Abstract
We examined the contribution of photoheterotrophic microbes—those capable of light-mediated assimilation of organic compounds—to bacterial production and amino acid assimilation along a transect from Florida to Iceland from 28 May to 9 July 2005. Bacterial production (leucine incorporation at a 20 nM final concentration) was on average 30% higher in light than in dark-incubated samples, but the effect varied greatly (3% to 60%). To further characterize this light effect, we examined the abundance of potential photoheterotrophs and measured their contribution to bacterial production and amino acid assimilation (0.5 nM addition) using flow cytometry. Prochlorococcus and Synechococcus were abundant in surface waters where light-dependent leucine incorporation was observed, whereas aerobic anoxygenic phototrophic bacteria were abundant but did not correlate with the light effect. The per-cell assimilation rates of Prochlorococcus and Synechococcus were comparable to or higher than those of other prokaryotes, especially in the light. Picoeukaryotes also took up leucine (20 nM) and other amino acids (0.5 nM), but rates normalized to biovolume were much lower than those of prokaryotes. Prochlorococcus was responsible for 80% of light-stimulated bacterial production and amino acid assimilation in surface waters south of the Azores, while Synechococcus accounted for on average 12% of total assimilation. However, nearly 40% of the light-stimulated leucine assimilation was not accounted for by these groups, suggesting that assimilation by other microbes is also affected by light. Our results clarify the contribution of cyanobacteria to photoheterotrophy and highlight the potential role of other photoheterotrophs in biomass production and dissolved-organic-matter assimilation.
The discovery of proteorhodopsin-containing bacteria (3), aerobic anoxygenic phototrophic (AAP) bacteria (17), and assimilation of dissolved organic matter (DOM) by cyanobacteria (26) suggests that photoheterotrophy may be common in the oceans. Several studies indicate that Prochlorococcus may be able to assimilate components of the DOM pool, in addition to its large contribution to primary production in oligotrophic oceans (10). Church et al. (5, 6) found that leucine incorporation was 48 to 114% higher in light incubations than in the dark in the North Pacific Gyre, where Prochlorococcus is well known to be abundant. Other studies had demonstrated that Prochlorococcus is responsible for a large fraction of dissolved methionine turnover (∼1 nM) and up to 30% of leucine incorporation (20 nM), although the effect of light was not examined (33, 34). This capacity to take up amino acids is consistent with the presence of amino acid transport systems, as revealed by the whole-genome sequence of Prochlorococcus (28). However, the role of Prochlorococcus in assimilating other organic compounds and the effect of light on this assimilation are still unknown.
Synechococcus may also assimilate components of the DOM pool, as well as being a major contributor to primary production in temperate and tropical waters (20). Synechococcus can take up methionine and another important organic sulfur compound, dimethylsulfoniopropionate (DMSP) (21, 31). Although Synechococcus cannot incorporate thymidine (11), axenic strains of this cyanobacterium are capable of utilizing urea (7, 24) and amino acids (4, 23, 26), albeit at low rates. In addition, genomic analyses of Synechococcus strain WH8102 revealed genes homologous to those for transporting amino acids, oligopeptides, and cyanate (27). There is also evidence for aminopeptidase activity in Synechococcus (22). However, Zubkov and colleagues (33) found that Synechococcus was responsible for only 3% of methionine turnover in a mesotrophic region of the Arabian Sea. More work is needed to determine the extent of photoheterotrophy by these cyanobacteria.
The goals of this study were to identify the microbial groups responsible for light-dependent leucine incorporation and to examine the effect of light on the uptake of amino acids added at tracer levels. We hypothesize that stimulation of bacterial production by light is due to photoheterotrophy by Prochlorococcus. Our results suggest this was in fact the case, but other groups of photoheterotrophic bacteria contributed to the light effect as well.
MATERIALS AND METHODS
Abundance of AAP bacteria, cyanobacteria, and total prokaryotes.
The experimental work was done during the North Atlantic Spring Bloom (NASB) project onboard the R/V Seward Johnson from 28 May to 2 July 2005. The cruise track included a transit leg across the Sargasso Sea, from Fort Pierce, FL, to Ponta Delgada, Azores, followed by a 5-week transect survey beginning at 45°N and 20°W. Surface seawater for sampling was collected daily from the ship's under-way system from about a 5-m depth during the first transect. Samples during the second leg were collected at various depths with a rosette of Niskin bottles mounted on a conductivity-temperature-depth profiler.
