Abstract
Heat-labile enterotoxin (LT) from enterotoxigenic Escherichia coli is a heterohexameric protein consisting of an enzymatically active A subunit, LTA, and a carrier pentameric B subunit, LTB. It is clear from the crystal structure of LTB that the N-terminal α1 helix lies outside the core structure. However, the function of the N-terminal α1 helix of LTB is unknown. The present work was carried out to investigate the effect of site-directed mutagenesis of the α1 helix on LTB synthesis. Six amino acids (PQSITE) located at positions 2–7 from the N terminus, including 4 aa from the α1 helix, were deleted by site-directed mutagenesis. The deletion resulted in complete inhibition of LTB expression in E. coli when expressed along with its signal sequence. A single amino acid deletion within the α1 helix also resulted in loss of expression. However, a single amino acid deletion outside the α1 helix did not affect LTB synthesis. Mutant proteins, whose synthesis was not detected in vivo, could be successfully translated in vitro by using the coupled transcription–translation system. Immunoblot analysis, Northern blot analysis, and in vitro transcription–translation data collectively indicate that the lack of synthesis of the mutant proteins is caused by the immediate degradation of the expressed product by cellular proteases rather than by faulty translation of mutant LTB mRNA. Coexpression of the LTA could not rescue the degradation of LTB mutants.
Keywords: gene expression, site-directed mutagenesis, coupled transcription–translation
Diarrhea caused by the enterotoxigenic Escherichia coli (ETEC) is a major cause of death in developing countries, especially among children, with an estimated mortality of 1.5 million cases every year (1–3; for review, see ref. 4). Approximately 20% of cases of traveler's diarrhea are caused by ETEC, and thus the organism spreads to the developed countries (4, 5). ETEC produces a number of virulence factors, such as enterotoxins and colonization factors. Of these, the heat-labile (LT) and heat-stable enterotoxins produced by the ETEC are the major virulence factors responsible for its pathogenicity (5, 6). The LT belongs to a family of bacterial proteins designated heat-labile enterotoxins and shares phenotypic and genotypic similarities with other members of the family such as cholera toxin produced by Vibrio cholerae (7, 8; for review, see ref. 9).
The mature toxin consists of a single A polypeptide (LTA) and five B polypeptides (LTB) (10–12). LTB and cholera toxin B show a high degree of homology, with 85% conservation of amino acids (13). The genes of the two LT subunits, eltA and eltB, are transcribed as a single polycistronic mRNA (14) and are expressed with signal peptides. The two subunits are synthesized as a precursor protein, and each subunit has its own ribosome-binding site (15–17). After cleavage of the signal peptide, the two subunits of LT are released into the periplasmic space where they spontaneously assemble into a mature holotoxin (18, 19). LTA is known to influence LTB oligomerization. In the course of LTB pentamerization, LTA associates noncovalently with the B oligomer. However, LTA cannot associate with fully assembled LTB subunits. Because of defects in folding, the unassembled B monomers are rapidly degraded (20). LTB is responsible for delivering catalytically active LTA to target cells by binding to GM1 ganglioside receptors and thus acts as a carrier molecule (10, 21–26). The strong GM1 ganglioside receptor-binding activity demonstrated by LTB makes it an important mucosal adjuvant, and thus LTB shows great promise for developing oral vaccines.
The crystal structure of LT has been solved and can serve as an excellent model system to study the structure–function relationship of different regions. The N-terminal α1 helix of LTB is a structure whose function is unknown. The crystal structure shows that the α1 helix lies outside the core structure and interacts with the β5 strand through a disulfide bond between Cys9 and Cys86 (27). However, a crystal structure by itself is not enough to define the role of a domain or an amino acid in protein expression and function.
The N terminus of several polypeptides has been shown to serve multiple functions, including protein expression, folding, and interaction with other molecules (28–31). The N-terminal region of ribosomal protein S7 is crucial for its interaction with the 3′ major domain of 16S rRNA (28). The α helix present at the N terminus of rhodanese is known to participate in initial folding, in the global stability of the protein, and in providing resistance to degradation (29). In the case of aldehyde dehydrogenase-1 and -2, the N terminus helps in folding and maintaining protein stability (31). The N- and C-terminal helices of cytochrome C are the first to form and serve as a docking surface to guide subsequent folding of the protein (32). Because a specific role for the N-terminal α1 helix of LTB has not yet been established, the present work was undertaken to understand its role in LTB expression and secretion.
Results
Deletion of the N-Terminal 6 Amino Acids from the α1 Helix of LTB Impairs Expression in E. coli.
To study the role of the N-terminal α1 helix in structure–function analysis, 6 N-terminal aa (PQSITE) from positions 2–7, including 4 aa from the α1 helix, were deleted (Fig. 1). It was anticipated that the mutant protein would have a better chance of achieving stable expression in its native environment through the secretory pathway. Therefore, the mutant ltb gene, along with its natural N-terminal signal sequence, was cloned in pMMB vector to study the effect of the mutation under its normal secretory pathway in E. coli DH5α. Total cell extract, culture supernatant, and periplasmic fractions were analyzed for expression of mutant LTB by immunoblotting with polyclonal anti-LTB antibody (Fig. 2). As evident from the figure, no expression of mutant protein Δ6 (N-terminal 6-aa deletion) from plasmid pLTBΔ6 was detected in the supernatant, the periplasmic fraction, or the total cell extract of induced cells of E. coli (lanes 1, 2, and 4, respectively). Expression of wild-type (WT) LTB from plasmid pMMB68 in the periplasmic fraction and total cell extract (lanes 6 and 8, respectively) could be detected. The recombinant LTB, when expressed as inclusion body, could be detected in immunoblot analysis (data not shown), suggesting that the deletion mutation of LTB did not affect its reactivity with polyclonal anti-LTB antibody used in this work.
