Abstract
Phylogenetic analysis of rRNA gene, recA, nodA, nifD, and nifH sequences suggested that nitrogen-fixing symbionts from two populations of Lupinus texensis acquired the capacity for nodule symbiosis separately from other rhizobia in the alphaproteobacteria. Their closest 16S rRNA relatives were the nonsymbiotic taxa Chelatococcus, Bosea, and Balneomonas.
Legume nodule symbionts are found in several lineages in the alphaproteobacteria and at least two lineages in the betaproteobacteria (5, 17). Four genera of alphaproteobacteria (Rhizobium, Mesorhizobium, Sinorhizobium, and Bradyrhizobium) constitute the predominant symbionts for most legume species in habitats throughout the world. However, additional nodule-forming lineages of alphaproteobacteria that may have a more restricted geographic distribution or limited host range have been discovered in recent years (8, 14, 16, 22, 28, 31, 32). This suggests that further sampling of other host taxa and locations is likely to uncover additional lineages of nodule bacteria.
The legume genus Lupinus includes approximately 275 species of herbs and shrubs (11). Studies of most Lupinus species within their native range (4, 15, 21, 24), as well as outside their native distribution (6, 26, 30), indicate that strains of Bradyrhizobium are the predominant nodule symbionts. We sampled nodule bacteria in the native range of Lupinus texensis in Texas (13) and found that none of the isolates resembled Bradyrhizobium symbionts associated with other Lupinus species. Twenty-eight root nodules were collected from multiple plants at two sites 20 km apart (Williamson County and Travis County), and one bacterial isolate per nodule was obtained as previously described (25). DNA purification and PCR experiments used standard protocols (5). Amplification of the 5′ intervening sequence region of the 23S rRNA gene (27) indicated that all 28 isolates had a unique size variant (473 bp) that differed from length variants found in North American Bradyrhizobium strains (510 or 537 bp) (18, 20, 21). A 431-bp 5′ portion of the 23S rRNA gene was sequenced in five Texas isolates (GenBank accession numbers EF191402 to EF191406), and each sequence was the same. Blast searches indicated that this sequence was distantly related to all currently known lineages of legume nodule symbionts. The closest match among rhizobia was Bradyrhizobium japonicum USDA 110 (80% similarity). Other genera of alphaproteobacterial and betaproteobacterial symbionts had similarities ranging from 62 to 77%.
To screen the remaining L. texensis nodule isolates, PCR assays were performed with a primer pair (LUTf1 and 23LUTr2; Table 1) matching unique portions of the L. texensis symbiont 5′ 23S rRNA sequence. All isolates yielded the same 167-bp amplification product. Other genera of nodule bacteria (Bradyrhizobium, Mesorhizobium, Sinorhizobium, and Rhizobium) failed to yield an amplification product with these primers.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | 5′→3′ nucleotide sequence | Product size (bp) | Target gene |
|---|---|---|---|
| LUTf1 | CGGAGGGTCAGATGAAGGGATTA | 167 | 23S rRNA |
| 23LUTr2 | GGTTCCGATGACGATCAGGTTGG | ||
| recAf | GGCAGTTCGGCAAGGGCTCGAT | 529 | recA |
| recAr | ATCTGGTTGATGAAGATCACCAT | ||
| nifD.f1 | TCGGACTTCCAGGAAAAGGACAT | 521 | nifD |
| nifD.r2 | CCGWACTTYTCTTCCATGTGGC | ||
| nifH.f2 | TAYGGNAARGGGGGGATYGGYAAGTC | 437 | nifH |
| nifH.r3 | TCGCCGGACATGACGATGTAGAT | ||
| LUT5af | AGAAAATAGCAAGAAAGAGGAGGT | 619 | nodA |
| LUTar1 | AAHATGGATKRGGACCGTCGTC |
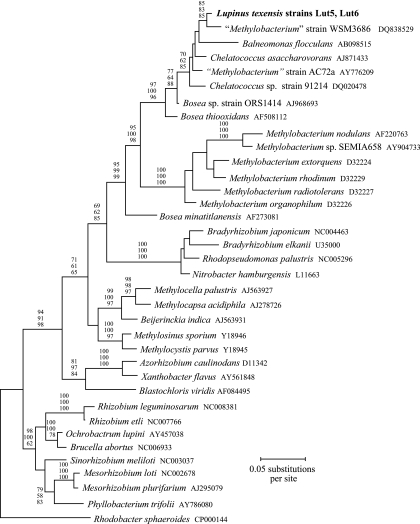