Samples for AAP bacteria and total prokaryote abundance were preserved and enumerated, following the protocol in Cottrell et al. (8). Each of 30 fields of view was subjected to the following four exposures: 4′,6′diamidino-2-phenylindole (60 ms); bacteriochlorophyll a (400 ms); chlorophyll a (1,500 ms); and phycoerythrin (50 ms). AAP bacteria were scored as 4′,6′diamidino-2-phenylindole and bacteriochlorophyll a positive but chlorophyll a and phycoerythrin negative. Prochlorococcus, Synechococcus, and picoeukaryotes were enumerated using flow cytometry and distinguished by their different size and pigment properties in unstained samples following common procedures (12, 25). For flow-cytometric analyses of total prokaryotes, samples were stained with SYTO 13 (final concentration, 5 μM) for 10 to 15 min at room temperature in the dark and discriminated following procedures described previously (30). Abundances were estimated with a FacsCalibur flow cytometer (Becton Dickinson, San Jose, CA) equipped with an air-cooled argon laser (488 nm, 25 mW).
Response of leucine incorporation and amino acid assimilation to irradiance.
To examine the effects of light on biomass production, [3H]leucine incorporation was measured in seawater incubated in the light and dark in a deck incubator with running seawater for 6 h. Experiments were conducted in triplicate with [3H]leucine (20 nM; specific activity of 173 Ci/mmol; Amersham) in 5.0 ml of seawater. Samples were placed in clear bags (Whirl Pack, Nasco Fort Atkinson, WI) and placed in the deck incubator covered by a clear acrylic sheet (Plexiglas XT colorless, 3 mm thick; Rohm & Haas), which partially screens out UV irradiance (50% transmission at 375 nm) (data not shown). Killed controls for light and dark treatments consisted of samples to which 5% trichloroacetic acid (TCA) was added before the addition of isotopes. At the end of the incubation, 1.5 ml was transferred from each bag to a 2-ml polypropylene centrifuge tube, terminated by the addition of 5% TCA, and processed using the microcentrifuge method (29). In addition, other 1.5-ml samples were incubated with 20 nM leucine for 1 h at the in situ temperature in the dark. Incubations were terminated and processed by the centrifuge method (29).
Amino acid assimilation by cyanobacteria.
We also examined assimilation of leucine (20 nM) and amino acids by cyanobacteria and other microbes separated by flow cytometry. The amino acid mixture consisted of 15 amino acids (TRK 440; specific activity, 40 Ci/mmol; Amersham) commonly found in protein but without asparagine, cysteine, glutamine, tryptophan, and methionine, added to a final concentration of 0.5 nM. Six replicate 4.5-ml samples from depths corresponding to the 30% light level were incubated in the deck incubator for 6 h under 30% light or in the dark. At the end of the incubation, samples were fixed with 2% paraformaldehyde, frozen in liquid nitrogen, and stored at −80°C before analysis by flow cytometry and sorting in the lab. Radiolabeled and stained cells were analyzed and sorted using a FacsCalibur flow cytometer following protocols previously described (33, 34).
RESULTS
Abundance of microbes in surface waters.
Total numbers of prokaryotes varied with geographic location (Table 1). Most of these prokaryotes were bacteria, as archaeal abundance was low in surface waters (data not shown). Prochlorococcus was present only between 27°N and 47°N, where it constituted on average about 4% of the total prokaryotic community in the surface layer, reaching the highest cell abundance of 8.4 × 104 cells ml−1 in the Sargasso Sea (Table 1). Abundance of Prochlorococcus declined substantially north of 45°N, to <1% of all cells. In contrast, Synechococcus was present in all surface waters, ranging from 0.5% to 15% of total cells. Abundance of AAP bacteria ranged from 1.3 × 104 cells ml−1 to 7.36 × 105 cells ml−1, constituting up to 50% of all cells. Picoeukaryote abundances were low at most stations ([0.1 to 2.9] × 104 cells ml−1) and reached maximum numbers (1.3 × 104 cells ml−1) at 50°N, where Synechococcus was most abundant.
TABLE 1.
