Abstract
The transcription initiation and elongation steps of protein-coding genes usually rely on unrelated protein complexes. However, the TFIIS elongation factor is implicated in both processes. We found that, in the absence of the Med31 Mediator subunit, yeast cells required the TFIIS polymerase II (Pol II)-binding domain but not its RNA cleavage stimulatory activity that is associated with its elongation function. We also found that the TFIIS Pol II-interacting domain was needed for the full recruitment of Pol II to several promoters in the absence of Med31. This work demonstrated that, in addition to its thoroughly characterized role in transcription elongation, TFIIS is implicated through its Pol II-binding domain in the formation or stabilization of the transcription initiation complex in vivo.
Keywords: RNA polymerase II, Saccharomyces cerevisiae, transcription regulation, Med31, Mediator subunit
The transcription of protein-coding eukaryotic genes by RNA polymerase II (Pol II) requires three successive steps: initiation, elongation, and termination. Transcription initiation of class II genes involves the binding of activators to regulatory sequences; the recruitment of RNA Pol II to the core promoter via interactions with activators, coactivators, and general transcription factors; and the initiation reaction per se with DNA strand opening and abortive initiation (1). A major coactivator target of transcriptional activators is the Mediator (2, 3). One of its activities is the recruitment and/or stabilization of Pol II at core promoters (4). After transcription initiation, Pol II enters elongation, during which it can be arrested because of the presence of specific DNA sequences that promote pausing, or because of obstacles such as DNA damage or bound proteins. To avoid or escape arrest, Pol II requires different elongation factors, including TFIIS (5). Evidence suggests that TFIIS could be implicated in both initiation and elongation.
In vitro, TFIIS can reactivate arrested elongation complexes by stimulating endonucleolytic cleavage by Pol II of the nascent RNA (5). TFIIS is composed of three domains that fold independently, as demonstrated by NMR analysis of its structure (6, 7). Cleavage-stimulating activity minimally requires the C-terminal two-thirds of the protein, that is, its domain II and III separated by a 15-aa linker (7, 8). Domain II forms a three-helix bundle followed by three short helices, with a basic patch on the third helix (α-3) that is essential for TFIIS binding to Pol II (7). Crystallographic analysis of a TFIIS–Pol II complex confirmed that this basic patch lies at the TFIIS–Pol II interface (7, 8). TFIIS binding to Pol II is required for domain III to reach the Pol II active site. Domain III forms a zinc ribbon that contains a conserved RSADE motif, responsible for the stimulation of RNA cleavage (7, 8).
TFIIS is also implicated in transcription initiation. First, the deletion of the DST1 gene, encoding TFIIS in yeast, is colethal with the deletion of the gene encoding the Med31 subunit of the Mediator complex (9). Second, TFIIS is recruited to the promoter of GAL1, and the deletion of DST1 reduces the recruitment of the transcription machinery on the promoter of GAL1 (10). However, the reported experiments did not investigate the generality of the TFIIS requirement in transcription initiation or the TFIIS elements required for this function.
Here, we analyze more precisely which activity of TFIIS was responsible for the colethality of dst1-Δ with med31-Δ. Remarkably, mutants of TFIIS impaired in its cleavage stimulatory activity could grow in a MED31 deleted strain. In line with these observations, domain II and the linker together, i.e., the Rpb1-interacting domain of TFIIS, were sufficient to rescue dst1-Δ med31-Δ colethality. We thus hypothesized that TFIIS could play a role in transcription initiation independent of its cleavage activity. We demonstrated that TFIIS is recruited at ADH1, VTC3, and MET17 promoters, and that the Pol II-binding activity of TFIIS was required for efficient recruitment of Pol II to these promoters in vivo in the absence of MED31. These data fit nicely with those obtained by Ranish and coworkers (11), indicating that, in vitro, TFIIS contributes to the formation of the Pol II preinitiation complexes independently of its role in elongation. Thus, we propose that TFIIS is required for optimal occupancy of Pol II at promoters of a subset of Saccharomyces cerevisiae genes.
Results
TFIIS Domain II and Linker Are Sufficient to Complement dst1-Δ med31-Δ Colethality.