A nearly full-length portion of the 16S rRNA gene (1,408 bp) was sequenced for two isolates (Lut5, Lut6). Both isolates had the same sequence (EF191407, EF191408). The maximum-likelihood (ML) tree showed that the L. texensis symbionts formed a lineage well separated from traditionally recognized genera of rhizobia (Fig. 1). The L. texensis symbionts were grouped along with a set of ecologically diverse taxa (Chelatococcus, Balneomonas, and Bosea thiooxidans) with high bootstrap support (96 to 100%). These taxa are not known to form symbiotic relationships with legumes. Chelatococcus asaccharovorans has been isolated from soil and water in various locations (1, 10). Balneomonas flocculans was collected from a hot spring in Japan (29). Bosea thiooxidans was isolated from agricultural soils in India (7). A second Bosea strain (ORS1414), found as a commensal inhabitant of legume nodules in Tunisia (although not capable of inducing nodule formation [36]), was also grouped in this lineage.
FIG. 1.
ML phylogenetic tree for 16S rRNA genes (1,408 bp) from L. texensis symbionts and other alphaproteobacteria. Values near branches are bootstrap percentages for maximum-parsimony (top), neighbor-joining (center), and ML (bottom) analyses.
Two nodule symbionts considered to be members of the genus Methylobacterium were also clustered in the group with the L. texensis symbionts and Chelatococcus, Balneomonas, and Bosea thiooxidans (Fig. 1). Strain WSM 3686 was isolated from the legume Lotononis in Zambia (DQ838529), and strain AC72a was sampled from Phaseolus vulgaris in Ethiopia (35). However, the identity of these two strains on the genus level is uncertain. Five species in the genus Methylobacterium that were included in the phylogenetic analysis formed a well-supported group (100% bootstrap value) that was clearly distinct from the lineage encompassing strains WSM 3686 and AC72a and the L. texensis symbionts (Fig. 1).
These results leave unresolved the proper genus name of the L. texensis nodule symbionts. The L. texensis symbionts had a 16S rRNA sequence closely affiliated with those of nonsymbiotic species classified into three separate genera (Balneomonas flocculans, 95.5% similarity; Bosea thiooxidans, 96.5% similarity; Chelatococcus asaccharovorans, 97.8% similarity). Because there was high bootstrap support for the placement of these taxa in a lineage distinct from Methylobacterium (Fig. 1), it appears to be inappropriate to classify the L. texensis symbionts in that genus.
Lateral transfer of portions of the 16S rRNA gene can be a potential source of phylogenetic distortions, by creating mosaic genes where different segments have different ancestries (33). However, several analyses failed to detect evidence for this as a factor affecting inferences about L. texensis symbiont 16S rRNA relationships. First, analysis of 16S rRNA sequences with GENECONV (23) did not identify any mosaic structure of the L. texensis symbiont 16S rRNA gene. Second, a split decomposition analysis (3) with SplitsTree (12) yielded a strictly dichotomously branching tree. Thus, there was no evidence for multiple alternative pathways of relationship indicative of mosaic 16S rRNA gene structure in the L. texensis symbionts. Finally, when 16S rRNA data were divided into two equal portions, both the 5′ and 3′ halves resulted in trees that grouped the L. texensis symbionts with Chelatococcus, Balneomonas, and Bosea thioxidans and placed this group apart from all other rhizobial genera, as in Fig. 1. Thus, substitutions throughout different portions of the 16S rRNA gene supported the same conclusions about relationships of the L. texensis symbionts.
