic guided study here. The selected tree classified correctly 63% of L. monocytogenes absence data (based on T. nodes 1 and 3) and 66% of the presence data (based on T. node 2). The variables most important in terms of improving classification (expressed in terms of capacity to reduce impurity in the overall classification structure [6], i.e., high relative improvement scores), as primary tree splitting variables as well as surrogate splitting variables, are given in Table 5. For the selected tree, the primary tree splitting variables were the most important variables (which is not always the case), and these expressed the degree of cropland in the watershed upstream of the sample site, as well as the nearest upstream distance to a dairy operation in the roadside-surveyed portion of the watershed. Hence, the classification tree indicated that a majority of the 32 L. monocytogenes-positive samples (n = 21, 66% of total positive) originated from sites where cropland area above the site was >37% and where the nearest upstream distance to a dairy operation was ≤2.2 km. Nineteen percent of the L. monocytogenes-positive samples were found at sites with ≤37% cropland coverage in the upstream watershed, irrespective of the NU_DAI variable. In contrast, 16% of the positive L. monocytogenes samples were found at sites with cropland coverage of >37% and the nearest upstream distance to a dairy operation exceeding 2.2 km. For the primary split of all the data (n = 314), the strongest surrogate and competitor variables were L5_DEV and NU_DAI, respectively. Both had modestly strong improvement scores relative to the primary variable improvement score; hence, they appeared to be viable prediction candidates in lieu of L5_CROP. For the node 2 split, the surrogate (L5_DEV) and competitor (NU_FOREST) variable provided, respectively, very low and modestly high improvement scores relative to the node 2 primary split score.
FIG. 4.
CART-based classification tree derived using (i) L. monocytogenes absence and presence information as target data and (ii) land use variables given in Table 1 as predictor variables. Each tree node represents the total number of observations (N) for each node and the number of nodal observations with either presence or absence. Terminal nodes (points in the tree at which no further data splitting occurs) are symbolized by bold rectangles. P, primary split condition (condition used to generate the given data split in the classification tree); S, surrogate split condition; and C, competitor split condition (a competitor variable split is one that provided the second-highest improvement score in the specific nodal splitting process). Percentages in brackets following the S and C split condition information represent, respectively, improvement scores as percentages of primary split improvement scores for given nodal observations (with the primary nodal split having the highest improvement score that could be produced from the available data).
TABLE 5.
Importance scores for variables used in CART with an importance greater than zeroa
| Variable | Importance score |
|---|---|
| L5_CROP | 100 |
| NU_DAI | 83 |
| NU_DEVHI | 60 |
| L5_DEV | 59 |
| L5_SHR | 48 |
| L5_FOR | 16 |
| NU_FOREST | 4 |
Predictor variables not included have an importance of zero. Importance is referenced to 100 (the most important variable in the selected CART classification tree as a primary splitting and surrogate splitting variable).
DISCUSSION
L. monocytogenes has been isolated from a variety of different sources and environmental matrices, although its isolation from surface waters has rarely been reported (16, 49). For a suite of different sized, mixed land-use catchment areas within the larger South Nation River watershed, we demonstrated that L. monocytogenes was repeatedly recovered in surface water over a 5-month sampling period, with frequencies comparable to previous findings from subsurface, surface, and saline waters (1, 9, 27, 50). This result emphasizes the fact that L. monocytogenes is able to survive in an environment characterized by spatially and temporally changing physical (e.g., temperature, solar irradiation, flow) and chemical (e.g., oxygen concentration, pH, nutrients) properties. Its ubiquity could be due in part to its ability to grow at a wide range of temperatures (25), form biofilms (35), and resist various environmental stresses (41).
The commonly used PALCAM medium, combined with a two-step enrichment procedure, yielded a high rate of false positives upon further confirmation, perhaps due to enterococci or bacilli (57). In conjunction with biochemical-based tests, the ALOA medium accurately discriminated L. monocytogenes from other Listeria species, whereas the API Listeria system proved to be insufficient for the identification of Listeria spp. obtained from complex environmental samples (37). Our genotype-based approach, based on the PCR amplification of three L. monocytogenes-specific virulence genes, was effective at identifying L. monocytogenes isolates. However, a few isolates did not exhibit any amplification of the iap or hlyA genes. Presumably, the lack of amplification of these genes may be due to subtle mismatches between the primer and the target DNA. Overall, the combination of ALOA growth characteristics and PCR detection of three virulence genes, recommended as an alternative to cultural and biochemical methods (18), proved to be the most suitable approach to accurately identify L. monocytogenes after the enrichment steps.