Abundances of microbes in surface waters sampled during the NASB expeditiona
| Station | Lat (°N) | Lon (°W) | Abundance (104 cells ml−1)
|
||||
|---|---|---|---|---|---|---|---|
| Prokaryotes | Prochlorococcus | Synechococcus | AAP | Picoeukaryotes | |||
| 1-1 | 29.3 | 76.1 | 46.1 (5) | 1.3 (2.8) | 0.9 (1.9) | 5.4 (2.4) | 0.1 (0.2) |
| 1-2 | 29.8 | 74.9 | 56.7 (7.6) | 0.3 (0.5) | 1.4 (2.4) | 2.9 (0.1) | 0.1 (0.2) |
| 1-3 | 30.1 | 71.2 | 53.1 (3.6) | 0.1 (0.1) | 1.2 (2.2) | 4.5 (0.2) | 0.1 (0.2) |
| 1-4 | 31.7 | 69.8 | 58.9 (7.8) | 0.2 (0.3) | 2.5 (4.3) | 4.2 (0.6) | 0.1 (0.2) |
| 1-5 | 32.6 | 67.0 | 63.3 (6) | 4.3 (6.8) | 1.9 (2.9) | 8.6 (0.2) | 0.1 (0.1) |
| 1-6 | 33.3 | 64.8 | 79.2 (11.6) | 2.5 (3.1) | 3.6 (4.5) | 5.4 (0.2) | 0.2 (0.2) |
| 1-7 | 34.1 | 62.0 | 77.8 (1.9) | 2.0 (2.6) | 1.3 (1.7) | 2.7 (0.2) | 0.1 (0.2) |
| 1-8 | 34.8 | 59.7 | 56.5 (6.3) | 1.9 (3.4) | 3.0 (5.2) | 3.7 (0.4) | 0.3 (0.5) |
| 1-9 | 35.3 | 56.8 | 64.3 (11) | 3.1 (4.9) | 1.6 (2.4) | 8.3 (0.3) | 0.1 (0.2) |
| 1-10 | 35.8 | 54.3 | 71.4 (12) | 3.0 (4.3) | 5.9 (8.3) | 4.1 (0.1) | 0.2 (0.3) |
| 1-11 | 36.3 | 51.3 | 78.7 (10) | 1.5 (1.9) | 4.1 (5.2) | 3.2 (0.1) | 0.3 (0.4) |
| 1-12 | 36.7 | 48.7 | 114.9 (14) | 6.8 (5.9) | 2.5 (2.2) | 7.3 (0.1) | 0.3 (0.2) |
| 1-13 | 37.1 | 45.6 | 81.6 (20) | 3.8 (4.6) | 1.2 (1.4) | 4.0 (0.5) | 0.1 (0.1) |
| 1-14 | 37.4 | 42.9 | 83.9 (18) | 8.4 (10.0) | 1.1 (1.3) | 3.8 (0.1) | 0.1 (0.1) |
| 1-15 | 37.6 | 40.1 | 87.8 (7.3) | 3.4 (3.9) | 1.0 (1.2) | 6.1 (0.0) | 0.2 (0.2) |
| 1-16 | 37.8 | 37.4 | 70.6 (13) | 0.1 (0.2) | 2.3 (3.2) | 9.9 (0.0) | 0.1 (0.2) |
| 1-17 | 37.9 | 34.6 | 85.3 (12) | 6.8 (8.0) | 1.2 (1.4) | 1.3 (1.3) | 0.2 (0.2) |
| 1-18 | 37.9 | 32.0 | 71.8 (23) | 6.5 (9.0) | 2.8 (3.9) | 7.4 (0.4) | 0.1 (0.2) |
| 1-19 | 37.9 | 29.0 | 81.1 (13) | 0.5 (0.6) | 0.3 (0.4) | 5.8 (0.7) | 0.1 (0.2) |
| 2-3 | 45.0 | 20.0 | 108.0 (12) | 1.5 (1.3) | 2.9 (2.6) | 4.4 (0.4) | 1.0 (0.9) |
| 2-5 | 48.0 | 20.0 | 82.3 (20) | 1.0 (1.0) | 8.4 (8.0) | 7.7 (0.2) | 2.3 (2.2) |
| 2-7 | 50.0 | 20.0 | 97.8 (11) | 0.2 (0.2) | 15.2 (15.0) | 7.3 (0.2) | 1.6 (1.6) |
| 2-13 | 52.9 | 20.0 | 192.0 (36) | 0.1 (0.1) | 6.3 (4.2) | 12.0 (0.1) | 1.3 (0.8) |
| 2-14 | 54.1 | 17.0 | 188.9 (24) | 0.1 (0.0) | 3.5 (2.1) | 12.0 (0.0) | 0.9 (0.5) |
| 2-23 | 57.0 | 16.0 | 194.0 (49) | 0.1 (0.0) | 5.4 (3.0) | NA (0.0) | 2.9 (1.6) |
| 2-27 | 58.0 | 15.5 | 238.1 (31) | 0.1 (0.0) | 1.8 (1.0) | NA (0.0) | 2.5 (1.4) |
| 2-40 | 61.1 | 19.0 | 131.2 (28) | 0.2 (0.1) | 4.1 (3.4) | NA (0.1) | 0.2 (0.2) |
| 2-48 | 64.5 | 25.0 | 289.0 (77) | 0.3 (0.1) | 0.9 (0.4) | NA (0.4) | 0.9 (0.4) |
Values in parentheses indicate standard deviations. Lat, latitude; Lon, longitude.