A 484-bp portion of the recA gene was sequenced in three isolates of the novel L. texensis symbionts. Two isolates (Lut5, Ltg2) had identical sequences, which differed by 96 bp from the sequence of isolate Lut6 (EF191415 to EF191417). Blast searches did not identify close matches to either sequence among known genera of nodule bacteria (or other bacteria). Comparison of the Ltg2/Lut5 recA sequence with those of 16 taxa of other rhizobia (from five genera, i.e., Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium) showed similarity percentages that ranged from 71% (B. japonicum USDA 110) to 76% (Rhizobium gallicum). Percent recA similarity for strain Lut6 ranged from 71% (B. japonicum USDA 110) to 76% (R. leguminosarum). recA sequences are not currently available for key taxa identified as relatives of the L. texensis symbionts based on the 16S rRNA data (Balneomonas, Chelatococcus, Bosea thiooxidans), so the specific topology of Fig. 1 cannot be confirmed with these data. Nevertheless, the results provide further evidence that the L. texensis symbionts are not closely related to known nodule bacteria.
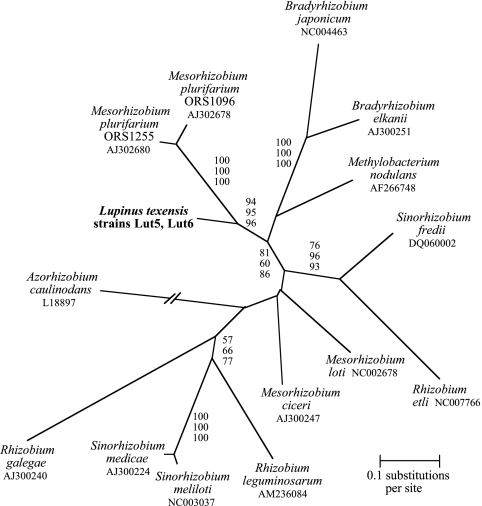
A complete sequence for the nodA gene (591 bp; Table 1 contains the primers used for amplification) was obtained for two of the novel L. texensis isolates. Both isolates had the same sequence (EF191413, EF191414). Because the origin of nod genes is not well resolved, an unrooted tree was used to depict relationships (Fig. 2). The tree topology indicated that the novel L. texensis symbiont formed a well-supported branch with Mesorhizobium plurifarium strains that were isolated from Acacia tortilis in Africa (2). These results imply that the L. texensis strains evolved to become legume nodule symbionts independently of the most closely related rhizobial taxon in the 16S rRNA tree (Methylobacterium nodulans). This conclusion is based on the observation that all of these taxa have a closer 16S rRNA relationship to nonsymbiotic taxa than they do to one another (Fig. 1) and the fact that M. nodulans and the L. texensis symbionts do not cluster as each other's closest relatives in the nodA tree (Fig. 2).
FIG. 2.
Unrooted phylogenetic tree derived from ML analysis of nodA sequences (591 bp) from L. texensis symbionts and other rhizobia. Values near branches are bootstrap percentages for maximum-parsimony (top), neighbor-joining (center), and ML (bottom) analyses.
