Abstract
Marine viruses are an important component of the microbial food web, influencing microbial diversity and contributing to bacterial mortality rates. Resistance to cooccurring cyanophages has been reported for natural communities of Synechococcus spp.; however, little is known about the nature of this resistance. This study examined the patterns of infectivity among cyanophage isolates and unicellular marine cyanobacteria (Synechococcus spp.). We selected for phage-resistant Synechococcus mutants, examined the mechanisms of phage resistance, and determined the extent of cross-resistance to other phages. Four strains of Synechococcus spp. (WH7803, WH8018, WH8012, and WH8101) and 32 previously isolated cyanomyophages were used to select for phage resistance. Phage-resistant Synechococcus mutants were recovered from 50 of the 101 susceptible phage-host pairs, and 23 of these strains were further characterized. Adsorption kinetic assays indicate that resistance is likely due to changes in host receptor sites that limit viral attachment. Our results also suggest that receptor mutations conferring this resistance are diverse. Nevertheless, selection for resistance to one phage frequently resulted in cross-resistance to other phages. On average, phage-resistant Synechococcus strains became resistant to eight other cyanophages; however, there was no significant correlation between the genetic similarity of the phages (based on g20 sequences) and cross-resistance. Likewise, host Synechococcus DNA-dependent RNA polymerase (rpoC1) genotypes could not be used to predict sensitivities to phages. The potential for the rapid evolution of multiple phage resistance may influence the population dynamics and diversity of both Synechococcus and cyanophages in marine waters.
Viruses that infect heterotrophic bacteria, cyanobacteria, and eukaryotic phytoplankton are abundant in marine ecosystems. It has been suggested that these viruses influence the abundance, diversity, and seasonal succession of their hosts (6, 20, 27, 37). While it has been difficult to obtain accurate in situ estimates of virus-induced host mortality (13, 27), some studies report that viruses may be responsible for up to or over 70% of the daily mortality of marine bacteria and phytoplankton (3, 37). If virus-induced mortality is high, viruses may also play a key role in nutrient cycling in marine environments. Viral lysis of host cells can increase the pool of dissolved organic matter and limit the flow of nutrients to higher trophic levels (6, 27, 35).
This study examined interactions between the marine cyanobacterium Synechococcus and cyanophages. Synechococcus spp. are abundant primary producers in coastal environments. Although cyanophages capable of infecting Synechococcus are also abundant, the mortality of Synechococcus due to cyanophage infection varies greatly, with estimates ranging from only 0.005% to up to 28% of the Synechococcus community per day (7, 13, 21, 28, 32). The low estimates of phage-induced mortality suggest that resistance to phage infection may be important in Synechococcus communities. Indeed, some Synechococcus isolates from coastal waters are resistant to cooccurring cyanophages (18, 32). McDaniel et al. (18) report that some Synechococcus strains isolated from the Gulf of Mexico were resistant to all 35 cyanophage isolates obtained from the same location. Other studies, however, have suggested that in offshore waters most Synechococcus cells are susceptible to cooccurring phages (20, 28). Currently, it is unknown whether and how phage resistance might influence the ecological impacts of cyanophages on Synechococcus communities.
Despite the observation that some Synechococcus isolates are resistant to phages, the mechanisms of resistance have not been investigated in detail. For other well-studied bacterium-phage systems, several different mechanisms of phage resistance or immunity have been described. The most common mechanism appears to be an alteration of host surface receptors which reduces or eliminates the ability of phages to attach to the host cell and establish an infection (1). Other mechanisms include the inability of a bacterium to support viral replication and the presence of restriction-modification systems in which host restriction endonucleases degrade viral DNA upon entry into the cell (11, 34). The presence of prophage in bacterial genomes can also provide immunity to infection by related phages. These lysogenic infections have been observed for some natural populations of Synechococcus (16-18); however, in at least one study, there was no evidence that lysogeny was responsible for the observed resistance to cyanophages (18).
In this study, we systematically selected for phage-resistant Synechococcus mutants by use of four Synechococcus strains and 32 cyanomyophages isolated from Rhode Island coastal waters. We first examined how readily phage resistance can be selected for in the lab. Next, we explored possible mechanisms conferring resistance in selected Synechococcus mutants. Finally, we examined the potential for the evolution of phage resistance to alter Synechococcus-cyanophage community interactions. Specifically, we asked whether resistance to one phage confers resistance to other phages, and if so, whether these pleiotropic effects were predictable based on the genetic similarity of the phages.
MATERIALS AND METHODS
Synechococcus and cyanophage isolates.