Effects of light on leucine incorporation.
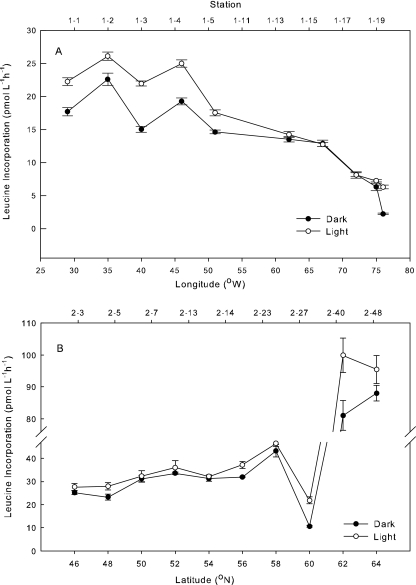
Leucine incorporation was greater in the light than in the dark in 19 out of 33 experiments. Rates were up to 6 pmol Leu liter−1 h−1 higher in the light near the Azores Islands (stations 1-13 to 1-19) (Fig. 1A), but there was no significant effect of light in the Sargasso Sea (30°N, 72°W to 32°N, 66°W). The light effect was also positive from 46°N to 64°N during the second transect, and rates of incorporation were on average 1 to 2 pmol Leu liter−1 h−1 higher with light from 46°N to 58°N (Fig. 1B). Further north, where bacterial production was high (>20 pmol Leu liter−1 h−1), light stimulated leucine incorporation the most. Rates were up to 100 pmol Leu liter−1 h−1 in 30%-light incubations of waters from the coast of Iceland (station 2-40) (Fig. 1B).
FIG. 1.
Bacterial production (leucine incorporation) along a transect from Fort Pierce, FL (76°W), to the Azores Islands (29°W) (A) or from the Azores (46°N) to Iceland (66°N) (B). Light incubations were done at 30% of surface irradiance. Error bars represent standard deviations.
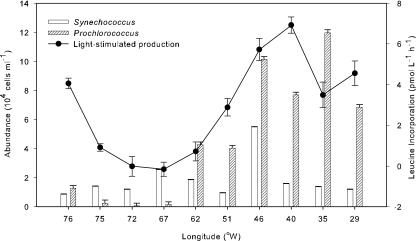
Light-stimulated leucine incorporation between Fort Pierce and the Azores was compared with the abundance of Prochlorococcus and Synechococcus (Fig. 2). Rates of light-induced incorporation were highest at the stations where Prochlorococcus was most abundant, accounting for up to 8% of the prokaryotic community, for example, at station 1-13. There was no significant light-stimulated production off the coast of Bermuda (32°N, 62°W), where the abundance of Synechococcus and Prochlorococcus was <2 × 104 cells ml−1. However, at 29°N, 76°W, light-stimulated leucine incorporation was high even though the abundance of Synechococcus and Prochlorococcus was low (<2 × 104 cells ml−1) (Fig. 2). The correlation between Prochlorococcus and light-stimulated production was significant (r = 0.70; P < 0.05; n = 10), while it was not for Synechococcus (r = 0.24; P > 0.05; n = 10). The correlation between abundance of AAP bacteria and light-stimulated production was also not significant (r = 0.15; P > 0.05; n = 10).
FIG. 2.
Abundance of cyanobacterial groups and light-stimulated leucine incorporation rate along the Fort Pierce-to-Azores transect from 76°W to 29°W. Light-stimulated leucine incorporation was calculated by subtracting the dark leucine incorporation from leucine incorporation at 30% of surface light. Error bars represent standard deviations.
Assimilation of leucine by cyanobacteria.
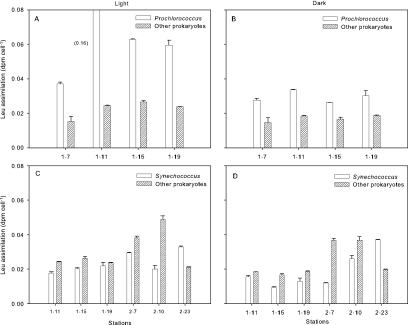
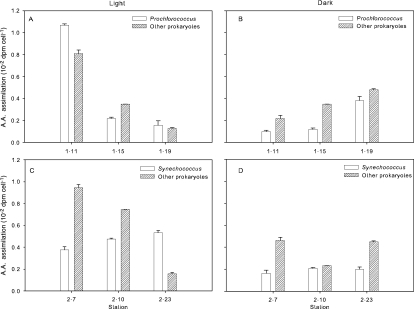
Cyanobacteria and other cells were separated by flow cytometry to determine leucine assimilation per cell (20 nM addition) (Fig. 3). Prochlorococcus had greater assimilation per cell than other prokaryotes at the stations where they were abundant (Fig. 3A and B). Assimilation per cell was up to threefold higher for Prochlorococcus than assimilation by the other groups we examined. Leucine assimilation by Prochlorococcus was on average 160% higher in the light than in the dark. Assimilation by prokaryotes other than cyanobacteria was also higher in the light than in the dark but only by 22% (Fig. 3A). Leucine assimilation by Synechococcus was not as high, but uptake per cell was comparable to that of other prokaryotes (Fig. 3C and D). Leucine assimilation by this cyanobacterial group was 44% higher in the light than in the dark.