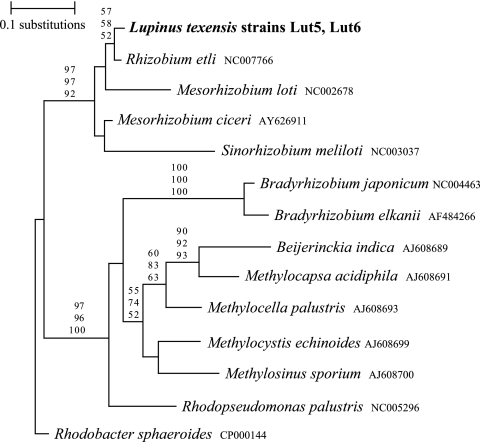
Partial sequences of the nifD (476 bp) and nifH (388 bp) genes were obtained for two isolates of the novel L. texensis symbionts, and for both genes, the two isolates proved to be identical (EF191409 to EF191412). The ML tree topologies for both the nifD and nifH genes were broadly similar but differed considerably from the ribosomal and nodA trees. Phylogenetic analysis of the partial nifD sequences (Fig. 3) showed that the L. texensis symbionts formed a group with Rhizobium, Mesorhizobium, and Sinorhizobium (92 to 97% bootstrap values). The nifH tree topology (not shown) also placed the novel L. texensis nodule bacteria together with Rhizobium, Sinorhizobium, and Mesorhizobium with high bootstrap support (86 to 92%). For both trees, Bradyrhizobium formed a well-separated lineage. A partition homogeneity test (9) indicated that both nif tree topologies were highly incongruent with the 16S rRNA tree (P < 0.001). To eliminate the possibility that these nif sequences came from another bacterial strain that somehow became mixed with the L. texensis strain cultures, a spontaneous streptomycin-resistant mutant was obtained for strain Lut5 and its partial nifD sequence was determined. The mutant had a nifD sequence identical to that of the Lut5 parent strain, and its 5′ 23S rRNA PCR product (obtained with lineage-specific primers LUTf1 and 23LUTr2) was also identical. Thus, there was no evidence that the parent strain was a mixed culture.
FIG. 3.
ML phylogenetic tree for partial nifD sequences (476 bp) from L. texensis symbionts and other alphaproteobacteria. Values near branches are bootstrap percentages for maximum-parsimony (top), neighbor-joining (center), and ML (bottom) analyses.
These results suggest that the novel L. texensis symbionts may have acquired their nif genes from a phylogenetically distant source through lateral transfer. Partition homogeneity tests also indicated that there was significant incongruence of both nifD and nifH phylogenetic trees from the nodA tree (P < 0.027 and P < 0.003, respectively).
Symbiotic behavior.
Inoculation experiments with L. texensis symbionts used standard protocols (34), and no nodules formed on uninoculated control plants in any experiment. When L. texensis plants were inoculated with three isolates (Lut3, Lut5, Lut6), all plants formed nodules (mean = 8 to 11 nodules per plant), and acetylene reduction assays indicate that there was substantial nitrogenase activity (0.6 to 1.1 μmol ethylene/plant/min). These values are within the range commonly found for Bradyrhizobium strains with various legume hosts (19, 20) and indicate that nodules were functional for nitrogen fixation. The identity of the nodule occupants was verified by reisolating one bacterial culture from a surface-sterilized nodule for each inoculation isolate and then sequencing the 5′ 23S rRNA region. The nucleotide sequences from the nodule isolates were identical to those from the inoculants. This confirms that the novel L. texensis strains were responsible for inducing nodule formation in this experiment.
Plants of Phaseolus vulgaris, Mimosa pudica, and Desmodium canadense all failed to form nodules when inoculated with these three isolates, while Cytisus scoparius and Macroptilium atropurpureum formed tiny nodules (mean = 0.1 and 7 nodules/plant, respectively). However, acetylene reduction assays failed to detect any nitrogenase activity in these plants. A bacterial culture was reisolated from a surface-sterilized nodule from both Cytisus and Macroptilium plants. PCR assays with the 23S rRNA primers specific to the novel L. texensis symbionts (LUTf1, 23LUTr2) indicated that the Cytisus and Macroptilium nodule isolates exhibited the same 167-bp product as the inoculant L. texensis strains.