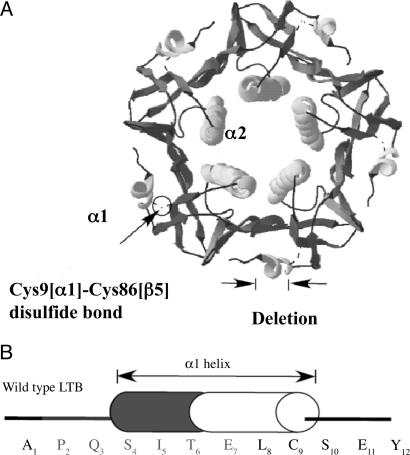
Fig. 1.
Ribbon structure of LTB pentamer and sequence of N-terminal α1 helix. (A) Ribbon representation of LT of enterotoxigenic E. coli. The structure is generated by using program SWISS-PDB Viewer (52). The position of the N-terminal deletion is marked by two arrows. The Cys9[α1]–Cys86[β5] disulfide bond is shown by a circle. Two helices (α1 and α2) are shown. (B) Amino acid sequence of the N terminus of the ltb gene. Cylinder represents the α1 helix. Corresponding amino acids represented by single letter code are given. The deleted residues are shown in gray.
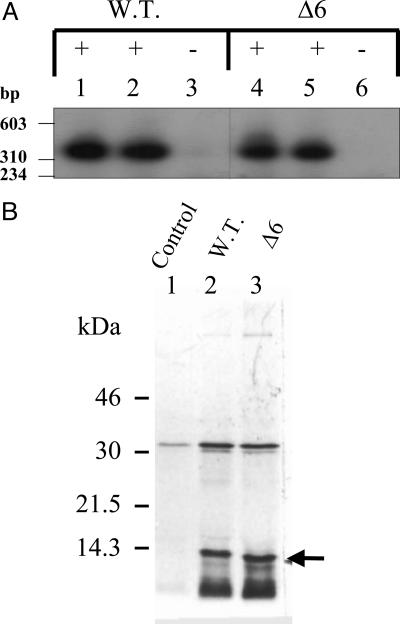
Fig. 2.
Analysis of expression of mutant LTB protein. Immunoblot analysis of expression of mutant LTB clone, pLTBΔ6. E. coli DH5α cells harboring pMMB68 [containing the wild-type (W.T.) LTB gene] and mutant LTB plasmid pLTBΔ6 were induced with IPTG. Different fractions of E. coli cell lysates were analyzed for expression with anti-LTB antibodies. Lanes 1 and 2 represent culture supernatant and periplasmic fraction of induced cells harboring mutant plasmid pLTBΔ6. Lanes 3 and 4 represent total cell extract from the induced cells harboring the pLTBΔ6. Lanes 5 and 6 represent culture supernatant and periplasmic fraction from cells harboring wild-type plasmid pMMB68 grown in the presence (+) of IPTG. Lanes 7 and 8 represent total cell extract of the same. + and − denote cells grown in the presence and absence of IPTG, respectively. Arrow points to the expressed protein. The migration of the molecular mass marker (kDa) is shown on the left.
WT and Mutant LTB mRNAs Are Efficiently Transcribed in E. coli.
The loss of expression could be caused either by faulty transcription or by faulty translation. It is likely that the mRNA of mutant LTB was not being transcribed at all or was rapidly degraded. To check whether the loss of expression was the result of an effect of the N-terminal deletion on ltb gene transcription, total RNA isolated from pMMB68 (WT) and deletion mutant (Δ6) clones expressed in E. coli DH5α was subjected to Northern blot analysis with radiolabeled ltb gene as a probe. As evident from Fig. 3A, the LTB mRNA transcript could be detected in the induced cultures of both the WT and the mutant clones (lanes 1 and 2, and 4 and 5, respectively). These results indicate that the transcription of ltb was not affected by the deletion. The mRNA appears to be highly stable as evident from the Northern blot as well as from its efficient translation by using an in vitro translation system described below.
Fig. 3.
Northern blot analysis and in vitro coupled transcription–translation analysis of mutant pLTBNΔ6 clone. (A) Northern blot analysis of total RNA isolated from E. coli DH5α cells harboring LTB plasmids, using a radiolabeled ltb gene probe. Lanes 1–3 represent RNA isolated from cells harboring wild-type (W.T.) LTB plasmid pMMB68. Lanes 4–6 represent RNA isolated from cells harboring mutant plasmid pLTBΔ6 (Δ6). + and − denote cells grown in the presence and absence of IPTG, respectively. Lanes 1 and 2, and 4 and 5 represent the respective samples in duplicate. Molecular mass markers (HaeIII-digested φX174 bacteriophage DNA) are shown on the left. (B) E. coli S30 extract for linear template was used for in vitro synthesis. Protein labeling was carried out in the presence of 1.85 × 106 Bq of [35S]methionine. Equimolar concentrations of wild-type and mutant Δ6 clones were used. The translated product was acetone-precipitated, separated on SDS/15% PAGE, and visualized by autoradiography. Lane 1, plasmid pMMB66EH (control); lane 2, pMMB68 (W.T.); and lane 3, pLTBΔ6 (Δ6 mutant). Arrow indicates translated LTB protein. The migration of the molecular mass marker (kDa) is shown on the left.
Mutant LTB Is Expressed in the E. coli in Vitro Translation System.
It is possible that the presence of the mutation at the 5′ end of ORF changed the secondary structure of the mRNA to the extent that it was no longer suitable for initiating translation. To address this question, an E. coli S30 extract system was used for in vitro coupled transcription–translation of linear WT pMMB68 and mutant (pLTBΔ6) templates (Fig. 3B). pMMB66EH vector alone (lane 1) was included as a negative control. In vitro translation analysis revealed that like the WT LTB (lane 2), the mutant Δ6 (lane 3) protein could also be synthesized by using this system. Because the E. coli cell extract does not have an intact periplasmic space, the expressed protein may not process for signal peptide removal from the mutant or WT LTB. That is why the in vitro translated protein was ≈13 kDa instead of 11.6 kDa, like in vivo expressed LTB. These findings indicate that the mutation did not block translation of the ltb mRNA. Based on these data, the lack of expression of the mutant protein in vivo was probably the result of a posttranscriptional and posttranslational defects and may be attributed to protein degradation.