Because TFIIS is composed of three domains that fold independently (6–8), we designed truncation mutants of TFIIS, using its structure as a guide (Fig. 1 A and B Left) to find which parts of the protein are required for complementation of the dst1-Δ med31-Δ colethality. These truncation mutants were transformed in a strain deleted for both MED31 and DST1, complemented by the MED31 gene on a URA3 centromeric plasmid [supporting information (SI) Table 1]. The various mutant strains were then tested for sensitivity to mycophenolic acid (MPA) or for growth on 5-fluorooroatic acid (5FOA). MPA is an inhibitor of guanine nucleotide biosynthesis. Transcriptional elongation defects due to the loss of TFIIS cleavage activity prevent growth on MPA (12), whereas the absence of growth on 5FOA revealed colethality of the dst1 truncation mutation with med31-Δ. The second line of Fig. 1B confirms that deletion of the entire DST1 gene is lethal in the med31-Δ context. We found that TFIIS capacity to stimulate the hydrolytic activity of Pol II was dispensable in the med31-Δ context, because complete truncation of amino acids 266–309 of TFIIS domain III did not lead to lethality (Fig. 1B, line 3) but renders the strain sensitive to MPA. Conversely, domain III alone was not sufficient to render the strain viable (Fig. 1B, line 4). This observation confirms that TFIIS cleavage activity is dispensable to complement dst1-Δ med31-Δ colethality.
Fig. 1.
The cleavage activity and N terminus of TFIIS are not necessary to complement the colethality of dst1-Δ with med31-Δ. (A) Molecular structure of the TFIIS–RNA Pol II complex (8). TFIIS is indicated in green and Pol II in gray. The position of the R200A mutated residue used in this study appears as a red ball. Orange balls outline residues essential for TFIIS cleavage activity. Black bars indicate TFIIS domains. Domain I is not represented because its structure was not solved in the complex with Pol II. (B) Test of DST1 truncation mutant growth on MPA and 5FOA medium at 30°C. (Left) Schematic representation of TFIIS deletion mutants used relative to the wild-type protein. The amino acid positions bordering the fragments are indicated on the left.
We thus investigated the requirement for TFIIS domains I and II in this assay. The deletion of TFIIS domain I (amino acids 1–132) did not lead to MPA sensitivity or to colethality (Fig. 1B, line 5). Accordingly, domain I alone was unable to complement either phenotype (Fig. 1B, line 6). However, domain II and the following linker were sufficient to restore growth in the double-deleted strain (Fig. 1B, line 7) but were unable to restore resistance to MPA. This minimal domain, named TFIIS− (133–265), could not be shortened further (Fig. 1B, lines 8 and 9). In conclusion, the minimal sequence needed to complement dst1-Δ med31-Δ colethality consisted of domain II and the linker to domain III.
The TFIIS-R200A Domain II Mutant Could Not Complement dst1-Δ med31-Δ Colethality and Was Not Impaired in Elongation.
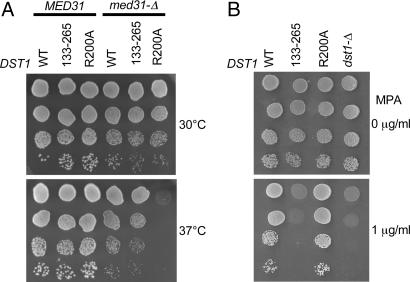
We found the TFIIS domain II plus linker to be required for complementation of the dst1-Δ med31-Δ colethality. Because the only previously known function of TFIIS domain II is Pol II binding in vitro through an interaction with Rpb1, we investigated the role of this function for dst1-Δ med31-Δ complementation. Structural and biochemical data suggest that a basic patch of TFIIS domain II is essential for Pol II binding (7, 8). We mutated the arginine 200 residue of the TFIIS basic patch to an alanine (dst1-R200A) and investigated its effect on complementation of the dst1-Δ med31-Δ colethal phenotype. As shown Fig. 2A Upper, the dst1-R200A med31-Δ double mutant strain grew as well as the DST1 med31-Δ strain at 30°C. However, when grown at 37°C, the dst1-R200A mutant was colethal with med31-Δ, whereas wild-type DST1 and dst1-(133–265) remained fully viable. This observation suggested that the Pol II-binding function of TFIIS is essential in the med31-Δ context.
Fig. 2.