L. texensis plants were grown for 44 days after inoculation with the same three isolates and with two Bradyrhizobium strains isolated from other Lupinus species (nine per treatment). The two Bradyrhizobium strains failed to form nodules on L. texensis, and the growth of plants inoculated with these strains was not different from that of uninoculated controls. Two isolates (Lut5 and Lut6) resulted in mean plant biomass values (0.43 ± 0.08 [standard error] and 0.46 ± 0.08 g) that were significantly higher than those of uninoculated controls (0.20 ± 0.03 g; P < 0.05 [analysis of variance]). A third isolate (Lut3) showed a smaller growth increase (0.28 ± 0.03 g), which was not significantly greater than that of the controls. Thus, the novel L. texensis bacteria can benefit their host legume under nitrogen-limiting conditions, but isolates vary in the capacity to improve plant growth.
When plants of three other species of Lupinus (L. perennis, L. succulentus, L. microcarpus) were inoculated with four L. texensis isolates (Lut3, Lut5, Lut6, and Ltg2), no nodules developed on any of the plants. All three of these Lupinus species did form nodules when inoculated with various strains of Bradyrhizobium isolated from North American Lupinus species. These results suggest that the capacity for nodule symbiosis with the novel L. texensis symbionts may not be widespread in the genus Lupinus and that L. texensis symbionts may be more or less specialized on their original host.
Conclusions.
We have shown that natural populations of L. texensis harbor a novel lineage of nitrogen-fixing nodule symbionts that are distantly related to the Bradyrhizobium strains utilized by other species of Lupinus. The fact that the closest known relatives of these strains in the 16S rRNA tree are nonsymbiotic taxa (Chelatococcus, Balneomonas, Bosea thioxidans; Fig. 1) implies that the L. texensis symbionts acquired the capacity for nodule symbiosis separately from other rhizobial groups of alphaproteobacteria. Further studies of sequence variation in relatives of this bacterial lineage and studies of their biogeographic distribution and host range are needed to better understand the origin of this symbiotic interaction.
Acknowledgments
We are grateful to J. Pfeil for expert assistance with sequencing.
Financial support was provided by NSF grant DEB-0235766.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Auling, G., H. J. Busse, T. Egli, T. El-Banna, and E. Stackebrandt. 1993. Description of the gram-negative, obligately aerobic, nitriloacetate (NTA)-utilizing bacteria as Chelatobacter heintzii, gen. nov., sp. nov., and Chelatococcus asaccharovorans, gen. nov., sp. nov. Syst. Appl. Microbiol. 16:104-112. [Google Scholar]
- 2.Ba, S., A. Willems, P. De Lajudie, P. Roche, H. Jeder, P. Quatrini, M. Neyra, M. Ferro, J. C. Prome, M. Gillis, C. Boivin-Masson, and J. Lorquin. 2002. Symbiotic and taxonomic diversity of rhizobia isolated from Acacia tortilis subsp. raddiana in Africa. Syst. Appl. Microbiol. 25:130-145. [DOI] [PubMed] [Google Scholar]
- 3.Bandelt, J. J., and A. W. Dress. 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1:242-252. [DOI] [PubMed] [Google Scholar]
- 4.Barrera, L. L., M. E. Trujillo, M. Goodfellow, F. J. Garcia, I. Hernandez-Lucas, G. Davila, P. van Berkum, and E. Martinez-Romero. 1997. Biodiversity of bradyrhizobia nodulating Lupinus spp. Int. J. Syst. Bacteriol. 47: 1086-1091. [DOI] [PubMed] [Google Scholar]