All L. monocytogenes isolates collected were characterized with respect to serology, EcoRI ribotypes, and AscI and ApaI pulsotypes. Even if accurate, serology is time-consuming and requires significant technical skills (39). Recently, PCR methods have been used as an alternative to serotyping (4, 11, 60). The multiplex PCR assay used here (10) was designed for the discrimination of the four major serovars isolated from food and clinical samples (1/2a, 1/2b, 1/2c, and 4b). Serovar groups 1/2c and 3b were not detected in this study, indicating that these infrequently detected food and clinical serovars (26) are also absent or infrequent in surface waters. Our results confirm previously established relationships between serovar group, ribotypes, and pulsotypes (29). Both ribotyping and PFGE demonstrated, on the basis of the Simpson index of discrimination (23), that surface water populations were diverse, although PFGE was more discriminant than ribotyping. The majority of isolates grouped into lineages I and II, defined on the basis of most molecular subtyping methods (58). Lineage II is thought to be overrepresented among natural and food environments isolates, in contrast to lineage I, which is thought to be overrepresented among human isolates (49). Indeed, it was hypothesized that with elevated levels of genetic diversity and recombination rates, lineage II isolates were more able to adapt to diverse host and environmental conditions than were highly clonal lineage I isolates (32). However, our results showed both lineages to be equally abundant in surface waters. Lineage III isolates, thought to be associated predominantly with animals (32), were less frequently detected, in agreement with previous studies (43). By means of PFGE, it was recently demonstrated that even if some subtypes were associated with specific sources (human, food, farm, or environment), others were widely distributed and recovered in several sources (17). Lineage distribution of L. monocytogenes water isolates varied with the season. Temperature was previously shown to influence the distribution of serovars in surface waters, with serovar 1/2a being significantly associated with cold periods and serovar 4b being significantly associated with higher air temperatures (51). These findings are in agreement with our results, where lineage II (i.e., serovar 1/2a) isolates were dominant during the fall whereas lineage I (i.e., serovar 4b) isolates were dominant in the summer. Fifty-nine of the 75 isolates matched food or clinical isolates in the PulseNet Canada National Listeria Database, and several of the ribotypes match those archived in the Pathogen Tracker database. Overall, the majority of L. monocytogenes isolates obtained from surface waters of this watershed match those widely associated with food-borne outbreaks and disease.
Different methods have been used to assess L. monocytogenes virulence properties; of these methods, analysis of restriction polymorphisms of virulence factors and in vitro virulence assays proved to correlate with pathogenic potential in animal models (44, 55). One of the proteins implicated in L. monocytogenes cell invasion is an internalin (InlA) that can carry up to nine mutations leading to the secretion of a truncated protein (33, 46). A PCR-RFLP assay has been developed to rapidly identify five of these polymorphisms (46). By use of this assay, nearly half of our isolates were found to exhibit the two polymorphism types that result in excretion of a truncated protein. Interestingly, these particular polymorphisms seemed to be lineage dependent, since they were recovered only for lineage II isolates. This result is in agreement with previous results from France, where isolates exhibited the same polymorphism type/lineage relationship, with the exception of isolate 7F (46), and it also supports a recent finding about inlA mutations being phylogenetically distinct between the different lineages (33). The invasion of Caco-2 human epithelial cells does not support what was previously found in France (46), since mean entry rates were equivalent for isolates exhibiting polymorphism types 1 (truncated and excreted internalin A) and 2. However, some type 2 and 4 isolates described in France also exhibited functional InlA and a high entry percentage in Caco-2 cells (46), and a new inlA mutation affecting lineage I strains is not detectable using the PCR-RFLP assay (33). Since we did not perform any inlA sequencing, it is difficult to conclude whether the high Caco-2 entry rates for some type 2 isolates were due to functional protein or if the low Caco-2 entry rates for some type 1 isolates were due to undetected truncated protein. Nevertheless, our results demonstrated that at least half of the water isolates carried genotypes compatible with the synthesis of a functional InlA, thus being potentially virulent.