Cultures of marine Synechococcus were obtained from the Woods Hole Collection of Cyanobacteria (Woods Hole Oceanographic Institute, Woods Hole, MA). Synechococcus strains WH7803, WH8012, WH8018, and WH8101 were used in this study. Strains WH7803 and WH8012 were originally isolated from the Sargasso Sea, while WH8018 and WH8101 were isolated from coastal waters adjacent to Woods Hole, MA (33). Synechococcus cultures were maintained in SN medium (33) at 20 to 23°C under constant illumination. The 32 cyanophage isolates used in this study were isolated from Narragansett Bay, RI, between 1999 and 2002 (15). These cyanophages were isolated using Synechococcus strains WH7803, WH8012, WH8018, and WH8113. Based on g20 gene sequences, these phages are genetically distinct myoviruses that span the diversity seen thus far for known myovirus isolates of Synechococcus spp. (15, 39). Viral stocks were stored at 4°C prior to use. The specific isolates used were S-RIM1 to -13, -15 to -21, -23 to -24, -26 to -32, and -34 to -36 (15). The g20 gene sequences of these cyanophages are available from GenBank under accession numbers AY259244 to AY259283.
Selection and purification of phage-resistant Synechococcus.
To select for phage-resistant Synechococcus, we combined 100 μl of Synechococcus strain WH7803, WH8012, WH8018, or WH8101 (0.5 × 108 to 2.0 × 108 cells ml−1) and 10 to 40 μl of phage stock (∼5 × 107 phages ml−1) in 24-well microtiter plates. Each Synechococcus strain was challenged with each of the 32 phage isolates. After 1 h of incubation, 1.5 ml of SN medium was added to the wells. Replicate wells were used for each phage-host strain combination. Plates were incubated at 20 to 23°C under constant illumination. Wells were visually monitored daily for cell lysis (clearing of the well relative to that seen for a control well containing cells but no virus) and then weekly after lysis was observed. Phage-resistant Synechococcus cells were recovered as regrowth in viral lysates after prolonged incubation (i.e., 4 to 10 weeks). To isolate single colonies from regrowth, cells were serially diluted in SN medium and then combined with 50 ml SN soft agar at 37°C and immediately poured into petri dishes (2). All agar was purified by multiple washes with dionized water prior to use (33). Plates were incubated in the dark for 24 h and then moved to a growth chamber at 23°C with diurnal cycles (14 h of light). After 4 to 12 weeks, single colonies were visible, and individual colonies were removed from the agar by use of a sterile pipette tip and transferred to SN medium in a 50-ml Erlenmeyer flask. These single colonies were used in all subsequent analyses. To sequentially select for resistance to additional phages, the process described above was repeated using phage-resistant single colonies. Each resistant strain was again screened against the 32 phage isolates.
To confirm that the single colonies were phage resistant, each single colony was tested for resistance to the phage used in the initial selection in 24-well microtiter plates as described above. Resistance was defined as cell growth in the presence of virus that was comparable to the growth of cells with no virus. To confirm that the strains were derived from a given ancestral Synechococcus strain, the DNA-dependent RNA polymerase gene (rpoC1) was amplified via PCR from each single colony as described by Palenik (22). PCR products were digested with two restriction endonucleases, MboI and RsaI (Invitrogen), according to the manufacturer's instructions. Digested products were separated by gel electrophoresis (2% agarose) and visualized by staining with ethidium bromide. All Synechococcus cultures maintained in our lab can be differentiated by the banding patterns generated by these two restriction enzymes.
Adsorption kinetics and one-step growth assays.
Adsorption kinetic assays were conducted as described by Suttle and Chan (29). Briefly, 5 × 105 phage were added to 1 ml of exponentially growing Synechococcus culture (5 × 107 cells ml−1). Three replicate assays were simultaneously conducted for each Synechococcus-phage pair. Immediately after the addition of phage and then at 15, 30, 45, and 60 min, a 10-μl sample was taken from each well and diluted 100-fold in cold SN medium. Samples were centrifuged for 5 min at 5,200 × g (4°C) to pellet cells and adsorbed phage; an aliquot of the supernatant was titered via plaque assay (15) to estimate the number of unadsorbed phage (Nu). The adsorption rate constant (k) was determined using the formula Nu = N0e−kCt (log transformed), where C is cell density and t is time measured in minutes (4). The value for k is the mean of three independent determinations for each Synechococcus-phage pair. For each resistant strain-phage pair tested, a control consisting of the ancestral strain paired with the same phage isolate was simultaneously tested. Standard t tests were used to compare the adsorption rate means between each resistant strain-phage pair and the control. In assays where the adsorption rates of more than one resistant strain were compared to the adsorption rate of the ancestral strain, a Dunnett's t test was used to correct for multiple comparisons. Regression analysis was used to determine if adsorption rates were significantly different from zero.