FIG. 3.
Leucine assimilation rates for Prochlorococcus (A and B) or Synechococcus (C and D) and prokaryotes other than cyanobacteria (Other prokaryotes). Cell samples were taken from depths corresponding to the 30% light level and incubated in the dark (B and D) or at 30% of surface light irradiance (A and C) with 20 nM [3H]leucine. Error bars indicate standard errors of three measurements.
Prochlorococcus had the highest per-cell assimilation rates, 2.6-fold higher than those of other prokaryotes (Table 2). Picoeukaryotes had the second highest per-cell rates (6.92 × 10−17 mol Leu cell−1 h−1), whereas Synechococcus had the lowest (2.50 × 10−17 mol Leu cell−1 h−1). In contrast, rates of leucine assimilation per cell volume for Prochlorococcus and other prokaryotes were not significantly different. Synechococcus and picoeukaryotes had much lower assimilation rates per cell volume than other prokaryotes and Prochlorococcus (Table 2).
TABLE 2.
Leucine assimilation per cell and per biovolume for picoplankton groups in surface waters from Fort Pierce, FL, to the Azores islands
| Group | Leu assimilation per cell, 10−17 mol cell−1 h−1 (SE) | Biovolume (μm−3)a | Leu assimilation per biovolume, 10−17 mol μm−3 h−1 (SE) | No. of samples |
|---|---|---|---|---|
| Other prokaryotesb | 2.65 (0.54) | 0.037 | 71.73 (14.5) | 7 |
| Prochlorococcus | 7.06 (3.83) | 0.131 | 53.93 (29.3) | 4 |
| Synechococcus | 2.50 (0.72) | 0.449 | 5.57 (1.6) | 5 |
| Picoeukaryotes | 6.92 (1.08) | 2.000 | 3.46 (0.5) | 2 |
Cell size for prokaryotes other than cyanobacteria was estimated by epifluorescence microscopy. Sizes for Prochlorococcus, Synechococcus and picoeukaryotes were taken from the work of Zubkov et al. (36).
Prokaryotes other than Prochlorococcus and Synechococcus.
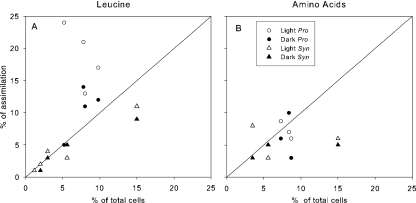
Prochlorococcus accounted for on average 18% of total leucine assimilation in the light, ranging from 13% to 24% (Fig. 4A). In the dark, leucine assimilation by Prochlorococcus accounted for only 5% to 14% of total assimilation. The percentage of total leucine assimilation by Synechococcus was much lower in both the light and dark, ranging from <1% to 10% of the total. Prochlorococcus had on average higher rates of assimilation than expected based on its abundance when incubated both at 30% light and in the dark (Fig. 4A). The contribution of Synechococcus to <10% of the total leucine assimilation was close to that expected based on its abundance, which was also on average <10% of the prokaryotic community (Fig. 4A).
FIG. 4.
Percent assimilation of [3H]leucine (A) or 3H-labeled amino acids (B) versus percentage of total prokaryotic abundance for the indicated cyanobacterial group. The diagonal represents the one-to-one line for Prochlorococcus (Pro) and Synechococcus (Syn).
We examined the contributions of Prochlorococcus, Synechococcus, and other potential photoheterotrophs to the light effect by combining data from the light-dark incubations and flow cytometry (Table 3). There was high variability among stations in rates of leucine incorporation and in cyanobacterial abundance. The average light-stimulated incorporation by the total community was 3.57 pmol Leu liter−1 h−1, while that for Prochlorococcus was on average 2.76 pmol Leu liter−1 h−1 and for Synechococcus 0.7 pmol Leu liter−1 h−1. About 80% of the light-stimulated incorporation can be accounted for by Prochlorococcus and 12% by Synechococcus (Table 3). The difference between the total leucine assimilation and assimilation by the cyanobacteria suggests that 38% of the light-stimulated rate was the result of microbes other than cyanobacteria.