With half of surface water L. monocytogenes isolates potentially virulent, and several pulsotypes detected here being identical to those from clinical and food isolates associated with sporadic and epidemic listeriosis, the resistance to multiple antibiotics detected in many isolates is a concern. L. monocytogenes is naturally resistant to nalidixic acid, cephalosporins, fosfomycin, and fluoroquinolones (52), is otherwise susceptible to a wide range of antibiotics, and is generally able to acquire resistance via conjugative transfer of plasmids (8). The sensitivity of our isolates to erythromycin, gentamicin, penicillin, and tetracycline is comparable to those in data obtained for L. monocytogenes from an estuarine environment (45). More generally, our findings are in general agreement with those previously published for food, human, animal, and environmental isolates (2, 42, 52, 59), with the exception of findings for florfenicol, to which all of our isolates were susceptible.
It has been hypothesized that cattle are a natural reservoir for L. monocytogenes and that beef or dairy farms could release large numbers of the bacterium into the surrounding environment (31). Two studies have attempted to relate L. monocytogenes distribution in the environment to agricultural practices and showed that the presence of L. monocytogenes seemed to be associated with the presence of cattle in the catchment area (51) and to the proximity of bovine farms (49). In the present work, we used data mining methods (CART) to uncover relationships between land use variables (expressed as potential surrogates of point and nonpoint pollution sources) and L. monocytogenes-positive water samples. It was found that crop land (e.g., cash and livestock cropping systems, such as corn, soybean, and wheat) coverage in the sample site catchment areas as well as the upstream proximity to dairy operations were the most important predictor variables. The cropping variable could be important, since liquid and solid manures derived from local livestock operations are frequently applied to land as fertilizer; in this study region, a majority of it is of bovine origin. Tile drainage is a critical driver of water flow in the region, and contaminated tile drain effluent can enter adjacent surface water courses (3). Upstream proximity to a dairy operation was another important variable, and the results of this study showed that a majority of the L. monocytogenes-positive water samples were located within a few kilometers downstream of a dairy operation. This finding implies that against the backdrop of other potential human and wildlife sources of contamination, dairy operations, which can consist of dairy barns, pasture lands, manure lagoons, and manured fields, are significant contaminant sources. This finding is consistent with those of previous studies (49, 51), suggesting that similar landscape attribute relationships might have affinities beyond those reported for the environmental setting discussed in this study. Overall, the seasonal distribution of L. monocytogenes in agricultural watersheds will likely depend on the timing of manure applications (spring being the most important period in temperate climates) and the access of animals on pasture to watercourses.
The present work demonstrated that L. monocytogenes is a widespread bacterium in a suite of different mixed-use catchment areas embedded in the greater South Nation River watershed. A significant fraction of L. monocytogenes isolates were found to be potentially virulent, associated with clinical and food isolates, and resistant to multiple antibiotics, and these could represent a potential source of infection for humans, wildlife, or livestock ingesting surface water. However, for health significance purposes, one needs to take into account the possibility that the infective dose of orally ingested L. monocytogenes might be high (e.g., 103 and 108 cells for immunocompromised and normal mice) (13). The L. monocytogenes occurrence in surface waters proved to be related to direct upstream land use, specifically, crop land and proximity to a dairy operation. This result emphasizes the need for identifying sources of microbial contamination for freshwater, in order to settle appropriate water management procedures and, thus, reduce the potential impacts such pathogens might have on human or zoonotic health.
Acknowledgments
This work was funded in part by the National Water Quality Surveillance Research Program through an agreement between Agriculture and Agri-Food Canada and Health Canada.