Expression of Mutant LTB in the Presence of Mg2+ Ions Rescues Its Degradation.
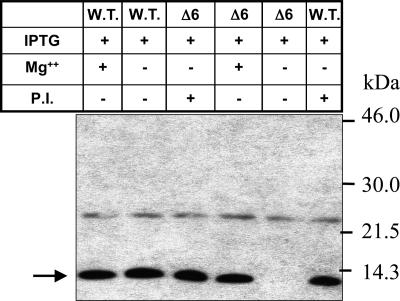
To address the possibility of degradation of the mutant protein soon after its synthesis in E. coli, the mutant plasmids were transformed into E. coli BL21(DE3)pLysS cells (lon and ompT protease-deficient) and analyzed for expression after induction with isopropyl β-d-thiogalactoside (IPTG) (Fig. 4). It is to be noted that the ltb is under the control of Tac promoter in pMMB68, and the T7 promoter/polymerase system is not used for the expression of ltb in E. coli BL21(DE3)pLysS. These cells were used solely for being lon and ompT protease-deficient. Like DH5α cells, no expression of the mutant LTB (Δ6) could be detected in these cells (lane 5). Because Mg2+ ions at higher concentrations (≥80 mM) are known to inhibit most of the proteases (33, 34), an attempt was made to express the mutant protein in the presence of 100 mM Mg2+ ions. The expression of mutant LTB could be detected in the induced E. coli BL21(DE3)pLysS cells in the presence of Mg2+ ions (lane 4) and in the medium supplemented with protease inhibitor mixture (lane 6). No significant change in the expression of WT LTB was observed in the presence of Mg2+ (lane 1) or protease inhibitor mixture (lane 3) in the medium (compare with lane 2). A similar effect of Mg2+ was observed on the expression of mutant LTB (Δ6) in E. coli DH5α cells (data not shown). These data suggest that failure to detect the mutant protein in the induced E. coli cells was indeed caused by degradation of the newly synthesized mutant protein by cellular proteases.
Fig. 4.
Expression of mutant lone pLTBΔ6 in E. coli BL21(DE3)pLysS cells. E. coli BL21(DE3)pLysS cells harboring pMMB68 [wild-type (W.T.), lanes 1 and 2] and mutant pLTBΔ6 (Δ6, lanes 3–5) were grown in LB medium with or without 100 mM MgCl2 or protease inhibitor mixture; they were induced with IPTG for 2 h. P.I. indicates the addition of complete protease inhibitor mixture (Roche, Mannheim, Germany) to the LB-ampicillin medium (2 tablets per 20 ml of medium). Total cell extract was immunoblotted with anti-LTB polyclonal antibody. + and − denote the presence and absence of the respective reagent to the medium. Uninduced or induced cultures are indicated by the absence (−) or presence (+) of IPTG, respectively. Arrow points to the expressed protein. The migration of the molecular mass marker (kDa) is shown on the right.
Analysis of Additional Deletion Mutations in the α1 Helix of LTB.
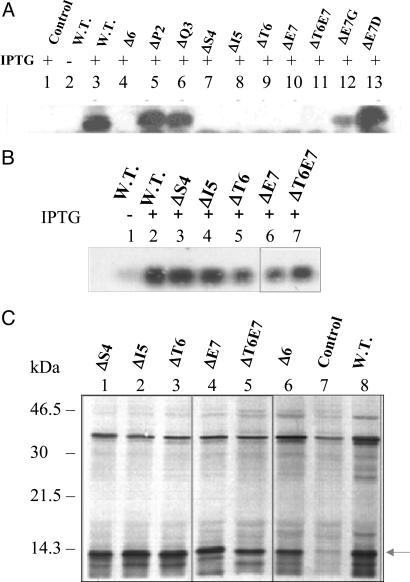
To identify the amino acids important for the structure–function relationship, amino acids from positions 2–7 were sequentially deleted from the N terminus of LTB to generate different mutants. Total cell extracts of E. coli DH5α cells transformed with the mutant LTB plasmids were analyzed for the expression of mutant proteins (Fig. 5A). Deletion mutant proteins ΔS4 (deletion of serine, lane 7), ΔI5 (deletion of isoleucine, lane 8), ΔT6 (deletion of threonine, lane 9), ΔE7 (deletion of glutamic acid, lane 10), and ΔT6E7 (double deletion of threonine and glutamic acid, lane 11) failed to express. However, deletion mutants ΔP2 (deletion of proline, lane 5), ΔQ3 (deletion of glutamine, lane 6), substitution mutant E7G (substitution of glycine for glutamic acid, lane 12) and E7D (substitution of aspartic acid for glutamic acid, lane 13) were expressed.
Fig. 5.
Analysis of expression of other LTB mutants. (A) Immunoblot analysis of expression. The E. coli DH5α cells harboring various mutant plasmids were induced at A600 = 0.4 for 5 h. The total cell extracts from different cultures were analyzed by Western blotting using anti-LTB polyclonal antibodies. Lane 1, control pMMB66EH; lanes 2 and 3, wild-type (W.T.) pMMB68; lane 4, pLTBΔ6; lane 5, pLTBΔP2; lane 6, pLTBΔQ3; lane 7, pLTBΔS4; lane 8, pLTBΔI5; lane 9, pLTBΔT6; lane 10, pLTBΔE7; lane 11, pLTBΔT6E7; lane 12, pLTBE7G; lane 13, pLTBT7D. + and − denote cells grown in the presence and absence of IPTG, respectively. (B) Northern blot analysis of LTB mutants. E. coli DH5α cells harboring the respective plasmids were induced for 5 h. Total RNA was isolated and subjected to Northern blot analysis using a radiolabeled ltb gene probe. Lanes 1 and 2, W.T. pMMB68; lane 3, pLTBΔS4; lane 4, pLTBΔI5; lane 5, pLTBΔT6; lane 6, pLTBΔE7; lane 7, pLTBΔT6E7. + and − denote cells grown in the presence and absence of IPTG, respectively. (C) In vitro translation of LTB mutants. E. coli S30 extract for linear template was used for in vitro synthesis. Protein labeling was carried out in the presence of 1.85 × 106 Bq of [35S] methionine. Equimolar concentrations of control and deletion plasmid DNA were used. The translated product was acetone-precipitated, separated on SDS/15% PAGE, and visualized by autoradiography. Lane 1, pLTBΔS4; lane 2, pLTBΔI5; lane 3, pLTBΔT6; lane 4, pLTBΔE7; lane 5, pLTBΔT6E7; lane 6, pLTBΔ6; lane 7, pMMB66EH (negative control); lane 8, pMMB68 (W.T.). The translated product is shown by an arrow.