Single-cell growth experiments were conducted to determine the latent periods and burst sizes of sensitive and resistant Synechococcus isolates by use of standard protocols (10, 29). Triplicate cultures of 2 × 107 cells ml−1 were inoculated with ∼4 × 105 to 8 × 105 phage ml−1 and incubated for 1 h to allow phage to adsorb. Samples were centrifuged for 5 min at 5,200 × g (4°C) to pellet cells and adsorbed phage. The pellet was washed with SN medium to remove all unadsorbed phage, resuspended in 1 ml of SN, and diluted 1:400 in SN medium. Phage was titered by plaque assay (15) from aliquots taken every 2 h for the first 12 h and then again at 20 and 25 h postinoculation.
Prophage induction.
All derived resistant strains were tested for the presence of prophage by use of the inducing agent mitomycin C (Sigma Chemical Co., St. Louis, MO). Exponentially growing cells were diluted in fresh SN medium, and four replicate samples of each strain were placed into wells on a 24-well microtiter plate. Plates were maintained as described above. Prior to the addition of mitomycin C, 100-μl aliquots of each sample were mixed with concentrated cells of the ancestral Synechococcus strains WH7803 or WH8018 and soft agar and poured on agar plates (15) to determine if phage was present in the initial culture. Mitomycin C was then added to a final concentration of 1 μg ml−1 to two of the four replicates. This concentration of mitomycin C was found to induce lysogenic cyanophage in other studies (17, 21). After a 24-h incubation at room temperature, 50-μl aliquots of each replicate mitomycin C treatment culture were used in plaque assays against the ancestral strains WH7803 or WH8018, as described above. Plates were checked weekly for the appearance of plaques. All cyanophages used in this study can form plaques on WH7803 or WH8018 cells on agar plates (15). If prophage induction occurs, more plaques should be present after the mitomycin C treatment.
BOR and phage infectivity assays.
To characterize the phenotypes (i.e., sensitivities to a particular phage) of the ancestral Synechococcus strains and the derived resistant strains, Synechococcus strains grown from single colonies were challenged with each of the 32 cyanomyophage isolates. Replicate samples of each Synechococcus-phage pair were incubated in 24-well microtiter plates as described above. Control wells containing host cells, but no phage, were included on each plate. Wells were visually monitored for cell lysis for up to 4 weeks. If a well cleared, the Synechococcus strain was considered susceptible to the phage. Cells that were not lysed after 4 weeks were considered to be resistant to the phage. In this way, each Synechococcus strain was assigned a breadth-of-resistance (BOR) profile, i.e., the range of phage isolates to which a strain is resistant. Similarly, each cyanophage isolate was assigned a phage infectivity profile, i.e., the range of Synechococcus strains that the phage can infect.
To examine the relationships between the ancestral and phage-resistant Synechococcus phenotypes, we calculated the similarity of the BOR profiles between all the strains. Pairwise similarity for two Synechococcus strains was defined as the proportion of phages (out of 32 tested) to which the two strains are either both sensitive or both resistant. If the strains have the same BOR profile (that is, they are resistant to the same phages), then their BOR similarity is 1. If the strains differ in their responses to 16 of the 32 phages, then their similarity is 0.5. Cluster analysis (group average method) was used to illustrate the BOR relationships among the Synechococcus strains (PRIMER v5: PrimerE-Ltd, Plymouth, United Kingdom). A one-way analysis of similarity statistic was used to test whether the BOR profiles of strains derived from the same ancestor (WH7803, WH8101, WH8012, or WH8018) were more similar to one another than to strains derived from a different ancestor. This test is similar to a one-way analysis of variance, but it uses a permutation test to account for the interdependence of the similarity values.
We also analyzed the phenotypic relationships among the 32 phage isolates by calculating the similarity of phage infectivity profiles. In this case, the pairwise similarity for two phage strains was the proportion of Synechococcus strains (out of 27 [4 ancestral and 23 resistant] strains tested) that the paired phages could either both infect or both not infect. If two phages could infect exactly the same set of Synechococcus strains, then their similarity was scored as 1. Cluster analysis was used to illustrate the infectivity profile relationships among the 32 phage strains. A RELATE test (PRIMER) examined whether the similarity between two phages' infectivity profiles was correlated with their genetic relatedness (as assayed by g20 sequences). This statistic is similar to a correlation statistic but uses a permutation test to account for the interdependent genetic distance and profile similarity values. The PAUP software package (30) was used to calculate uncorrected pairwise genetic distances between g20 nucleotide sequences as previously described (15).
RESULTS
Selection of phage-resistant Synechococcus strains.