A. Scott provided excellent technical assistance. We thank several anonymous reviewers whose efforts greatly improved the manuscript.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Arvanitidou, M., A. Papa, T. C. Constantinidis, V. Danielides, and V. Katsouyannopoulos. 1997. The occurrence of Listeria spp. and Salmonella spp. in surface waters. Microbiol. Res. 152:395-397. [DOI] [PubMed] [Google Scholar]
- 2.Aureli, P., A. M. Ferrini, V. Mannoni, S. Hodzic, C. Wedell-Weergaard, and B. Oliva. 2003. Susceptibility of Listeria monocytogenes isolated from food in Italy to antibiotics. Int. J. Food Microbiol. 83:325-330. [DOI] [PubMed] [Google Scholar]
- 3.Ball Coelho, B. R., R. C. Roy, E. Topp, and D. R. Lapen. 2007. Tile water quality following liquid swine manure application into standing corn. J. Environ. Qual. 36:580-587. [DOI] [PubMed] [Google Scholar]
- 4.Borucki, M. K., and D. R. Call. 2003. Listeria monocytogenes serotype identification by PCR. J. Clin. Microbiol. 41:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brackett, R. E. 1999. Incidence, contributing factors, and control of bacterial pathogens in produce. Postharvest Biol. Technol. 15:305-311. [Google Scholar]
- 6.Breiman, L., J. Freidman, R. Olshen, and C. Stone. 1984. Classification and regression trees. Wadsworth International, Pacific Grove, CA.
- 7.Bruce, J. L. 1996. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 50:71-81. [Google Scholar]
- 8.Charpentier, E., and P. Courvalin. 1999. Antibiotic resistance in Listeria spp. Antimicrob. Agents Chemother. 43:2103-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colburn, K. G., C. A. Kaysner, C. Abeyta, and M. M. Wekell. 1990. Listeria species in a California coast estuarine environment. Appl. Environ. Microbiol. 56:2007-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doumith, M., C. Buchrieser, P. Glaser, C. Jacquet, and P. Martin. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doumith, M., C. Jacquet, P. Gerner-Smidt, L. M. Graves, S. Loncarevic, T. Mathisen, A. Morvan, C. Salcedo, M. Torpdahl, J. A. Vazquez, and P. Martin. 2005. Multicenter validation of a multiplex PCR assay for differentiating the major Listeria monocytogenes serovars 1/2a, 1/2b, 1/2c, and 4b: toward an international standard. J. Food Prot. 68:2648-2650. [DOI] [PubMed] [Google Scholar]
- 12.Dowe, M. J., E. D. Jackson, J. G. Mori, and C. R. Bell. 1997. Listeria monocytogenes survival in soil and incidence in agricultural soils. J. Food Prot. 60:1201-1207. [DOI] [PubMed] [Google Scholar]
- 13.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenlon, D. R. 1985. Wild birds and silage as reservoirs of Listeria in the agricultural environment. J. Appl. Bacteriol. 59:537-543. [DOI] [PubMed] [Google Scholar]
- 15.Fenlon, D. R., J. Wilson, and W. Donachie. 1996. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J. Appl. Bacteriol. 81:641-650. [DOI] [PubMed] [Google Scholar]
- 16.Fenlon, D. R. 1999. Listeria monocytogenes in the natural environment, p. 21-38. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety. Marcel Decker, Inc., New York, NY.
- 17.Fugett, E. B., D. Schoonmaker-Bopp, N. B. Dumas, J. Corby, and M. M. Wiedmann. 2007. Pulsed-field gel electrophoresis (PFGE) analysis of temporally matched Listeria monocytogenes isolates from human clinical cases, foods, ruminant farms, and urban and natural environments reveals source-associated as well as widely distributed PFGE types. J. Clin. Microbiol. 45:865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasanov, U., D. Hughes, and P. M. Hansbro. 2005. Methods for the isolation and identification of Listeria spp. and Listeria monocytogenes: a review. FEMS Microbiol. Rev. 65:2707-2716. [DOI] [PubMed] [Google Scholar]
- 19.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, C. H., B. F. Vogel, and L. Gram. 2006. Prevalence and survival of Listeria monocytogenes in Danish aquatic and fish processing environments. J. Food Prot. 69:2113-2122. [DOI] [PubMed] [Google Scholar]
