Abstract
The Tipula abdominalis larval hindgut microbial community presumably facilitates digestion of the lignocellulosic diet. The microbial community was investigated through characterization of bacterial isolates and analysis of 16S rRNA gene clone libraries. This initial study revealed novel bacteria and provides a framework for future studies of this symbiosis.
Insects are the largest taxonomic group of animals on earth. Although a few thorough studies have shown that insects host an environment with high microbial diversity (3-5, 9, 11, 17, 24), less than 1% of described insect species have been examined for microorganisms (10). Tipula abdominalis is an aquatic crane fly ubiquitous in aquatic riparian environments. T. abdominalis larvae are shredders, a functional feeding group of insects that consume coarse particulate organic matter, primarily leaf litter. In small riparian stream ecosystems, leaf litter comprises the majority of carbon and energy inputs (26); however, many organisms are unable to degrade this lignocellulosic material, which is difficult to digest due to highly structured plant polysaccharide polymers (cellulose, hemicellulose, and lignin) and which has low nutritional value due to a high C/N ratio (15). By converting lignocellulose into a form that other organisms can use, T. abdominalis larvae influence the bioavailability of carbon and energy within the ecosystem.
The larva itself is not capable of tissue-level synthesis of cellulolytic enzymes (22), and it was proposed that the larvae benefit nutritionally from microbially mediated digestion of leaf lignocellulose, providing simple fermentation products which can be used by the larvae (13). Scanning electron microscopy studies demonstrated a dense and morphologically diverse microbial community in the hindgut of T. abdominalis larvae (12). This microbial community was investigated for phylogenetic diversity and enzymatic activity towards model plant polymer substrates.
Larva collection and dissection.
Larvae were collected from 2nd-order streams (26) in Michigan. Hindguts were extracted and transferred to a reduced buffered salt solution (BSS) (14). Whole hindguts were homogenized in 1 ml BSS. Alternatively, loosely associated microorganisms were removed by vigorous vortex washing (three times) of the hindgut wall in BSS. Washed hindgut walls were homogenized in 1 ml BSS.
Bacterial isolation and characterization.
Gut homogenates were serially diluted and plated onto tryptic soy agar (Difco) and R2 agar (18) and incubated for up to 3 weeks at 22°C. Colonies were subcultured until pure cultures were obtained. Sequencing of the 16S rRNA genes from the cultured isolates was performed at MIDI Labs (Newark, DE). Fifty-nine isolates represented four classes (Fig. 1). Fifteen and 19 isolates had ≤97% and 100% 16S rRNA gene sequence similarity, respectively, to known organisms in databases.
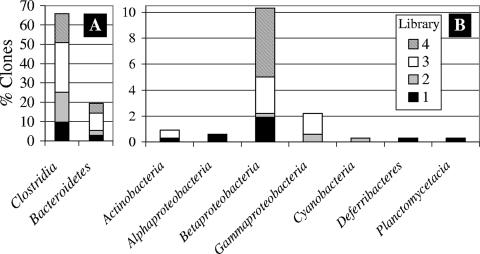
FIG. 1.
Phylogenetic distribution of clones by class. Note the change in scale: the y-axis scale runs from 0 to 70 in panel A and from 0 to 10 in panel B.
Isolates were screened for different enzymatic activities (hydrolysis of substrate) on the following model substrates as described previously: carboxymethylcellulose (CMC) (27); starch (Difco 272100); xylan (16); polygalacturonate (PGA) (23); and methylumbelliferyl conjugates of cellobiopyranoside (MUC), arabinofuranoside (MUA), glucoside (MUG), mannopyranoside (MUM), and xyloside (MUX) (20). Five (8.5%) isolates could hydrolyze all model substrates used in the current study, and 35 (60%) isolates could hydrolyze one or more of them. Isolates capable of degrading the methylumbelliferyl conjugates demonstrate enzymatic activity for degrading plant carbohydrate polymer ends produced by partial digestion of lignocellulose and may assist the digestion of leaf litter in the T. abdominalis larval hindgut.
Five groups of isolates had identical partial 16S rRNA gene sequences, but different enzymatic activities towards the study model substrates (Table 1). All five groups had high (>97%) sequence similarity to previously described bacteria. This difference in physiological characteristics would not have been observed if solely molecular techniques were employed. These results exemplify the importance of culture-dependent research in conjunction with molecular techniques. General evaluations of microbial diversity estimate that only 1 to 10% of known prokaryotic phylotypes have been cultured. Symbionts of the termite gut have been studied for over a century, yet only a small portion have been cultured (2).
TABLE 1.