As per earlier observations, it was suspected that the lack of expression in the various mutants was caused by degradation of the expressed protein product immediately after its synthesis. To rule out the possibility of faulty transcription and translation, Northern blot analysis was carried out with total RNA isolated from the induced E. coli DH5α cells harboring mutant LTB plasmids. The mutant proteins ΔS4 (lane 3), ΔI5 (lane 4), ΔT6 (lane 5), ΔE7 (lane 6), and ΔT6E7 (lane 7), whose expression was not detected by Western blotting, revealed the presence of abundant mRNA (Fig. 5B). The E. coli S30 extract system for linear template was used for in vitro coupled transcription–translation of all mutant LTB constructs in the presence of [35S]methionine. It was observed that the mRNAs for mutant proteins, ΔS4, ΔI5, ΔT6, ΔE7, ΔT6E7, and Δ6, whose expression was not detected in vivo, could be successfully translated by using the E. coli in vitro translation system (Fig. 5C, lanes 1–6, respectively). These results further confirm that the failure to express the mutant proteins was not caused either by faulty transcription or by faulty translation.
Coexpression of LTA and Mutant LTB to Generate Holotoxin in E. coli.
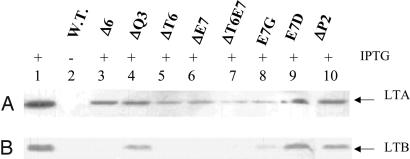
To check whether coexpression of LTA could alter the expression pattern of mutant LTB proteins, the LTA gene was excised out of the plasmid pLTA-LTB and cloned into plasmids harboring mutant LTB genes. Periplasmic fractions of induced E. coli DH5α cultures were analyzed by immunoblotting with anti-LTA antibody (Fig. 6A). Expression of LTA (lanes 1 and 3–10) was detected in all of the induced recombinant clones. The same blot, when stripped and reblotted with anti-LTB antibody, did not result in the detection of the mutant Δ6 (lane 3), ΔT6 (lane 5), ΔE7 (lane 6), and ΔT6E7 (lane 7) (Fig. 6B). However, expression of WT (lane 1), ΔQ3 (lane 4), E7G (lane 8), E7D (lane 9), and ΔP2 (lane 10) could be detected. Thus, coexpression of the LTA did not alter the expression pattern of the various mutant LTB proteins (compare Figs. 5A and 6).
Fig. 6.
Immunoblot analysis of periplasmic expression of LTA-mutant LTB holotoxin in E. coli. E. coli cells harboring plasmids containing both the LTA and LTB chains were induced at A600 = 0.4 for 5 h. Periplasmic fractions were analyzed for the presence of LTA or LTB subunit of holotoxin by using anti-LTA antibody and anti-LTB antibody, respectively. (A) Western blot analysis with anti-LTA antibody. (B) The same blot was then striped, washed, and immunoblotted with anti-LTB antibody. Lanes 1 and 2, pLTA-LTB; lane 3, pLTA-LTBΔ6; lane 4, pLTA-LTBΔQ3; lane 5, pLTA-LTBΔT6; lane 6, pLTA-LTBΔE7; lane 7, pLTA-LTBΔT6E7; lane 8, pLTA-LTBE7G; lane 9, pLTA-LTBE7D; lane 10, pLTA-LTBΔP2. + and − denote cells grown in the presence and absence of IPTG, respectively. Arrows indicate LTA (A) and LTB (B).
Discussion
Our data indicate that the deletion of 6 aa from the N-terminal α1 helix of LTB resulted in a complete loss of expression. Immunoblotting and Northern blot analysis, together with in vitro coupled transcription–translation data, suggest that the loss of expression was not because of faulty transcription and translation but because of degradation of the expressed protein. Mg2+ ions at a higher concentration (≥80 mM) are known to inhibit most of the proteases (33, 34), and therefore, the presence of Mg2+ could prevent the degradation of mutant LTB in E. coli cells. Thus, restoration of expression of mutant LTB in E. coli cells in the presence of Mg2+ ions further confirms that the mutant LTB was getting degraded soon after its synthesis (in the absence of Mg2+ ions).
It is evident from this work that the N-terminal α1 helix plays an important role in LTB expression in its natural environment. It is well established that LTB precursors are translocated to the periplasm in an unfolded state and fold into native structure once the export is completed (19, 20, 35, 36). Because the export is directed by the signal sequence, the deletion of α1 helix of LTB is unlikely to affect its translocation to the periplasm. It is, therefore, expected that the mutant protein lacking the α1 helix is unable to assemble into appropriately folded structure in the periplasm and is subsequently degraded. Crystal structure data suggest that the N-terminal α1 helix interacts with the β5 strand through a Cys9–Cys86 disulfide bond (27). The exposed hydrophobic surface of the β barrel (β2, β3, β4 and β1, β5, β6) is masked by the solvent-exposed N-terminal α1 helix. It is likely that in the absence of the α1 helix, this property remains unsatisfied, making mutant LTB unable to fold into its native structure and thus rendering LTB susceptible to degradation by cellular proteases.