The TFIIS-R200A mutation hampers complementation of dst1 colethality with med31, but not sensitivity to MPA. (A) Growth of dst1-(133–265) and dst1-R200A mutated strains relative to wild-type DST1 in MED31 or med31-Δ context. Suspensions of the strains were serially diluted, spotted on glucose-rich medium (YPD), and grown at 30°C or 37°C for 2 days. (B) Sensitivity to MPA of dst1-(133–265) and dst1-R200A strains compared with wild-type strain. Suspensions of the strains were serially diluted, spotted on synthetic complete medium, and grown at 37°C in the presence or absence of mycophenolate for 6 days.
Reciprocally, we tested whether dst1-R200A mutation decreased TFIIS transcription elongation activity. At 37°C, wild-type yeast strains are killed by lower MPA concentrations than those required to hamper growth at 30°C. We plated the dst1-R200A mutant strain on complete synthetic medium with 1 μg/ml MPA at 37°C (Fig. 2B). Contrary to the dst1-Δ strain, dst1-R200A grew as well as the DST1 wild-type strain, indicating that dst1-R200A was not defective in TFIIS transcription elongation activity.
Requirement of R200 Residue of TFIIS and Med31 for Pol II Recruitment at a Subset of Yeast Promoters.
To find genes affected in their transcription in the dst1-R200A context, we turned to global transcriptome analysis. DST1 med31-Δ and dst1-R200A med31-Δ cells were cultured at 30°C in yeast extract/peptone/dextrose (YPD) glucose-rich medium in exponential growth phase, then half of the culture was shifted to 37°C for 30 min. mRNAs were analyzed by using DNA microarrays as described (13). Ninety-two genes were expressed at least 2-fold less in the dst1-R200A med31-Δ strain compared with the DST1 med31-Δ strain when grown at 30°C, and 82 genes were less expressed in the dst1-R200A med31-Δ strain compared with the DST1 med31-Δ strain at 37°C (SI Tables 2 and 3). Shifting cells to 37°C increased only slightly the gene expression defect in the dst1-R200A med31-Δ strain. At 37°C, the transcription of 18 genes increased ≥2-fold in the mutant compared with the wild type. The maximum induction was only 3.3-fold for the BIK1 gene (SI Tables 4 and 5). Because the number of genes that were induced and the magnitude of the effect on mRNA levels were rather low, we did not investigate the significance of this observation further.
We analyzed more thoroughly three genes that were affected in the dst1-R200A med31-Δ strain: ADH1, MET17, and VTC3. We measured by RT-PCR the mRNA quantities in dst1-R200A med31-Δ at 30°C and 37°C. We found that mRNA levels decreased 1.5- and 2.5-fold for ADH1 at the permissive and restrictive temperatures, respectively; 2.1- and 3.8-fold for MET17; and 2.4- and 2.3-fold for VTC3, in good agreement with the microarray data.
TFIIS Is Recruited to the Promoter of ADH1, MET17, and VTC3.
We looked for TFIIS recruitment on these genes through ChIP. We used an N-terminal HA-tagged version of TFIIS, the fusion being expressed from the chromosome (a gift from M. Wery and P. Thuriaux, Commissariat à l'Energie Atomique, Saclay, France). Primer pairs were designed to amplify the promoters or the coding sequences of ADH1, MET17, and VTC3 (Fig. 3A). Primers pairs amplifying GAL1 sequences were used for background noise estimation, because GAL1 is not transcribed in the YPD glucose medium. We saw enrichment of TFIIS above background on the promoters and ORFs of all three genes (Fig. 3B). Because the presence of the HA tag could alter the function of TFIIS, we also performed ChIP experiments with anti-TFIIS antibodies (SI Fig. 5). The results were essentially the same, even though the background level was higher. In the case of MET17 and VTC3, the amount of TFIIS on the promoter was greater than or equal to the amount on the ORF, suggesting a role for TFIIS in transcription initiation in addition to that in transcription elongation on these genes.
Fig. 3.