When Synechococcus strains WH7803, WH8012, WH8018, and WH8101 were challenged with 32 genetically diverse myovirus isolates, a lytic (susceptible) interaction was observed for 101 out of the 128 Synechococcus-phage pairs. From these 101 susceptible interactions, regrowth of cells was observed for 50 of the Synechococcus-phage pairs, indicating the presence of phage-resistant strains (Table 1). After single-colony purification, a total of 23 phage-resistant strains were selected for further characterization. These strains were chosen for characterization because collectively they exhibited resistance to a diverse set of phage isolates. Each of the 23 strains was resistant to the phage used in the initial selection and, based on rpoC1 sequences, was derived from the ancestral Synechococcus strain (i.e., regrowth was not due to contamination by a different Synechococcus strain). Each derived resistant strain was given a designation based on the ancestral Synechococcus strain and the S-RIM number of the phage isolate used to select for resistance. For example, WH7803R8 was derived from WH7803 cells that had been selected for resistance to S-RIM8. Two of the 23 resistant strains (WH8101R3R32 and WH7803R8R21) underwent two sequential selection events. For example, in the case of WH8101R3R32, cells were first selected for resistance to S-RIM3 and then were selected for resistance to S-RIM32. One of the phage-resistant strains (WH8018R3R2R6) underwent three selection events.
TABLE 1.
Selection of phage-resistant Synechococcus mutants
| Synechococcus strain | No. of susceptible interactions (n = 32 phage isolates) | No. (%) of resistant mutants observed | No. of resistant mutants characterized | Mean no. of additional phages resisted (± SD)a |
|---|---|---|---|---|
| Batch culture strains | ||||
| WH7803 | 30 | 12 (40) | 7 | 7.8 (± 3.5) |
| WH8012 | 19 | 9 (47) | 2 | 6.0 (± 2.8) |
| WH8018 | 29 | 17 (59) | 9 | 10.0 (± 2.0) |
| WH8101 | 23 | 12 (52) | 5 | 4.5 (± 1.3) |
| Single-colony strainsb | ||||
| WH7803sca | 30 | 9 (30) | ||
| WH8101sca | 22 | 13 (59) | ||
| WH8101scb | 22 | 16 (73) |
Double and triple mutants not included.
sca, single colony a; scb, single colony b.
To select for resistant strains, we used Synechococcus cultures that had been maintained in culture for years. Thus, these cultures potentially contained a large amount of genetic diversity that was preserved during transfer to new medium every few weeks. This diversity could influence the number of phage-resistant mutants detected in our selection experiments and lead to an overestimate of the ease with which resistance can be acquired. To test this hypothesis, we plated and regrew the ancestral strains again from single colonies. These single colonies were then used to select for phage resistance. The number of phage-resistant strains observed for the cultures derived from a single colony was approximately the same as that observed for the original cultures (Table 1). This result reveals that phage resistance can arise during growth from a single colony to stationary phase in a flask.
Mechanisms of resistance.
After single-colony purification, phages were never detected in any of the cultures of phage-resistant strains. In addition, no mitomycin C-inducible prophages were detected in any of the cultures. It appears unlikely that the observed phage resistance is due to lysogeny. Moreover, the fact that the strains remained resistant after single-colony purification and in the absence of phage suggests that phage resistance was due to a genetic mutation rather than a physiological response.
Adsorption kinetic assays were performed using five phages (S-RIM1, S-RIM3, S-RIM6, S-RIM7, and S-RIM8) on the four ancestral Synechococcus strains and eight derived phage-resistant Synechococcus strains (Table 2). In all eight resistant host-phage pairs, the phage did not bind as well to the resistant strain as to the ancestral strain. The mean adsorption rate for susceptible Synechococcus-phage pairs ranged from 0.23 × 10−9 to 1.6 × 10−9 ml min−1. The phage adsorption rate for derived resistant strains was not different (P > 0.05) from that for the ancestral strains when both strains were susceptible to a phage, with one exception: S-RIM7 attaches better to WH7803R4 than it does to WH7803 (Table 2). In contrast, for seven of the eight resistant Synechococcus-phage pairs tested, the adsorption rate was significantly lower (P < 0.05) than the adsorption rate for the corresponding susceptible control pair (Table 2). For the eighth pair (WH7803R7 and S-RIM7), the difference in adsorption rates was marginally significant (P = 0.051).
TABLE 2.