- 5.Barrett, C. F., and M. A. Parker. 2006. Coexistence of Burkholderia, Cupriavidus and Rhizobium sp. nodule bacteria on two Mimosa species in Costa Rica. Appl. Environ. Microbiol. 72:1198-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottomley, P. J., H. H. Cheng, and S. R. Strain. 1994. Genetic structure and symbiotic characteristics of a Bradyrhizobium population recovered from a pasture soil. Appl. Environ. Microbiol. 60:1754-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, S. K., A. K. Mishra, B. J. Tindall, F. A. Rainey, and E. Stackebrandt. 1996. Oxidation of thiosulfate by a new bacterium, Bosea thiooxidans (strain BI-42) gen. nov., sp. nov.: analysis of phylogeny based on chemotaxonomy and 16S ribosomal DNA sequencing. Int. J. Syst. Bacteriol. 46:981-987. [DOI] [PubMed] [Google Scholar]
- 8.Dreyfus, B., J. L. Garcia, and M. Gillis. 1988. Characterization of Azorhizobium caulinodans gen. nov., sp. nov., a stem-nodulating, nitrogen-fixing bacterium isolated from Sesbania rostrata. Int. J. Syst. Bacteriol. 38:89-98. [Google Scholar]
- 9.Farris, J. S., M. Kallersjo, A. G. Kluge, and C. Bult. 1995. Testing significance of incongruence. Cladistics 10:315-319. [Google Scholar]
- 10.Gundlapally, S. R., and F. Garcia-Pichel. 2006. The community and phylogenetic diversity of biological soil crusts in the Colorado Plateau studies by molecular fingerprinting and intensive cultivation. Microb. Ecol. 52:345-357. [DOI] [PubMed] [Google Scholar]
- 11.Hughes, C., and R. Eastwood. 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. USA 103:10334-10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huson, D. H. 1998. Splits Tree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 13.Isely, D. 1998. Native and naturalized Leguminosae (Fabaceae) of the United States. Brigham Young University, Provo, UT.
- 14.Jaftha, J. B., B. W. Strijdom, and P. L. Steyn. 2002. Characterization of pigmented methylotrophic bacteria which nodulate Lotononis bainesii. Syst. Appl. Microbiol. 25:440-449. [DOI] [PubMed] [Google Scholar]
- 15.Jarabo-Lorenzo, A., R. Perez-Galdona, J. Donate-Correa, R. Rivas, E. Velasquez, M. Hernandez, F. Temprano, E. Martinez-Molina, T. Ruiz- Argueso, and M. Leon-Barrios. 2003. Genetic diversity of bradyrhizobial populations from diverse geographic origins that nodulate Lupinus spp. and Ornithopus spp. Syst. Appl. Microbiol. 26:611-623. [DOI] [PubMed] [Google Scholar]
- 16.Moreira, F. M. S., L. Cruz, S. M. de Faria, T. Marsh, E. Mart'nez-Romero, F. de Oliveira Pedrosa, R. M. Pitard, and J. P. W. Young. 2006. Azorhizobium doebereinerae sp. nov. microsymbiont of Sesbania virgata (Caz.) Pers. Syst. Appl. Microbiol. 29:197-206. [DOI] [PubMed] [Google Scholar]
- 17.Moulin, L., A. Munive, B. Dreyfus, and C. Boivin-Masson. 2001. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411:948-950. [DOI] [PubMed] [Google Scholar]
- 18.Parker, M. A. 1999. Relationships of bradyrhizobia from the legumes Apios americana and Desmodium glutinosum. Appl. Environ. Microbiol. 65:4914-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker, M. A., and A. Lunk. 2000. Relationships of bradyrhizobia from Platypodium and Machaerium (Papilionoideae tribe Dalbergieae) on Barro Colorado Island, Panama. Int. J. Syst. Evol. Microbiol. 50:1179-1186. [DOI] [PubMed] [Google Scholar]
- 20.Parker, M. A., J. L. Doyle, and J. J. Doyle. 2004. Comparative phylogeography of Amphicarpaea legumes and their root-nodule symbionts in Japan and North America. J. Biogeogr. 31:425-434. [Google Scholar]