LTB is synthesized by membrane-associated polysomes, where the nascent chain is inserted into the membrane before the termination of polypeptide synthesis (37). Thus, the N terminus of the protein is the first to come in contact with the periplasmic milieu and has a better chance of folding after the removal of the signal peptide. No proteins are known to associate with LTB during the process of folding. It is believed that LTB is not assisted by folding factors or chaperones in in vivo folding (38). Hence, the information carried by the primary amino acid sequence should accomplish proper folding of the protein. However, DsbA protein does help in disulfide bond formation (20, 39). Refolding studies on cytochrome C (32) and RNase A (40) indicate that the N-terminal or C-terminal helices are the first to fold and serve as a docking surface (template) to guide subsequent folding reactions. Likewise, the N-terminal α1 helix may be providing structural stability for the nucleation of protein folding, and in its absence, the mutant Δ6 LTB is targeted for degradation. On the other hand, one can argue that it is the intrachain disulfide bond between Cys9 and Cys86 that is important for maintaining the integrity of the structure. The major deletion of the α1 helix may make the disulfide bond formation difficult because Cys9 is part of the α1 helix, which would make it impossible for the protein to fold into its native structure, and thus the misfolded mutant Δ6 LTB gets degraded. However, if the disruption of the α1 helix only affected disulfide bond formation and not subsequent protein folding, the mutant protein would have been present in the total cell extract. Earlier studies carried out in the presence of reducing agents also suggest that disulfide bond formation is not necessary for maintaining the integrity of the α1 helix (20, 41). The failure to observe the presence of the mutant protein in the total cell extract indicates that the α1 helix deletion possibly heightens the protease sensitivity of mutant LTB. Degradation of proteins resulting from misfolding is widely acknowledged (42–44). Our results are in line with the earlier reports on bovine rhodanese, wherein the N-terminal α helix of the enzyme was reported to contribute to the global stability of the protein when expressed in E. coli (29).
Studies with LTB mutants with a single amino acid deletion from positions 2–7 from the N-terminal region of LTB indicate that the amino acids at positions 4–6 are crucial for protein stability. Deletion of Pro2 or Gln3 that lie outside the N-terminal α1 helix (at the N″ and N′ positions, respectively) did not affect expression. However, deletion of amino acids Ser4, Ile5, Thr6, or Glu7, which are part of the α1 helix, completely abolished expression. Because the amino acids Ser4–Ser10 are involved in hydrogen bonding, any single deletion would be expected to cause disruption of hydrogen bonding and would thus destabilize the helix. As expected, substitution of Glu7 with Asp within the α1 helix did not abolish the expression because the same amino acid is present at the 7th position in cholera toxin B (13). Although glycine is known to destabilize the helix (45), no effect on the expression was observed when Glu7 was substituted with Gly. The data clearly indicate that any single amino acid deletion within the α1 helix is not tolerated. It is logical to consider that the deletion of a single amino acid from a given position within the helix should be compensated by an adjacent amino acid, restoring the necessary main-chain hydrogen bonds. However, this may not be the case with LTB. The adjacent amino acid may not be favorable at that position because certain amino acids prefer to stay at specific positions, such as at the N terminus, at the C terminus, or in the middle of the helix (45).
LT is transcribed as a single polycistronic mRNA (14). In E. coli, both the subunits are synthesized and exported to the periplasm, where the LTB binds to the LTA to form the holotoxin (18, 19). Streatfield et al. (46) have suggested that an “intramolecular folding factor” mediates coordinate assembly of A and B subunits of the toxin. Although the presence of LTA is not obligatory for LTB pentamerization, because E. coli strains devoid of LTA are capable of assembling LTB into a pentamer (47, 48), LTA accelerates LTB subunit pentamerization in vivo (20). The C terminus of the LTA interacts with the LTB subunit and stabilizes the assembly intermediate. Therefore, coexpression of LTA and LTB was performed to check whether LTA could prevent the degradation of a folding-defective mutant LTB. LTA is known to assist folding and assembly of LTB (20, 44). However, this too, could not prevent the degradation of mutant LTB proteins in E. coli.
Thus, the present work has clearly demonstrated that the integrity of the N-terminal α1 helix of LTB is essential for its stability and may play a role in the initial folding of the protein, thus protecting it from degradation by cellular proteases. It is likely that the folding of LTB starts at the N terminus because of the formation of the α1 helix, which provides structural stability for the folding of the protein into a ternary structure. Further studies are needed to assess directly the role of the N-terminal α1 helix in the folding of LTB into a stable conformation.
Methods
Bacterial Strains and Plasmids.
E. coli DH5α cells [F− SupE44, ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1] and E. coli BL21(DE3)pLysS cells [F− ompT hsdSB (rB− mB−)gal dcm (DE3) pLysS(CmR), lon and omptT protease-deficient] were purchased from Novagen (San Diego, CA). Plasmid pMMB68 was kindly provided by J. Holmgren (University of Goteborg, Sweden). Plasmid pDF82 harboring the LTA gene, lta of heat-labile enterotoxin, was a gift from H. S. Mason (Boyce Thompson Institute, New York, NY).
Bacterial Medium and Chemicals.
All of the chemicals were of molecular biology grade. Bacto tryptone, yeast extract, and Bacto agar were procured from Difco Laboratories (Sparks, MD). Ampicillin was purchased from Sigma (St. Louis, MO). All restriction enzymes, DNA-modifying enzymes, and DNA markers were purchased from New England Biolabs (Ipswich, MA) and used as recommended by the suppliers. [α-32P]dCTP (specific activity, 1.11 × 104 Bq/mmol at 3.7 × 108 Bq/ml) and [35S]methionine (specific activity, 4.3475 × 1013Bq/mmol at 4.07 × 108 Bq/ml) were purchased from PerkinElmer (Waltham, MA). The ECL-Western blotting kit and multiprime DNA labeling kit were procured from Amersham (Piscataway, NJ). The TRIzol-RNA isolation kit and E. coli S30 cell extract were obtained from Invitrogen (Carlsbad, CA) and Promega (Madison, WI), respectively.