- 21.Hein, I., D. Klein, A. Lehner, A. Bubert, E. Brandl, and M. Wagner. 2001. Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res. Microbiol. 152:37-46. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Y.-T., S.-U. Chen, M.-Z. Wu, C.-Y. Chen, W.-S. Hsieh, B.-N. Tsao, C.-J. Horng, and P.-R. Hsueh. 2006. Molecular evidence for vertical transmission of listeriosis, Taiwan. J. Med. Microbiol. 55:1601-1603. [DOI] [PubMed] [Google Scholar]
- 23.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchison, M. L., L. D. Walters, S. M. Avery, B. A. Synge, and A. Moore. 2004. Levels of zoonotic agents in British livestock manures. Lett. Appl. Microbiol. 39:207-214. [DOI] [PubMed] [Google Scholar]
- 25.Junttila, J. R., S. I. Niemela, and J. Hirn. 1988. Minimum growth temperatures of Listeria monocytogenes and non-haemolytic Listeria. J. Appl. Bacteriol. 65:321-327. [DOI] [PubMed] [Google Scholar]
- 26.Kerouanton, A., A. Brisabois, E. Denoyer, F. Dilasser, J. Grout, G. Salvat, and B. Picard. 1998. Comparison of five typing methods for the epidemiological study of Listeria monocytogenes. Int. J. Food Microbiol. 43:61-71. [DOI] [PubMed] [Google Scholar]
- 27.Korhonen, L. K., M. Niskanen, H. Heinonen-Tanski, P. J. Martikainen, L. Salonen, and I. Taipalinen. 1996. Groundwater quality in wells in central rural Finland: a microbiological and radiochemical survey. Ambio 25:343-349. [Google Scholar]
- 28.Lu, Z., D. Lapen, A. Scott, A. Dang, and E. Topp. 2005. Identifying host sources of fecal pollution: diversity of Escherichia coli in confined dairy and swine production systems. Appl. Environ. Microbiol. 71:5992-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukinmaa, S., K. Aarnisalo, M.-L. Suihko, and A. Siitonen. 2004. Diversity of Listeria monocytogenes isolates of human and food origin studied by serotyping, automated ribotyping and pulse-field gel electrophoresis. Clin. Microbiol. Infect. 10:562-568. [DOI] [PubMed] [Google Scholar]
- 30.Miettinen, H., and G. Wirtanen. 2005. Prevalence and location of Listeria monocytogenes in farmed rainbow trout. Int. J. Food Microbiol. 104:135-143. [DOI] [PubMed] [Google Scholar]
- 31.Nightingale, K. K., Y. H. Schukken, C. R. Nightingale, E. D. Fortes, A. J. Ho, Z. Her, Y. T. Grohn, P. L. McDonough, and M. Wiedmann. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70:4458-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nightingale, K. K., K. Windham, and M. Wiedmann. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nightingale, K. K., K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norton, D.-M., M. A. McCamey, K. L. Gall, J. M. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl. Environ. Microbiol. 67:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norwood, D. E., and A. Gilmour. 1999. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 86:576-582. [DOI] [PubMed] [Google Scholar]
- 36.Pagotto, F., E. Daley, J. Farber, and D. Warburton. 2001. Isolation of Listeria monocytogenes from all food and environmental samples. HPB method MFHPB-30. Health Canada, Ottawa, Ontario, Canada.
- 37.Paillard, D., V. Dubois, R. Duran, F. Nathier, C. Guittet, P. Caumette, and C. Quentin. 2003. Rapid identification of Listeria species by using restriction fragment length polymorphism of PCR-amplified 23S rRNA gene fragments. Appl. Environ. Microbiol. 69:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paillard, D., V. Dubois, R. Thiebaut, F. Nathier, E. Hoogland, P. Caumette, and C. Quentin. 2005. Occurrence of Listeria spp. in effluents of French urban wastewater treatment plants. Appl. Environ. Microbiol. 71:7562-7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palumbo, J. D., M. K. Borucki, R. E. Mandrell, and L. Gorski. 2003. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J. Clin. Microbiol. 41:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.PCI Geomatics. 2002. Using PCI software. PCI Geomatics, Richmond Hill, Ontario, Canada.