Adsorption rate (k) means of susceptible and resistant Synechococcus-phage interactions
| Phage isolate | Synechococcus straina | S/R interactionb | Adsorption rate mean (k) | t test P valuec |
|---|---|---|---|---|
| S-RIM1 | WH7803 | S | 5.0 × 10−10 | |
| WH7803R8 | S | 6.1 × 10−10 | 0.155 | |
| WH7803R8R21 | S | 5.5 × 10−10 | 0.615 | |
| S-RIM3 | WH8018 | S | 2.3 × 10−10 | |
| WH8018R3 | R | NSd | 0.007 | |
| WH8018 | S | 5.3 × 10−10 | ||
| WH8018R3R2R6 | R | NSd | 0.003 | |
| WH8101 | S | 7.0 × 10−10 | ||
| WH8101R3 | R | 1.9 × 10−10 | 0.011 | |
| WH8101 | S | 7.0 × 10−10 | ||
| WH8101R3R32 | R | 1.5 × 10−10 | 0.009 | |
| S-RIM6 | WH7803 | S | 3.0 × 10−10 | |
| WH7803R8 | S | 4.2 × 10−10 | 0.346 | |
| WH8018 | S | 4.3 × 10−10 | ||
| WH8018R3R2R6 | R | NSd | 0.014 | |
| S-RIM7 | WH7803 | S | 8.2 × 10−10 | |
| WH7803R4 | S | 1.6 × 10−9 | 0.02 | |
| WH7803R7 | R | NSd | 0.051 | |
| WH7803R8 | S | 1.2 × 10−9 | 0.224 | |
| WH8012 | S | 9.5 × 10−10 | ||
| WH8012R7 | R | NSd | 0.002 | |
| S-RIM8 | WH7803 | S | 4.2 × 10−10 | |
| WH7803R8 | R | NSd | 0.0002 |
All cultures were grown from single colonies except WH8012.
S, susceptible; R, resistant.
The mean adsorption rate of phage on the ancestral susceptible strain was compared to the mean adsorption rate of the same phage on the resistant strain.
NS, not significantly different from zero according to regression analysis.
One-step growth assay.
One-step growth experiments were conducted for two different viruses (S-RIM1 and S-RIM8) on the susceptible host WH7803. The latent period for S-RIM1 and S-RIM8 was between 6 and 8 h, with most lysis occurring by 12 h. The burst sizes for S-RIM1 and S-RIM8 were estimated to be 63 and 86 phages per cell, respectively. No viral replication was observed in one-step growth assays using the phage isolate S-RIM8 on resistant strains WH7803R8 and WH7803R8R21.
BOR profiles.
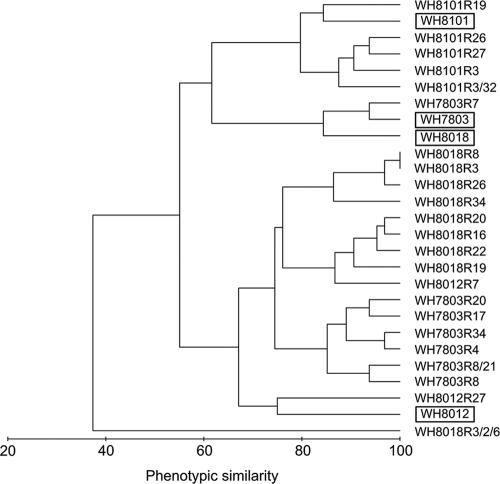
Selection for resistance against a single phage led to resistance to additional genetically distinct phages. On average, a strain became resistant to 7.8 (± 3.2) phages (range, 1 to 13). Of the 23 phage-resistant strains tested, only two share the same BOR profile: WH8018R8 and WH8018R3 were resistant to the same set of phages (Fig. 1). The Synechococcus strains that underwent two or three sequential selection events gained resistance to additional phages. The strain that underwent three consecutive selection events (WH8018R2R3R6) was resistant to 31 of the 32 phage isolates tested.
FIG. 1.
Cluster diagram of BOR profiles for the 4 ancestral Synechococcus strains (in boxes), for 20 strains that were selected for resistance to one phage, for 2 strains selected for resistance to two phages, and for 1 strain selected for resistance to three phages. Phenotypic similarity between two Synechococcus strains is defined as the percentage of phages to which the strains are either both sensitive or both resistant. See the text for details.
The BOR profiles were constrained by the identity of the ancestral strain (Fig. 1; analysis of similarity R = 0.595; P = 0.001). Resistant strains derived from the same ancestor (e.g., WH7803, WH8018, WH8012, or WH8101) have phenotypes more similar than do strains derived from different ancestors. In a few cases, the acquisition of resistance to one phage resulted in the loss of resistance to other phages. For example, in WH7803-derived strains, the six strains that gained resistance to S-RIM8 lost resistance to S-RIM5 and S-RIM26.
Phage infectivity profiles.