PCR Amplification.
Mutations at the N terminus of LTB were introduced by PCR using the following oligonucleotides as forward primers: Δ6, 5′-CCAGAGCTCTATGTTCGGAATATCAC-3′; ΔP2, 5′-CCAGAGCTCAGTCTATTACAGAACTATGTTC-3′; ΔQ3, 5′-CCAGAGCTCCTTCTATTACAGAACTATGTTCG-3′; ΔS4, 5′-CCAGAGCTCCTCAGATTACAGAACTAT G T TCGGAATATCAC-3′; ΔI5, 5′-CCAGAGCTCCTCAGTCTACAGAACTATGTTCGGAATATCAC-3′; ΔT6, 5′-CCAGAGCTCCTCAGTCTATTGAACTATGTTCGGAATATCAC 3′; ΔE7, 5′-CCAGAGCTCCTCAGTCTATTACACTAT G T T C G GAATATCACAAC-3′; ΔT6E7, 5′-CCAGAGCTCCTCAGTCTATTCTATGTTCGGAATATCACAACACAC-3′; E7G, 5′-CCAGAGCTCCTCAGTCTATTACAGGCCTATGTTCGGAATATCACAC-3′; E7D, 5′-CCAGAGCTCCTCAGTCTATTACAGATCTATGTTCGGAATATCACAAC-3′.
The oligonucleotide 5′-TATAAAGCTTCCTAGCATTAGAC 3′ was used as a reverse primer. The oligonucleotides were synthesized by Rama Biotechnologies (Hyderabad, India) and BioSynthesis, Inc. (Lewisville, TX). The restriction sites SacI and HindIII (underlined sequences) were introduced in the primers for cloning purposes. Specific mutations were introduced into the LTB gene by PCR amplification using the above-mentioned primers, with Taq polymerase enzyme (Promega) in 1× reaction buffer and 1.5 mM MgCl2. PCR involved denaturation at 92°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 1 min for 30 cycles with a thermal cycler 480 (PerkinElmer). Sequencing of the mutant ltb gene was performed on both of the strands to confirm the induced mutations. Sequencing primers 5′-GTGTGGAATTGTGAGCGG-3′ and 5′-TGATAACCATTTCTCTTT-3′ were used to sequence the cloned gene from the 5′ and 3′ ends, respectively.
Cloning Strategies.
Plasmid DNA isolation, ligation, and bacterial transformation were carried out essentially as described by Sambrook et al. (49).
Cloning of mutant ltb gene into pMMB vector.
pMMB68 is a derivative of pMMB66EH vector (GenBank accession no. X15234) in which ltb gene has been cloned at EcoRI–HindIII sites. The PCR-amplified product and the pMMB68 vector DNA were digested with SacI and HindIII restriction enzymes. Digestion of the plasmid pMMB68 with the above enzymes resulted in the removal of the ltb gene, leaving its signal sequence intact. SacI–HindIII-digested, PCR-amplified mutant ltb gene was then ligated with the digested plasmid containing the LTB signal sequence and transformed into E. coli-competent cells.
Cloning of lta gene in translation frame with mutant ltb gene.
lta gene was amplified with pDF82 as a template. Oligonucleotides (containing an EcoRI site, underlined) 5′-CAGAATTCCGATGAAAAATATAACTT-3′ and 5′-CCGAATTCTGTT A T ATATG-3′ were used as forward and reverse primers, respectively. The EcoRI-digested, PCR-amplified product was cloned at EcoRI site in plasmid pMMB68 (harboring WT ltb gene). This cloning resulted in the LTA and LTB subunits to be in the same translational frame. The resultant plasmid was named pLTA-LTB.
SacI–MluI-digested product from pLTA-LTB vector containing Tac promoter, LTA with its own signal sequence, and the LTB signal sequence, was ligated with the SacI–MluI-digested vector of different LTB mutants generated earlier (pLTBΔ6, pLTBΔP2, pLTBΔQ3, pLTBΔT6, pLTBΔE7, pLTBΔT6E7, pLTBE7G, and pLTBE7D). The recombinant plasmids were transformed into E. coli-competent cells. The recombinant clones thus obtained were named pLTA-LTBΔ6, pLTA-LTBΔP2, pLTA-LTBΔQ3, pLTA-LTBΔT6, pLTA-LTBΔE7, pLTA-LTBΔT6E7, pLTA-LTBE7G, and pLTA-LTBE7D.
Expression of Mutant ltb Gene and Preparation of Cellular Fractions.
Expression of the recombinant LTB and preparation of different cellular fractions were carried out as described earlier (50). E. coli cells harboring mutant or WT plasmids were grown in 50 ml of LB medium in the presence of ampicillin (50 μg/ml) at 37°C in a gyratory shaker until A600 reached 0.4. The cells were then induced with 1 mM IPTG and grown for 5 h at 37°C. The culture was chilled on ice, and the cells were pelleted at 4,000 × g for 10 min at 4°C (Sorvall centrifuge, SS34 rotor; Thermo, Waltham, MA). The cells were directly suspended in reducing sample buffer and boiled for 5 min before loading onto SDS/PAGE. Periplasmic fraction was prepared by adding 5 μl of lysozyme (2 mg/ml) and 100 μl of ice-cold buffer (100 mM sodium phosphate buffer, pH 7.6/12.5 mM EDTA/0.3 M sucrose) to the cells. The fraction was incubated on ice for 20 min with occasional gentle shaking and then centrifuged at 8,000 × g for 10 min in a microcentrifuge at 4°C (220.59v rotor; Hermle Labortechnïk, Wehingen, Germany). The collected supernatant represented the periplasmic fraction.
SDS/PAGE and Western Blotting.