TABLE 3.
Contributions to leucine assimilation (20 nM addition) by cyanobacteria and other microbial groupsa
| Group | Leucine assimilation, pmol liter−1 h−1 (SE)
|
% by groupc (SE) | No. of samples | ||
|---|---|---|---|---|---|
| Light | Dark | Light − darkb (SE) | |||
| Total community | 25.92 (3.67) | 22.50 (3.76) | 3.42 (0.85) | 100 (25) | 7 |
| Prochlorococcus | 4.63 (1.11) | 1.88 (0.27) | 2.75 (1.08) | 80 (30) | 4 |
| Synechococcus | 1.46 (0.96) | 1.05 (0.52) | 0.41 (0.62) | 12 (7) | 7 |
| Otherd | 21.82 (0.89) | 20.52 (0.48) | 1.30 (0.75) | 38 (8) | 7 |
Values are averages for all stations.
Difference in leucine incorporation between light and dark incubations.
Percentage of the difference between light and dark incubations accounted for by the indicated group.
Difference between total community and cyanobacteria (Prochlorococcus and Synechococcus). Values were calculated per sample, and then percentages were averaged for all stations.
Amino acid assimilation by cyanobacteria.
Uptake of a 0.5 nM mixture of 15 amino acids by cyanobacteria was compared with uptake by other prokaryotes (Fig. 5). Assimilation per cell for all groups was on average higher in the light than in the dark. Prochlorococcus and Synechococcus were able to assimilate low concentrations of the amino acids at per-cell rates comparable to those of other prokaryotes. The average assimilation per cell in the light for both cyanobacterial groups was 0.04 ± 0.01 dpm cell−1, while for other prokaryotes it was 0.06 ± 0.02 dpm cell−1. Synechococcus assimilation rates were twofold higher in the light (Fig. 5C) than in the dark (Fig. 5D).
FIG. 5.
Rates of amino acid (A.A.) assimilation per cell for Prochlorococcus, Synechococcus, and other prokaryotes. Samples were incubated in the dark (B and D) or at 30% of surface light irradiance (A and C). Error bars indicate standard errors of three measurements.
The percentage of total amino acid assimilation by Prochlorococcus was closer to what was expected based on its abundance than was the case for leucine assimilation (Fig. 4B), as indicated by the values being closer to the one-to-one line. Prochlorococcus contributed up to 24% of total leucine assimilation but only 10% of total amino acid assimilation. Total amino acid assimilation by Synechococcus was similar to that by Prochlorococcus (Fig. 4B). However, the contribution to total amino acid assimilation by Synechococcus was higher than its average contribution to the total assimilation of leucine (Fig. 4A and B).
DISCUSSION
Our study adds to several lines of evidence that demonstrate the potential for Prochlorococcus to utilize amino acids (5, 6, 33, 35). Church et al. (5) observed that leucine incorporation responded to irradiance and attributed the effect to Prochlorococcus because of its high abundance in the North Pacific Gyre, although the contribution of Prochlorococcus to light-enhanced assimilation was not quantified. Zubkov et al. (33, 35) found that Prochlorococcus assemblages were capable of assimilating both methionine and leucine at ∼1-nM and 5-nM concentrations, respectively, but the effect of light was not examined. We were able to measure the specific contribution of Prochlorococcus to light-stimulated bacterial production and amino acid assimilation in the North Atlantic Ocean and found evidence that other microbes are also involved in the light-stimulated assimilation of these compounds.
Synechococcus, another abundant phototroph in the upper ocean (19), might also be able to assimilate some organic compounds. Zubkov et al. (33) found that the contribution of Synechococcus to methionine uptake was <5% in the Arabian Sea, and methionine uptake per Synechococcus cell was only 30% of the activity of other bacterial cells. In contrast, our results indicate that Synechococcus was able to take up leucine (20 nM) and a mixture of amino acids (0.5 nM) at rates comparable to those for other bacteria, consistent with results in previous studies (26, 32). Malmstrom et al. (21) demonstrated that Synechococcus was able to assimilate DMSP at rates higher than those for other bacteria and accounted for about 20% of DMSP assimilation in the northwest Atlantic Ocean and the Gulf of Mexico. Vila-Costa et al. (31) showed that not only Synechococcus but also Prochlorococcus and many eukaryotic phytoplankton, including diatoms, also take up DMSP.