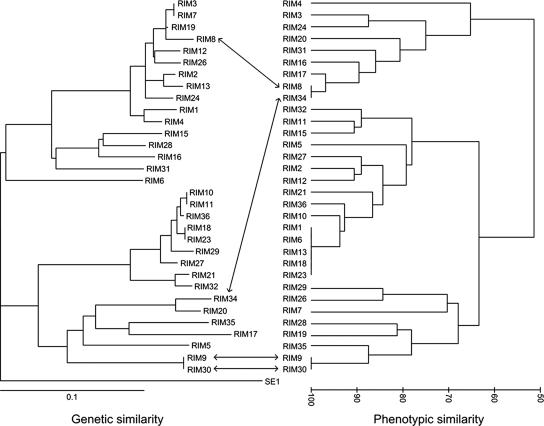
On average, the S-RIM phages could infect 17.0 (±6.7) (range, 5 to 26) out of the 27 different Synechococcus strains (4 ancestral strains and 23 derived resistant strains). As with the Synechococcus BOR profiles, there was a high diversity of phage infectivity profiles: 26 out of 32 of the profiles were unique (Fig. 2). Five profiles were identical (S-RIM1, S-RIM6, S-RIM13, S-RIM18, and S-RIM23), and these belonged to phages that infect all 27 Synechococcus strains except for the triply selected strain WH8018R3R2R6. We found no relationship between phage phenotype (i.e., the ability to infect particular strains) and phage genotype (based on g20 gene sequences); there was no correlation between the similarity of two phages' infectivity profiles and their genetic relatedness (Fig. 2; RELATE ρ = 0.035; P = 0.245).
FIG. 2.
Comparison of the genetic and phenotypic similarities among the 32 cyanomyophages in this study. A neighbor-joining tree (left) illustrates the genetic similarities among phage isolates based on pairwise distances between g20 amino acid sequences (15). A cluster diagram (right) illustrates phenotypic similarities between phage isolates; phenotypic similarity is defined as the percentage of Synechococcus strains that the paired phages could either both infect or both not infect. See the text for further details. The bottom arrows illustrate a case where genotypic and phenotypic similarities are both high, whereas the top arrows highlight two phages that have a high phenotypic similarity but a low genetic similarity.
DISCUSSION
In this study, we have shown that Synechococcus strains can acquire resistance to a genetically diverse set of cyanomyophage isolates. Phage resistance arose in 50% of the susceptible host-phage pairs. Resistance was stable through single-colony isolation as well as through several transfers in batch culture, suggesting that the resistance is due to heritable genetic changes in the bacterial cells. Furthermore, mitomycin C-inducible prophages were not detected in any of the resistant strains; thus, within the constraints of this assay, lysogeny does not appear to be responsible for the observed resistance.
The results from adsorption kinetic assays suggest that the genetic mutations conferring phage resistance in these Synechococcus strains alter phage receptor sites. For all resistant host-phage pairs, phage could adsorb faster to the susceptible ancestral strains than to the derived resistant strains. Moreover, in six out of eight resistant host-phage pairs, the adsorption rate on resistant hosts was not significantly different from zero, suggesting that phage could not adsorb to these cells. The adsorption rate means for susceptible Synechococcus-phage pairs in this study (0.23 × 10−9 to 1.6 × 10−9 ml min−1) were lower than the means previously reported for Synechococcus phages (3.9 × 10−9 to 6.1 × 10−9 ml min−1) (13, 29, 36) but fall within the range observed for other phage systems (4, 5). Resistance in many bacteriophage-Escherichia coli systems has been shown to be conferred by host mutations that alter the structure of a receptor site, change the exposure of a site, or lead to the complete loss of a receptor (1, 10). While phage resistance in cyanobacteria has not been extensively studied, there is evidence that resistance in the filamentous cyanobacterium Anabaena is due to mutations that alter the structure of the O antigen component of the lipopolysaccharide (LPS), thus preventing phage adsorption (38).
The number and identities of host receptors used by cyanophages for attachment to Synechococcus are unknown (13). Our results, however, suggest extensive pleiotropy associated with phage resistance mutations in Synechococcus. On average, selection for resistance to one phage resulted in resistance to eight other phages. In addition, each time Synechococcus strain WH7803 gained resistance to S-RIM8, it simultaneously lost resistance to S-RIM5 and S-RIM26. Thus, it appears that different phage isolates use some of the same host receptors. There are other examples of different phages sharing receptors: bacteriophages T3, T4, and T7 all absorb to the LPS of E. coli B, and a mutation leading to changes in the LPS can confer resistance to all three of these phages (10). Likewise, the E. coli outer membrane protein A (OmpA) is used as a receptor by a diverse suite of phages (19, 23).
While our results suggest that some cyanophage isolates use the same host receptors, it also appears that Synechococcus can become resistant to the same phage in multiple ways. If there were only a few mutations that lead to cyanophage resistance, then we would expect a small number of BOR phenotypes among the resistant strains of Synechococcus. Instead, many of the strains share resistance to the same phage but differ in their interactions with other phages. Only 2 of the 23 resistant strains shared the same BOR phenotype (i.e., were resistant to the same set of phages). Complex patterns of phage cross-resistance were also observed among ompA mutants of E. coli (19). T-even phages were found to attach to different areas of the OmpA protein; therefore, mutations altering one region of the protein provided resistance to some phages but not to others. In addition, a few phages interacted with several regions of the protein, and thus mutations changing any of these regions could independently confer resistance (19). Independent mutations at different loci can also confer resistance to the same phage. For example, in E. coli K-12, mutations in either an outer membrane protein or an inner membrane protein inhibit bacteriophage adsorption (8, 12). In a recent screen of E. coli genes, mutations in any of nine LPS biosynthesis genes conferred resistance to T7 bacteriophage by preventing adsorption (24). These examples illustrate several ways in which independent mutations could confer resistance to one phage and at the same time lead to the complex patterns of cross-resistance we observed.