The samples were analyzed on 12% or SDS/15% polyacrylamide gel as described by Laemmli (51). The proteins were electrotransferred onto nitrocellulose membrane at 35 mA overnight. Nonspecific sites were blocked by incubating the membrane in 1% nonfat milk powder in 50 mM PBS (pH 7.4) for 1 h. The blot was then incubated with goat anti-LTB polyclonal antibodies (Reagent Bank, National Institute of Immunology, New Delhi, India) followed by washing with PBS buffer containing 0.05% Tween 20. The blot was then incubated with anti-goat IgG horseradish peroxidase conjugate (Vector Laboratories, Burlingame, CA) for 1 h followed by three washes with 0.05% Tween 20/PBS buffer. The immunoreactive bands were visualized by using 4-chloro-1-naphthol (5 mg/ml) and hydrogen peroxide (1 μl/ml) or by the ECL-Western blotting kit. Stripping buffer (100 mM β-mercaptoethanol/2% SDS/62.5 mM Tris·Cl, pH 6.7) was used when immunoblotting with a different antibody was to be carried out. The blot was washed three times with 80 mM PBS (pH 7.3) after stripping.
Northern Blot Analysis.
Uninduced and induced cultures of E. coli cells harboring mutant or WT plasmid were grown for 5 h, and total RNA was prepared by using an RNA midi kit (Qiagen, Valencia, CA) or TRIzol reagent. The RNA was electrophoresed in a 1.2% formaldehyde gel at 50 mA for 2 h and transferred to nylon membrane (GeneScreen; PerkinElmer) by the capillary transfer method as described by Sambrook et al. (49). The ltb probe was prepared by radiolabeling the ltb fragment with [α-32P]dCTP, using a random primer labeling kit (Amersham). The blot was hybridized with radiolabeled probe for 2 h at 68°C followed by washes with 2× SSC/0.5% SDS at room temperature for 5 min, 2× SSC/0.1% SDS at room temperature for 15 min, 0.1× SSC/0.5% SDS at 67°C for 1 h, and 0.1× SSC at room temperature for 5 min. The blot was then subjected to autoradiography.
In Vitro Translation.
E. coli S30 extract for the linear template from Promega was used for in vitro translation. S30 extract was mixed with template DNA (≈4 μg), and protein labeling was carried out by using 1.5 μl of [35S]methionine (4.3475 × 1013 Bq/mmol at 4.07 × 108 Bq/ml) in a 50-μl reaction mix at 37°C for 2 h. Five microliters of the reaction mix was then precipitated with 20 μl of acetone, centrifuged at 10,000 × g for 10 min, air-dried, electrophoresed on SDS/polyacrylamide gel, and visualized by autoradiography.
Acknowledgments
We thank Dr. T. E. Dever, National Institutes of Health, Bethesda, MD, for critically reading the manuscript and for valuable suggestions. We acknowledge the technical assistance provided by Mr. Ram Bodh and Mr. Ved Prakash. This work was supported by a Council for Scientific and Industrial Research, India, research fellowship (to P.V.A.) and by the Department of Biotechnology, India.
Abbreviations
- IPTG
isopropyl β-d-thiogalactoside
- LT
heat-labile enterotoxin
- LTA
heat-labile enterotoxin A subunit
- LTB
heat-labile enterotoxin B subunit.
Footnotes
The authors declare no conflict of interest.
References
- 1.Kosek M, Bern C, Guerrant RL. Bull World Health Org. 2003;81:197–204. [PMC free article] [PubMed] [Google Scholar]
- 2.Qadri F, Svennerholm A-M, Faruque ASG, Sack RB. Clin Microbiol Rev. 2005;18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ericsson CD. Int J Antimicro Agents. 2003;21:116–124. doi: 10.1016/s0924-8579(02)00282-0. [DOI] [PubMed] [Google Scholar]
- 4.Qadri F, Khan AI, Faruque AS, Begum YA, Chowdhury F, Nair GB, Salam MA, Sack DA, Svennerholm AM. Emerg Infect Dis. 2005;11:1104–1107. doi: 10.3201/eid1107.041266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boedeker EC. Curr Opin Gastroenterol. 2005;21:15–19. [PubMed] [Google Scholar]
- 6.Nair GB, Takeda Y. Microbial Pathogen. 1998;24:123–131. doi: 10.1006/mpat.1997.0177. [DOI] [PubMed] [Google Scholar]
- 7.Holmes RK, Jobling MG, Connell TD. In: Handbook of Natural Toxins: Bacterial Toxins and Virulence Factors in Disease. Moss J, Iglewski B, Vaughan M, Atu A, editors. Vol 8. New York: Dekker; 1995. pp. 225–256. [Google Scholar]
- 8.Karasawa T, Ito H, Tsukamoto T, Yamasaki S, Kurazono H, Faruque SM, Nair GB, Nishibuchi M, Takeda Y. Infect Immun. 2002;70:7153–7155. doi: 10.1128/IAI.70.12.7153-7155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spangler BD. Microbiol Rev. 1992;56:622–647. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmgren J. Nature. 1981;292:413–417. doi: 10.1038/292413a0. [DOI] [PubMed] [Google Scholar]
- 11.Hirst TR. In: A Sourcebook of Bacterial Protein Toxins. Alouf JE, Freer JH, editors. London: Academic; 1991. pp. 75–100. [Google Scholar]
- 12.Gill DM, Clements JD, Robertson DC, Finkelstein RA. Infect Immun. 1981;33:677–682. doi: 10.1128/iai.33.3.677-682.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domenighini M, Pizza M, Jobling MG, Holmes RK, Rappuoli R. Mol Microbiol. 1995;15:1165–1167. doi: 10.1111/j.1365-2958.1995.tb02289.x. [DOI] [PubMed] [Google Scholar]