Uptake of amino acids by Prochlorococcus and Synechococcus during our study was a relatively small fraction (2% to 10%) of total uptake by the community. However, the abundance of these cyanobacteria was also low, reaching only 10% of prokaryotes in the waters we sampled. In oceanic regimes where the abundance is higher, amino acid uptake by cyanobacteria is potentially large. The assimilated 3H-labeled amino acids in our experiments were probably used for biomass synthesis and incorporated into macromolecules, most likely protein, because the formaldehyde treatment has the same effect on cells as TCA (16). Cyanobacteria may rely on external sources for some amino acids (auxotrophy), but Prochlorococcus and Synechococcus have all of the genes necessary to synthesize amino acids (27, 28). Regardless, dissolved free amino acid utilization could account for a high percentage of the bacterial carbon and nitrogen demand (13, 14, 18) and thus could be a large component of DOM fluxes in the oceans (15).
Light stimulated leucine assimilation even where Prochlorococcus and Synechococcus abundances were low, suggesting that other microbes are involved in the light effect. The unexplained light-stimulated leucine incorporation was probably not due to uptake by picoeukaryotes, which accounted for only about 2.6% of total uptake. More likely, the unexplained light effect was due to AAP bacteria (17) and proteorhodopsin-bearing bacteria (3). During our study, AAP bacterial abundances reached 50% of total prokaryotic abundance in some surface waters, but there was no correlation between AAP bacteria and light-enhanced production. However, Alonzo-Saez et al. (2) found significant light enhancement in leucine incorporation by Roseobacter bacteria, some of which are phototrophs (1). Proteorhodopsin has been found in several bacterial groups, including SAR11 and SAR86 (3, 9). During our study, SAR11 bacteria made up more than 40% and SAR86 up to 17% of all prokaryotes (data not shown). Proteorhodopsin-containing bacteria, perhaps AAP bacteria, and other potential photoheterotrophic prokaryotes could explain the large fraction of light-induced assimilation unaccounted for by cyanobacteria.
Evaluating the effect of light on leucine and amino acid incorporation provides insight into light-driven heterotrophic biomass production and DOM assimilation in oceanic ecosystems. Where they are abundant, Prochlorococcus and Synechococcus might play a more important role in the cycling of DOM than previously thought. However, during our study, DOM uptake by cyanobacteria and picoeukaryotes was not enough to account for the total stimulation by light. These results emphasize the potential role of other photoheterotrophic microbes in light-stimulated uptake of amino acids and possibly other compounds. The results from this study stress the need for further work to identify the different microbial groups responsible for light-affected processes.
Acknowledgments
We thank our NASB colleagues for their support during the Florida-Azores-Iceland expedition.
Support for this project was provided by grants from the National Science Foundation (OCE-0452377 and MCB-0453993) to D.L.K. and M.T.C.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Allgaier, M., H. Uphoff, A. Felske, and I. Wagner-Dobler. 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 69:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso-Saez, L., J. M. Gasol, T. Lefort, J. Hofer, and R. Sommaruga. 2006. Effect of natural sunlight on bacterial activity and differential sensitivity of natural bacterioplankton groups in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 72:5806-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Béjà, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 4.Chen, T. H., T. L. Chen, L. M. Hung, and T. C. Huang. 1991. Circadian-rhythm in amino-acid-uptake by Synechococcus Rf-1. Plant Physiol. 97:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church, M. J., H. W. Ducklow, and D. M. Karl. 2004. Light dependence of [3H]leucine incorporation in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 70:4079-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church, M. J., H. W. Ducklow, R. M. Letelier, and D. M. Karl. 2006. Temporal and vertical dynamics in picoplankton photoheterotrophic production in the subtropical North Pacific Ocean. Aquat. Microb. Ecol. 45:41-53. [Google Scholar]
- 7.Collier, J. L., B. Brahamsha, and B. Palenik. 1999. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145:447-459. [DOI] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., A. Mannino, and D. L. Kirchman. 2006. Aerobic anoxygenic phototrophic bacteria in the mid-Atlantic Bight and the North Pacific gyre. Appl. Environ. Microbiol. 72:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Torre, J. R., L. M. Christianson, O. Béjà, M. T. Suzuki, D. M. Karl, J. Heidelberg, and E. F. DeLong. 2003. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc. Natl. Acad. Sci. USA 100:12830-12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eiler, A. 2006. Evidence for the ubiquity of mixotrophic bacteria in the upper ocean: implications and consequences. Appl. Environ. Microbiol. 72:7431-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuhrman, J. A., and F. Azam. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters—evaluation and field results. Mar. Biol. 66:109-120. [Google Scholar]