Several studies have shown that the community compositions of Synechococcus (based on RNA polymerase rpoC1 sequences) and cyanophages (based on capsid assembly protein g20 sequences) vary seasonally (15, 20, 31). Based on these findings, Muhling et al. (20) suggested that cyanophages may be responsible for the succession of host genotypes over seasonal time scales. Our study suggests that using such conserved marker sequences is likely to obscure the majority of cyanophage-Synechococcus interactions. We showed that Synechococcus strains with the same rpoC1 sequence could have very different sensitivities to phages. After selection for phage resistance, 26 host strains could be distinguished based on sensitivities to 32 phages, yet all of these strains belong to one of only four rpoC1 genotypes. In this case, rpoC1 sequences could not be used to predict sensitivities to phages. Likewise, the genetic similarity of phages based on g20 sequences did not predict phage infectivity phenotypes. Although the g20 gene product is probably not directly involved in phage adsorption, the lack of correlation between g20 sequences and infectivity phenotypes suggests that g20 sequences are not good predictors of tail fiber homology and/or that phages with diverse tail fibers can bind to the same host receptor. Recent analyses of complete genomes suggest that cyanophages may contain diverse tail fibers and that lateral gene transfer events also may be common (14, 25, 26). Thus, until the genetic and structural elements responsible for virus-host interactions are known, it may be difficult to use conserved molecular markers to conclusively relate changes in the genetic compositions of natural viral and host populations.
The impact that cyanophages have on marine bacterial populations, whether by imposing mortality or by maintaining diversity, will be influenced by the composition of host phenotypes in a population. The abundance and persistence of resistant phenotypes will depend in part on the physiological cost of resistance. Studies with other systems have demonstrated that phage resistance can impose a fitness cost, as changes in cell surface molecules that act as phage receptors alter the ability of the host to take up key nutrients (1). At least some of the phage-resistant strains in this study appear to incur a fitness cost (9), suggesting a mechanism by which both sensitive and resistant strains may coexist in a population.
Our results show that in a laboratory setting Synechococcus strains can acquire phage resistance that appears to be due to genetic mutations that alter the ability of phages to adsorb to cells. Furthermore, we were able to sequentially select for resistance to additional phages. After selection for resistance to three phages, one strain (WH8018R3R2R6) was resistant to 31 of the 32 diverse phage isolates tested. The high degree of pleiotropy among phage resistance mutations suggests that interactions among cyanophages and Synechococcus in natural populations may be highly complex. A mutation that confers resistance to one particular phage is likely to alter whether other, genetically distinct cyanophages can infect that host cell. As a result, the evolution of phage resistance may be a significant factor affecting the population dynamics of Synechococcus and cyanophages in natural communities.
Acknowledgments
This work was supported by the RWU Research Foundation and an NSF Biological Oceanography grant (OCE-0314523) to M.F.M. and an NSF Biological Oceanography grant (OCE-0315645) and a Junior Investigator Award from the Gordon and Betty Moore Foundation to J.B.H.M.
We thank F. Pierciey, A. Shepard, and J. Torbett for assistance in the laboratory; F. Valois and J. Waterbury for Synechococcus cultures; and J. Lennon and C. Amrich for critical review of the manuscript.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Bohannan, B. J. M., and R. E. Lenski. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3:362-377. [Google Scholar]
- 2.Brahamsha, B. 1996. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 62:1747-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brussaard, C. P. 2004. Viral control of phytoplankton populations—a review. J. Eukaryot. Microbiol. 51:125-138. [DOI] [PubMed] [Google Scholar]
- 4.Bull, J. J., M. R. Badgett, R. Springman, and I. J. Molineux. 2004. Genome properties and the limits of adaptation in bacteriophages. Evolution 58:692-701. [DOI] [PubMed] [Google Scholar]
- 5.De Paepe, M., and F. Taddei. 2006. Viruses' life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4:e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 7.Garza, D. R., and C. A. Suttle. 1998. The effect of cyanophages on the mortality of Synechococcus spp. and selection for UV resistant viral communities. Microb. Ecol. 36:281-292. [DOI] [PubMed] [Google Scholar]
- 8.Kiino, D. R., and L. B. Rothmandenes. 1989. Genetic analysis of bacteriophage N4 adsorption. J. Bacteriol. 171:4595-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lennon, J. T., S. A. M. Khatana, M. F. Marston, and J. B. H. Martiny. 28 June 2007. Is there a cost of virus resistance in marine cyanobacteria? ISME J. doi: 10.1038/ismej.2007.37. [DOI] [PubMed]
- 10.Lenski, R. E. 1988. Dynamics of interactions between bacteria and virulent bacteriophage. Adv. Microb. Ecol. 10:1-44. [Google Scholar]
- 11.Levin, B. R., and R. E. Lenski. 1985. Bacteria and phage: a model system for the study of the ecology and co-evolution of hosts and parasites, p. 227-242. In D. Rollinson and R. M. Anderson (ed.), Ecology and genetics of host-parasite interactions. Academic Press, London, United Kingdom.