- 14.Dallas WS, Gill DM, Falkow S. J Bacteriol. 1979;139:850–858. doi: 10.1128/jb.139.3.850-858.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mekalanos JJ, Swartz DJ, Pearson GD, Harford N, Croyne F, deWilde M. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 16.Spicer EK, Noble JA. J Biol Chem. 1982;257:5716–5721. [PubMed] [Google Scholar]
- 17.Dallas WS, Falkow S. Nature. 1980;288:499–501. doi: 10.1038/288499a0. [DOI] [PubMed] [Google Scholar]
- 18.Hirst TR, Hardy SJ, Randall LL. J Bacteriol. 1983;153:21–26. doi: 10.1128/jb.153.1.21-26.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofstra H, Witholt B. J Biol Chem. 1985;260:16037–16044. [PubMed] [Google Scholar]
- 20.Hardy SJ, Holmgren J, Johansson S, Sanchez J, Hirst TR. Proc Natl Acad Sci USA. 1988;85:7109–7113. doi: 10.1073/pnas.85.19.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fishman PH. J Membr Biol. 1982;69:85–97. doi: 10.1007/BF01872268. [DOI] [PubMed] [Google Scholar]
- 22.Finkelstein RA, Burks MF, Zupan A, Dallas WS, Jacob CO, Ludwig DS. Rev Infect Dis. 1987;9:S490–S502. doi: 10.1093/clinids/9.supplement_5.s490. [DOI] [PubMed] [Google Scholar]
- 23.De Wolf MJ, Fridkin M, Epstein M, Kohn LD. J Biol Chem. 1981;256:5481–5488. [PubMed] [Google Scholar]
- 24.De Wolf MJ, Fridkin M, Kohn LD. J Biol Chem. 1981;256:5489–5496. [PubMed] [Google Scholar]
- 25.De Wolf MJ, Van Dessel G, Lagrou A, Hilderson HJ, Dierick W. Biochim Biophys Acta. 1985;832:165–174. doi: 10.1016/0167-4838(85)90328-0. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig DS, Holms RK, Schoolnik GK. J Biol Chem. 1985;260:12528–12534. [PubMed] [Google Scholar]
- 27.Sixma TK, Kalk KH, van Zanten BA, Dauter Z, Kingma J, Witholt B, Hol WG. J Mol Biol. 1993;230:890–918. doi: 10.1006/jmbi.1993.1209. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto A, Usui M, Yamasaki N, Yamada N, Kuwano E, Tanaka I, Kimura M. Eur J Biochem. 1999;266:591–598. doi: 10.1046/j.1432-1327.1999.00901.x. [DOI] [PubMed] [Google Scholar]
- 29.Trevino RJ, Tsalkova T, Kramer G, Hardesty B, Chirgwin JM, Horowitz PM. J Biol Chem. 1998;273:27841–27847. doi: 10.1074/jbc.273.43.27841. [DOI] [PubMed] [Google Scholar]
- 30.Merlino A, Graziano G, Mazzarella L. Proteins. 2004;57:692–701. doi: 10.1002/prot.20270. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Weiner H. Protein Sci. 2001;10:1490–1497. doi: 10.1110/ps.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rumbley J, Hoang L, Mayne L, Englander SW. Proc Natl Acad Sci USA. 2001;98:105–112. doi: 10.1073/pnas.98.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oleksyszyn J, Powers JC. Methods Enzymol. 1994;244:423–441. doi: 10.1016/0076-6879(94)44032-8. [DOI] [PubMed] [Google Scholar]
- 34.Maurizi MR, Thompson MW, Singh SK, Kim SH. Methods Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 35.Randall LL, Hardy SJS. Cell. 1986;46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- 36.Eilers M, Schatz G. Cell. 1988;42:481–483. doi: 10.1016/0092-8674(88)90458-8. [DOI] [PubMed] [Google Scholar]
- 37.Hofstra H, Witholt B. J Biol Chem. 1984;259:15182–15187. [PubMed] [Google Scholar]
- 38.Amin T, Larkins A, James RF, Hirst TR. J Biol Chem. 1995;270:20143–20150. doi: 10.1074/jbc.270.34.20143. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Webb H, Hirst TR. Mol Microbiol. 1992;6:1949–1958. doi: 10.1111/j.1365-2958.1992.tb01368.x. [DOI] [PubMed] [Google Scholar]
- 40.Udgaonkar JB, Baldwin RL. Nature. 1988;335:694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- 41.Hedges PA, Hardy SJ. FEBS Lett. 1996;381:63–66. doi: 10.1016/0014-5793(96)00082-8. [DOI] [PubMed] [Google Scholar]
- 42.Goldberg AL, St John AC. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- 43.Goldberg AL, Dice JF. Annu Rev Biochem. 1974;43:835–869. doi: 10.1146/annurev.bi.43.070174.004155. [DOI] [PubMed] [Google Scholar]
- 44.Bross P, Corydon TJ, Andresen BS, Jorgensen MM, Bolund L, Gregersen N. Hum Mutat. 1999;14:186–198. doi: 10.1002/(SICI)1098-1004(1999)14:3<186::AID-HUMU2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 45.Richardson JS, Richardson DC. In: Prediction of Protein Structure and the Principles of Protein Conformation. Fasman GD, editor. New York: Plenum; 1989. pp. 1–91. [Google Scholar]
- 46.Streatfield SJ, Sandkvist M, Sixma TK, Bagdasarian M, Hol WG, Hirst TR. Proc Natl Acad Sci USA. 1992;89:12140–12144. doi: 10.1073/pnas.89.24.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirst TR, Sanchez J, Kaper JB, Hardy SJ, Holmgren J. Proc Natl Acad Sci USA. 1984;81:7752–7756. doi: 10.1073/pnas.81.24.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neill RJ, Ivins BE, Holmes RK. Science. 1983;221:289–291. doi: 10.1126/science.6857285. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. pp. 1.21–1.82. [Google Scholar]
- 50.Mukhija R, Rupa P, Pillai D, Garg LC. Gene. 1995;165:303–306. doi: 10.1016/0378-1119(95)00525-b. [DOI] [PubMed] [Google Scholar]
- 51.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]