Putative identity of isolates and their enzymatic activity on model plant polymers
| Isolatea | % Similarity | Cultured strain match | Enzymatic activity onb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CMC | Starch | Xylan | PGA | MUA | MUX | MUC | MUG | MUM | |||
| Proteobacteria | |||||||||||
| 1C49L | 100 | Caulobacter leidyia | − | − | − | − | − | + | + | + | − |
| 2C74 | 100 | Caulobacter leidyia | − | − | + | − | − | + | + | + | +w |
| 3C72 | 100 | Caulobacter leidyia | − | − | − | − | − | + | + | + | +w |
| 4C16 | 100 | Methylobacterium aminovorans | − | − | − | − | − | − | − | − | − |
| 8C22P | 99.1 | Methylobacterium zatmanii | − | − | − | − | − | − | − | − | − |
| 5C5 (3) | 100 | Methylobacterium hispanicum | + | − | − | − | − | − | − | − | − |
| 9C110 | 98.4 | Serratia fonticola | + | − | − | + | + | + | + | + | − |
| Firmicutes | |||||||||||
| 10C22 | 100 | Bacillus circulans | + | − | − | − | − | − | − | − | − |
| 11C46 | 100 | Bacillus circulans | − | + | − | − | + | + | + | + | − |
| 12C70 | 100 | Bacillus cohnii | − | − | − | − | − | − | + | + | − |
| 13C100 | 99.8 | Bacillus firmus | +w | + | − | − | − | − | − | − | − |
| 14C111 | 99.8 | Bacillus firmus | + | − | − | − | − | − | +w | − | − |
| 15C59 (2) | 99.8 | Bacillus fusiformis | − | − | − | − | − | − | − | − | − |
| 20C75 | 96.6 | Bacillus sphaericus | − | − | − | − | − | − | − | − | − |
| 17C67 | 100 | Bacillus silvestris | − | − | − | − | − | − | − | − | − |
| 18C68 | 100 | Bacillus silvestris | − | − | + | − | − | − | − | − | − |
| 21C27 | 100 | Bacillus megaterium | − | − | − | − | − | + | + | + | − |
| 22C25 | 100 | Bacillus weihenstephanensis | − | − | − | − | − | − | − | − | − |
| 23C26 | 99.8 | Bacillus pumilus | − | − | − | − | + | +w | + | + | − |
| 24C57 | 99.2 | Bacillus thuringiensis | + | + | − | − | − | + | + | − | − |
| 27C64 (5) | 99.1 | Paenibacillus amylolyticus | + | + | + | + | + | + | + | + | + |
| 29C58s | 97.6 | Paenibacillus amylolyticus | − | − | − | − | − | − | − | − | − |
| 31C28 | 96.2 | Paenibacillus agaridevorans | − | + | + | − | + | + | + | + | + |
| 32C23 | 95 | Paenibacillus agaridevorans | + | +w | + | − | + | + | + | + | + |
| 33C53L (2) | 98 | Paenibacillus glycanilyticus | − | − | − | − | + | + | + | + | + |
| 35C82 | 100 | Staphylococcus aureus | − | − | − | − | − | − | − | − | − |
| 36C7 (3) | 99 | Staphylococcus saprophyticus | − | − | − | − | − | − | − | − | − |
| 39C61W (2) | 100 | Staphylococcus warneri | − | − | − | − | − | − | − | − | − |
| 41C21 | 100 | Bacillus muralis | − | − | − | − | + | + | + | − | − |
| Actinobacteria | |||||||||||
| 42C60W | 99.2 | Georgenia muralis | +w | − | − | − | + | + | + | − | +w |
| 43C17 | 93.5 | Arthrobacter nicotianae | − | − | − | + | + | + | + | + | + |
| 44C3 (8) | 95 | Leifsonia poae | − | − | − | − | − | − | − | − | − |
| 19C65S1 | 99.4 | Microbacterium foliorum | − | + | − | − | + | + | + | − | − |
| 52C2 (2) | 97 | Microbacterium lacticum | + | − | − | − | − | − | − | − | − |
| 54C108 | 97 | Microbacterium lacticum | + | − | + | + | − | − | − | − | − |
| 55C94 (2) | 99.8 | Micrococcus luteus | − | − | − | − | − | − | − | − | − |
| 57C60O | 100 | Rhodococcus luteus | − | − | − | − | − | − | − | − | − |
| 58C40 | 99.4 | Sanguibacter suarezii | + | + | − | − | + | + | + | + | − |
| 59C99 | 98.6 | Sanguibacter suarezii | − | − | − | − | + | − | + | + | − |
Numbers in parentheses represent the number of isolates with identical partial 16S rRNA gene sequences and identical enzymatic profiles for the substrates tested in this study.