- 12.Likhacheva, N. A., V. V. Samsonov, V. V. Samsonov, and S. P. Sineoky. 1996. Genetic control of the resistance to phage C1 of Escherichia coli K-12. J. Bacteriol. 178:5309-5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann, N. H. 2003. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev. 27:17-34. [DOI] [PubMed] [Google Scholar]
- 14.Mann, N. H., M. R. J. Clokie, A. Millard, A. Cook, W. H. Wilson, P. J. Wheatley, A. Letarov, and H. M. Krisch. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187:3188-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marston, M. F., and J. L. Sallee. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl. Environ. Microbiol. 69:4639-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDaniel, L., L. A. Houchin, S. J. Williamson, and J. H. Paul. 2002. Lysogeny in marine Synechococcus. Nature 415:496. [DOI] [PubMed] [Google Scholar]
- 17.McDaniel, L., and J. H. Paul. 2005. Effect of nutrient addition and environmental factors on prophage induction in natural populations of marine Synechococcus species. Appl. Environ. Microbiol. 71:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniel, L. D., M. delaRosa, and J. H. Paul. 2006. Temperate and lytic cyanophages from the Gulf of Mexico. J. Mar. Biol. Assoc. UK 86:517-527. [Google Scholar]
- 19.Morona, R., M. Klose, and U. Henning. 1984. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J. Bacteriol. 159:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhling, M., N. J. Fuller, A. Millard, P. J. Somerfield, D. Marie, W. H. Wilson, D. J. Scanlan, A. F. Post, I. Joint, and N. H. Mann. 2005. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ. Microbiol. 7:499-508. [DOI] [PubMed] [Google Scholar]
- 21.Ortmann, A. C., J. E. Lawrence, and C. A. Suttle. 2002. Lysogeny and lytic viral production during a bloom of the cyanobacterium Synechococcus spp. Microb. Ecol. 43:225-231. [DOI] [PubMed] [Google Scholar]
- 22.Palenik, B. 1994. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl. Environ. Microbiol. 60:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power, M. L., B. C. Ferrari, J. Littlefield-Wyer, D. M. Gordon, M. B. Slade, and D. A. Veal. 2006. A naturally occurring novel allele of Escherichia coli outer membrane protein A reduces sensitivity to bacteriophage. Appl. Environ. Microbiol. 72:7930-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qimron, U., B. Marintcheva, S. Tabor, and C. C. Richardson. 2006. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl. Acad. Sci. USA 103:19039-19044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan, M. B., M. L. Coleman, P. Weigele, F. Rohwer, and S. W. Chisholm. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:790-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan, M. B., D. Lindell, J. A. Lee, L. R. Thompson, J. P. Bielawski, and S. W. Chisholm. 2006. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4:1344-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 28.Suttle, C. A., and A. M. Chan. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suttle, C. A., and A. M. Chan. 1993. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar. Ecol. Prog. Ser. 92:99-109. [Google Scholar]
- 30.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA.
- 31.Wang, K., and F. Chen. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol. 34:105-116. [Google Scholar]
- 32.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterbury, J. B., S. W. Watson, F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can. Bull. Fish. Aquat. Sci. 214:71-120. [Google Scholar]
- 34.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 35.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781-788. [Google Scholar]
- 36.Wilson, W. H., N. G. Carr, and N. H. Mann. 1996. The effect of phosphate status on the kinetics of cyanophage infection in the oceanic cyanobacterium Synechococcus sp. WH7803. J. Phycol. 32:506-516. [Google Scholar]
- 37.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, X. D., I. Khudyakov, and C. P. Wolk. 1997. Lipopolysaccharide dependence of cyanophage sensitivity and aerobic nitrogen fixation in Anabaena sp. strain PCC 7120. J. Bacteriol. 179:2884-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong, Y., F. Chen, S. W. Wilhelm, L. Poorvin, and R. E. Hodson. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]