- 12.Gasol, J. M., and P. A. del Giorgio. 2000. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64:197-224. [Google Scholar]
- 13.Hoch, M., and D. L. Kirchman. 1995. Ammonium uptake by heterotrophic bacteria in the Delaware estuary and adjacent coastal waters. Limnol. Oceanogr. 40:886-897. [Google Scholar]
- 14.Jørgensen, N. O. G., N. Kroer, R. B. Coffin, X. H. Yang, and C. Lee. 1993. Dissolved free amino-acids, combined amino-acids, and DNA as sources of carbon and nitrogen to marine-bacteria. Mar. Ecol. Prog. Ser. 98:135-148. [Google Scholar]
- 15.Keil, R. G., and D. L. Kirchman. 1999. Utilization of dissolved protein and amino acids in the northern Sargasso Sea. Aquat. Microb. Ecol. 18:293-300. [Google Scholar]
- 16.Kiene, R. P., and L. J. Linn. 1999. Filter-type and sample handling affect determination of organic substrate uptake by bacterioplankton. Aquat. Microb. Ecol. 17:311-321. [Google Scholar]
- 17.Kolber, Z. S., C. L. Van Dover, R. A. Niederman, and P. G. Falkowski. 2000. Bacterial photosynthesis in surface waters of the open ocean. Nature 407:177-179. [DOI] [PubMed] [Google Scholar]
- 18.Kroer, N., N. O. G. Jørgensen, and R. B. Coffin. 1994. Utilization of dissolved nitrogen by heterotrophic bacterioplankton: a comparison of three ecosystems. Appl. Environ. Microbiol. 60:4116-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, W. K. W. 1998. Annual average abundance of heterotrophic bacteria and Synechococcus in surface ocean waters. Limnol. Oceanogr. 43:1746-1753. [Google Scholar]
- 20.Li, W. K. W., D. V. S. Rao, W. G. Harrison, J. C. Smith, J. J. Cullen, B. Irwin, and T. Platt. 1983. Autotrophic picoplankton in the tropical ocean. Science 219:292-295. [DOI] [PubMed] [Google Scholar]
- 21.Malmstrom, R. R., R. P. Kiene, M. Vila, and D. L. Kirchman. 2005. Dimethylsulfoniopropionate (DMSP) assimilation by Synechococcus in the Gulf of Mexico and northwest Atlantic Ocean. Limnol. Oceanogr. 50:1924-1931. [Google Scholar]
- 22.Martinez, J., and F. Azam. 1993. Aminopeptidase activity in marine chroococcoid cyanobacteria. Appl. Environ. Microbiol. 59:3701-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montesinos, M. L., A. Herrero, and E. Flores. 1997. Amino acid transport in taxonomically diverse cyanobacteria and identification of two genes encoding elements of a neutral amino acid permease putatively involved in recapture of leaked hydrophobic amino acids. J. Bacteriol. 179:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47:989-996. [Google Scholar]
- 25.Olsen, R., E. Zettler, and M. D. DuRand. 1993. Phytoplankton analysis using flow cytometry, p. 175-186. In P. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 26.Paerl, H. W. 1991. Ecophysiological and trophic implications of light-stimulated amino-acid utilization in marine picoplankton. Appl. Environ. Microbiol. 57:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 28.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 29.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in sea water using 3H-leucine. Microb. Food Webs 6:107-113. [Google Scholar]
- 30.Troussellier, M., C. Courties, P. Lebaron, and P. Servais. 1999. Flow cytometric discrimination of bacterial populations in seawater based on SYTO 13 staining of nucleic acids. FEMS Microbiol. Ecol. 29:319-330. [Google Scholar]
- 31.Vila-Costa, M., R. Simo, H. Harada, J. M. Gasol, D. Slezak, and R. P. Kiene. 2006. Dimethylsulfoniopropionate uptake by marine phytoplankton. Science 314:652-654. [DOI] [PubMed] [Google Scholar]
- 32.Willey, J. M., and J. B. Waterbury. 1989. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl. Environ. Microbiol. 55:1888-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zubkov, M. V., B. M. Fuchs, G. A. Tarran, P. H. Burkill, and R. Amann. 2003. High rate of uptake of organic nitrogen compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Appl. Environ. Microbiol. 69:1299-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zubkov, M. V., and G. A. Tarran. 2005. Amino acid uptake of Prochlorococcus spp. in surface waters across the South Atlantic Subtropical Front. Aquat. Microb. Ecol. 40:241-249. [Google Scholar]
- 35.Zubkov, M. V., G. A. Tarran, and B. M. Fuchs. 2004. Depth related amino acid uptake by Prochlorococcus cyanobacteria in the Southern Atlantic tropical gyre. FEMS Microbiol. Ecol. 50:153-161. [DOI] [PubMed] [Google Scholar]
- 36.Zubkov, M. V., M. A. Sleigh, P. H. Burkhill, and R. J. G. Leakey. 2000. Picoplankton community structure on the Atlantic Meridional Transect: a comparison between seasons. Prog. Oceanogr. 45:369-386. [Google Scholar]