+, hydrolysis of substrate; −, no observable hydrolysis of substrate; +w, weak hydrolysis of substrate.
Clone libraries.
Bacterial DNA was extracted from hindgut wall homogenates (21) and purified using Sephadex G-200 spin columns (25). 16S rRNA genes were amplified from purified community genomic DNA using bacterial domain forward primer 27F (5′-AGAGTTTGATCMTGGCTCAG) and universal reverse primer 1492R (5′-GGTTACCTTGTTACGACTT) with puReTaq Ready-To-Go PCR beads (Amersham Biosciences). Each PCR began at 94°C for 3.5 min, followed by 15 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min, and finishing with 4 min at 72°C. Fifteen-cycle PCR products were cloned directly into the vector for library construction. Clone libraries were generated using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). The anterior hindguts of two larvae were pooled to construct one library (library 1), and one library was constructed per hindgut for libraries 2, 3, and 4. Clones were sequenced at Iowa State University's DNA Sequencing Facility (Ames, IA).
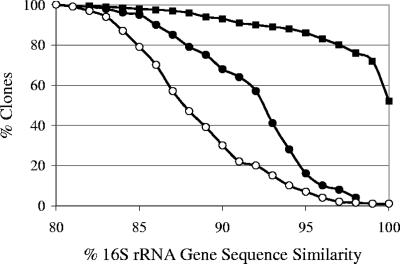
Clone sequences were analyzed for chimeras using Greengenes software tools (7, 8). DOTUR was used to determine the similarity of the clones to one another at various sequence similarities, as well as to calculate diversity indices (19). Diversity indices approached the maximum for all libraries. Coverage values for all libraries were greater than 0.5, indicating that the most prevalent phylogenetic groups had been sampled (Table 2). The percentage of sequence similarity to previously reported sequences was determined by comparing the 16S rRNA gene sequences to the RDP database (6) using the program RDPquery, version 2.5 (http://simo.marsci.uga.edu/public_db/rdp_query.htm). From 305 clones, 122 operational taxonomic units (OTUs) sharing ≥97% sequence similarity (OTU0.97) were identified, representing 9 classes (Fig. 1). The majority of clones had highest sequence similarity to Clostridia and Bacteroidetes, representing 65% and 19% of the total clones, respectively. Clones were compared to one another, as well as to previously described uncultured and cultured bacteria, at various percent sequence similarities. Clones were more similar to one another than to previously described sequences and more similar to uncultured than cultured bacteria (Fig. 2). At ≥97% sequence similarity, 80% of clones were similar to another clone from this collection, while only 8% and 2% were similar to previously described uncultured and cultured bacteria, respectively.
TABLE 2.
Diversity indices calculated from the hindgut-derived 16S rRNA gene clone libraries at OTU0.97
| Library or Na | Result for diversity index:
|
|||||
|---|---|---|---|---|---|---|
| N | Sb | Shannon (H) | H/Hmaxc | Coveraged | Chao1e | |
| 1 | 46 | 32 | 3.365 | 0.97 | 0.54 | 53 |
| 2 | 59 | 32 | 3.033 | 0.88 | 0.61 | 83 |
| 3 | 124 | 73 | 4.084 | 0.95 | 0.63 | 134 |
| 4 | 76 | 36 | 3.237 | 0.90 | 0.68 | 105 |
| N | 305 | 122 | 4.348 | 0.91 | 0.8 | 175.36 |
N, total number of clones.
S, number of OTUs.
Hmax, ln(S).
Calculated from Good's equation: coverage = 1 − (n1/N).
Chao1 = S + (n1)2/(2n2), where n1 is the number of singletons and n2 is the number of doubletons.
FIG. 2.
Percentage of clones similar to another clone from this study (closed squares) or previously described uncultured (closed circles) or cultured (open circles) bacteria at x% sequence similarity.
In summary, our data indicate that the hindgut bacterial community is phylogenetically diverse. Many of the members are more related to one another than to previously described bacteria. Isolates demonstrated enzymatic activities that are physiologically relevant to the hindgut environment and digestion of a lignocellulosic diet. While these isolates demonstrate the ability to degrade model plant polymers, it will be important in future studies to link the isolate activity in vitro with function in vivo in the larval gut. Leaf litter degradation by shredders is an important component of the stream ecosystem, and those insects consuming lignocellulose-rich diets have developed numerous mechanisms for surviving on a nutrient-poor resource. One mechanism employed is the establishment of a gut microbial consortium capable of lignocellulose degradation, and identification of microorganisms with such capabilities is essential to understanding carbon and energy cycling in stream environments.