- 21.Parker, M. A., and D. A. Kennedy. 2006. Diversity and relationships of bradyrhizobia from legumes native to eastern North America. Can. J. Microbiol. 52:1148-1157. [DOI] [PubMed] [Google Scholar]
- 22.Rivas, R., E. Velasquez, A. Willems, N. Vizcaino, N. S. Subba-Rao, P. F. Mateos, M. Gillis, F. B. Dazzo, and E. Martinez-Molina. 2002. A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (L.f.) Druce. Appl. Environ. Microbiol. 68:5217-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 24.Simms, E. L., D. Lee Taylor, J. Povich, R. P. Shefferson, J. L. Sachs, M. Urbina, and Y. Tausczik. 2006. An empirical test of partner choice mechanisms in a wild legume-rhizobium interaction. Proc. Biol. Sci. 273:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spoerke, J. M., H. H. Wilkinson, and M. A. Parker. 1996. Nonrandom genotypic associations in a legume-Bradyrhizobium mutualism. Evolution 50:146-154. [DOI] [PubMed] [Google Scholar]
- 26.Stepkowski, T., L. Moulin, A. Krzyzanska, A. McInnes, I. J. Law, and J. Howieson. 2005. European origin of Bradyrhizobium populations infecting lupins and serradella in soils of Western Australia and South Africa. Appl. Environ. Microbiol. 71:7041-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sterner, J. P., and M. A. Parker. 1999. Diversity and relationships of bradyrhizobia from Amphicarpaea bracteata based on partial nod and ribosomal sequences. Syst. Appl. Microbiol. 22:387-392. [DOI] [PubMed] [Google Scholar]
- 28.Sy, A., E. Giraud, P. Jourand, N. Garcia, A. Willems, P. De Lajudie, Y. Prin, M. Neyra, M. Gillis, C. Boivin-Masson, and B. Dreyfus. 2001. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 183:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda, M., I. Suzuki, and J. I. Koizumi. 2004. Balneomonas flocculans gen. nov., sp. nov., a new cellulose-producing member of the α-2 subclass of Proteobacteria. Syst. Appl. Microbiol. 27:139-145. [DOI] [PubMed] [Google Scholar]
- 30.Thorsson, A. T., H. Sverrisson, and K. A. Jonsson. 2000. Genotyping Icelandic isolates of rhizobia based on rDNA-RFLP. Icel. Agric. Sci. 13:17-25. [Google Scholar]
- 31.Trujillo, M. E., A. Willems, A. Abril, A. M. Planchuelo, R. Rivas, D. Ludena, P. F. Mateos, E. Martinez-Molina, and E. Velasquez. 2005. Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl. Environ. Microbiol. 71:1318-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valverde, A., E. Velasquez, F. Fernandez-Santos, N. Vizcaino, R. Rivas, P. Mateos, E. Martinez-Molina, J. M. Igual, and A. Willems. 2005. Phyllobacterium trifolii sp. nov., nodulating Trifolium and Lupinus in Spanish soils. Int. J. Syst. Evol. Microbiol. 55:1985-1989. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Y., and Z. Zhang. 2000. Comparative sequence analyses reveal frequent occurrence of short segments containing an abnormally high number of non-random base variations in bacterial rRNA genes. Microbiology 146:2845-2854. [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson, H. H., J. M. Spoerke, and M. A. Parker. 1996. Divergence in symbiotic compatibility in a legume-Bradyrhizobium mutualism. Evolution 50:1470-1477. [DOI] [PubMed] [Google Scholar]
- 35.Wolde-Meskel, E., Z. Terefework, A. Frostegard, and K. Lindstrom. 2005. Genetic diversity and phylogeny of rhizobia isolated from agroforestry legume species in southern Ethiopia. Int. J. Syst. Evol. Microbiol. 55:1439-1452. [DOI] [PubMed] [Google Scholar]
- 36.Zakhia, F., H. Jeder, A. Willems, M. Gillis, B. Dreyfus, and P. de Lajudie. 2006. Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb. Ecol. 51:375-393. [DOI] [PubMed] [Google Scholar]