- 41.Pearson, L. J., and E. H. Marth. 1990. Listeria monocytogenes: threat to a safe food supply: a review. J. Dairy Sci. 73:912-928. [DOI] [PubMed] [Google Scholar]
- 42.Prazak, A. M., E. A. Murano, I. Mercado, and G. R. Acuff. 2002. Antimicrobial resistance of Listeria monocytogenes isolated from various cabbage farms and packing sheds in Texas. J. Food Prot. 65:1796-1799. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, A., K. Nightingale, G. Jeffers, E. Fortes, J. M. Kongo, and M. Wiedmann. 2006. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152:685-693. [DOI] [PubMed] [Google Scholar]
- 44.Roche, S. M., P. Velge, E. Bottreau, C. Durier, N. Marquet-van der Mee, and P. Pardon. 2001. Assessment of the virulence of Listeria monocytogenes: agreement between a plaque-forming assay with HT-29 cells and infection of immunocompetent mice. Int. J. Food Microbiol. 68:33-44. [DOI] [PubMed] [Google Scholar]
- 45.Rodas-Suárez, O. R., J. F. Flores-Pedroche, J. M. Betancourt-Rule, E. I. Quiñones-Ramírez, and C. Vázquez-Salinas. 2006. Occurrence and antibiotic sensitivity of Listeria monocytogenes strains isolated from oysters, fish, and estuarine waters. Appl. Environ. Microbiol. 72:7410-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rousseaux, S., M. Olier, J. P. Lemaître, P. Piveteau, and J. Guzzo. 2004. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruecker, N. J., S. L. Braithwaite, E. Topp, T. Edge, D. R. Lapen, G. Wilkes, W. Robertson, D. Medeiros, C. W. Sensen, and N. F. Neumann. 2007. Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl. Environ. Microbiol. 73:3945-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauders, B. D., D. Pettit, B. Currie, P. Suits, A. Evans, K. Stellrecht, D. M. Dryja, D. Slate, and M. Wiedmann. 2005. Low prevalence of Listeria monocytogenes in human stool. J. Food Prot. 68:178-181. [DOI] [PubMed] [Google Scholar]
- 49.Sauders, B. D., M. Z. Durak, E. Fortes, K. Windham, Y. Schukken, A. J. Lembo, Jr., B. Akey, K. K. Nightingale, and M. Wiedmann. 2006. Molecular characterization of Listeria monocytogenes from natural and urban environments. J. Food Prot. 69:93-105. [DOI] [PubMed] [Google Scholar]
- 50.Schaffter, N., and A. Parriaux. 2002. Pathogenic-bacterial water contamination in mountainous catchments. Water Res. 36:131-139. [DOI] [PubMed] [Google Scholar]
- 51.Schaffter, N., J. Zumstein, and A. Parriaux. 2004. Factors influencing the bacteriological water quality in mountainous surface and groundwaters. Acta Hydrochim. Hydrobiol. 32:225-234. [Google Scholar]
- 52.Srinivasan, V., H. M. Nam, L. T. Nguyen, B. Tamilselvam, S. E. Murinda, and S. P. Oliver. 2005. Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Path. Dis. 2:201-211. [DOI] [PubMed] [Google Scholar]
- 53.Steele, M., and J. Odumeru. 2004. Irrigation water as source of foodborne pathogens on fruit and vegetables. J. Food Prot. 67:2839-2849. [DOI] [PubMed] [Google Scholar]
- 54.Steinberg, D., and P. Colla. 1995. CART: tree-structured non-parametric data analysis. Salford Systems, San Diego, CA.
- 55.Van Langendonck, N., E. Bottreau, S. Bailly, M. Tabouret, J. Marly, P. Pardon, and P. P. Velge. 1998. Tissue culture assays using Caco-2 cell line differentiate virulent from non-virulent Listeria monocytogenes strains. J. Appl. Microbiol. 85:337-346. [DOI] [PubMed] [Google Scholar]
- 56.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vlaemynck, G., V. Lafarge, and S. Scotter. 2000. Improvement of the detection of Listeria monocytogenes by the application of ALOA, a diagnostic, chromogenic isolation medium. J. Appl. Microbiol. 88:430-441. [DOI] [PubMed] [Google Scholar]
- 58.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-532. [PubMed] [Google Scholar]
- 59.Yücel, N., S. Citak, and M. Önder. 2005. Prevalence and antibiotic resistance of Listeria species in meat products in Ankara, Turkey. Food Microbiol. 22:241-245. [Google Scholar]
- 60.Zhang, W., and S. J. Knabel. 2005. Multiplex PCR assay simplifies serotyping and sequence typing of Listeria monocytogenes associated with human outbreaks. J. Food Prot. 68:1907-1910. [DOI] [PubMed] [Google Scholar]