Nucleotide sequence accession numbers.
Isolate 16S rRNA gene sequences were submitted to GenBank (1) under accession no. AY504427 to AY504477 and AY497196 to AY497203. For clones, one sequence per OTU0.99 was submitted under accession no. EF176774 to EF176920.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2005. GenBank. Nucleic Acids Res. 33:D34-D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breznak, J. A. 2000. Ecology of prokaryotic microbes in the guts of wood- and litter-feeding termites, p. 209-232. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbioses, ecology. Kluwer, Dordrecht, The Netherlands.
- 3.Breznak, J. A., and A. Brune. 1994. Role of microorganisms in the digestion of lignocellulose. Annu. Rev. Entomol. 39:453-487. [Google Scholar]
- 4.Brune, A. 2005. Symbiotic associations between termites and prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an online electronic resource for the microbiological community, 3rd ed. Springer-SBM, New York, NY.
- 5.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, NY.
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeSantis, T. Z., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillon, R. J., and V. M. Dillon. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49:71-92. [DOI] [PubMed] [Google Scholar]
- 10.Kane, M. D., and U. G. Mueller. 2002. Insights from insect-microbe symbioses, p. 289-313. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life. Wiley-Liss, Inc., New York, NY.
- 11.Kane, M. D., and N. E. Pierce. 1994. Diversity within diversity: molecular approaches to studying microbial interactions with insects, p. 509-524. In B. S. B. Schiewater, G. P. Wagner, and R. DeSalle (ed.), Molecular ecology and evolution: approaches and applications. Birkhauser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 12.Klug, M. J., and S. Kotarski. 1980. Bacteria associated with the gut tract of larval stages of the aquatic cranefly Tipula abdominalis (Diptera: Tipulidae). Appl. Environ. Microbiol. 40:408-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawson, D. L., and M. J. Klug. 1989. Microbial fermentation in the hindgut of two stream detritivores. J. North Am. Benthol. Soc. 8:85-91. [Google Scholar]
- 14.Leadbetter, J. R., and J. A. Breznak. 1996. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov. isolated from the hindgut of the termite Reticulitermes flavipes. Appl. Environ. Microbiol. 62:3620-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin, M. M., J. S. Martin, J. J. Kukor, and R. W. Merritt. 1980. The digestion of protein and carbohydrate by the stream detritivore, Tipula abdominalis (Diptera, Tipulidae). Oceologia 46:360-364. [DOI] [PubMed] [Google Scholar]
- 16.Mondou, F., F. Shareck, R. Morosoli, and D. Kluepfel. 1986. Cloning of the xylanase gene of Streptomyces lividans. Gene 49:323-330. [DOI] [PubMed] [Google Scholar]
- 17.Moran, N. A. 2001. The coevolution of bacterial endosymbionts and phloem-feeding insects. Ann. Missouri Bot. Garden 88:35-44. [Google Scholar]
- 18.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharrock, K. R. 1988. Cellulase assay methods: a review. J. Biochem. Biophys. Methods 17:81-106. [DOI] [PubMed] [Google Scholar]
- 21.Shinzato, N., T. Matsumoto, I. Yamaoka, T. Oshima, and A. Yamagishi. 1999. Phylogenetic diversity of symbiotic methanogens living in the hindgut of the lower termite Reticulitermes speratus analyzed by PCR and in situ hybridization. Appl. Environ. Microbiol. 65:837-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinsabaugh, R. L., A. E. Linkins, and E. F. Benfield. 1985. Cellulose digestion and assimilation by three leaf-shredding aquatic insects. Ecology 66:1464-1471. [Google Scholar]
- 23.Starr, M. P., A. K. Chatterjee, P. B. Starr, and G. E. Buchanan. 1977. Enzymatic degradation of polygalacturonic acid by Yersinia and Klebsiella species in relation to clinical laboratory procedures. J. Clin. Microbiol. 6:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanada, Y., and H. K. Kaya. 1993. Insect pathology. Academic Press, London, United Kingdom.
- 25.Tsai, Y.-L., and B. H. Olson. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vannote, R., G. Minshall, K. W. Cummins, J. Sedell, and C. Cushing. 1980. The river continuum concept. Can. J. Fish. Aquat. Sci. 37:130-137. [Google Scholar]
- 27.Wood, W. A., and S. T. Kellogg. 1988. Biomass part A: cellulose and hemicellulose, vol. 160. Academic Press, San Diego, CA.