TFIIS is recruited to the promoter and coding sequence of three different genes. (A) Schematic promoter and coding region organization of ADH1, MET17, and VTC3 showing the location of primer pairs used in ChIP analysis. Scale is of 200 base pairs for one graduation. A promoter-specific primer pair (labeled Pr) and an ORF-specific primer pair (labeled ORF) are used for each gene. The location of the major transcription start site (TSS) relative to ATG of genes is indicated in base pairs. TSS are located at −61, −49, −43, −38, −33, −31, −20, and −12 for ADH1, at −52, −43, −40, and −38 for MET17 (33). Their position is unknown for VTC3. Diamonds indicate activator-binding sequences (taken from Saccharomyces Genome Database). Small parallelograms indicate the position of the TATA box (34). The arrows indicate the position of the TSS. Only confirmed neighbor genes are represented (Saccharomyces Genome Database). (B) ChIP analysis of 3HA-TFIIS binding to promoter or ORF of ADH1, MET17, and VTC3. Analysis of nontranscribed GAL1 ORF served as control for TFIIS background binding. Cells were grown to exponential phase in glucose-rich medium (YPD) at 30°C before cross-linking.
TFIIS Domain II Is Required for the Recruitment of Pol II to the Promoter of ADH1, MET17, and VTC3.
The growth defect observed in the dst1-R200A mutant could result from lower Pol II recruitment or stability on affected promoters. Thus Pol II occupancy on ADH1, MET17, and VTC3 genes was studied by ChIP. Cells were grown in YPD glucose-rich medium at 30°C, and the culture was shifted to 37°C for 30 min before cross-linking. Pol II was immunoprecipitated with 8WG16 anti-C-terminal domain (CTD) antibody. Fig. 4A shows that, taking the error margin into account, Pol II recruitment to promoters or ORFs was not modified in any TFIIS mutant strains compared with wild-type strain in a MED31 wild-type background. However, deletion of MED31 decreased the levels of Pol II on the promoters and ORFs by a factor of 2–4 (compare black histogram values of Fig. 4 A and B), showing that the ADH1, MET17, and VTC3 genes depend on Med31 for highest occupancy. Pol II recruitment to promoters or ORFs did not change significantly in dst1- (1–265) med31-Δ compared with the DST1 med31-Δ strain, confirming that the cleavage activity of TFIIS domain III does not influence Pol II occupancy of ADH1, MET17, and VTC3, even in the med31-Δ mutant context (Fig. 4B). On the contrary, recruitment of Pol II was diminished 2-fold on promoters and coding sequences in the dst1-R200A med31-Δ strain compared with the DST1 med31-Δ strain. At 30°C, similar results were obtained (SI Fig. 6), in agreement with mRNA levels. We concluded that the correct transcription of ADH1, MET17, and VTC3 did not require TFIIS cleavage activity, but that Pol II recruitment or stabilization on promoters of these genes depends on TFIIS Pol II-binding activity in the absence of Med31.
Fig. 4.
ChIP analysis of the occupancy of promoter and ORF of ADH1, MET17, and VTC3 in DST1 and MED31 mutant contexts. (A) ChIP analysis of Pol II in DST1, dst1-(1–265), dst1-R200A, and MED31 strains grown in rich glucose medium at 37°C. Immunoprecipitation signal over input signal is represented in arbitrary units. (B) ChIP analysis of Pol II in DST1, dst1-(1–265), and dst1-R200A in med31-Δ background grown in rich glucose medium at 37°C. (C) ChIP analysis of TBP in DST1, dst1-(1–265), dst1-R200A, and med31-Δ strains grown in rich glucose medium at 37°C. (D) ChIP analysis of TFIIS binding to promoter or ORF of ADH1, MET17, and VTC3 in med31-Δ DST1 and med31-Δ dst1-R200A strains. Cells were cultured to log phase in glucose-rich medium at 30°C and then shifted at 37°C for 30 min before cross-linking. Anti-TFIIS polyclonal antibodies were used for immunoprecipitation.
Recruitment of TATA Box-Binding Protein (TBP) and TFIIS Independent of Pol II to ADH1, MET17, and VTC3.
Because Pol II occupancy was diminished at ADH1, MET17, and VTC3 promoters and ORFs in dst1-R200A med31-Δ cells, we wanted to know whether other components of the preinitiation complex followed the same pattern. TBP recruitment was analyzed by ChIP, using anti-TBP antibodies (a gift from A. Weil, Vanderbilt University, Nashville, TN), in DST1 med31-Δ and dst1-R200A med31-Δ cells after a shift of 30 min to 37°C. Fig. 4C showed that no defect in TBP recruitment could be seen.
TFIIS recruitment was analyzed by ChIP, by using anti-TFIIS antibodies (a gift from C. Kane, University of California, Berkeley, CA), in DST1 med31-Δ and dst1-R200A med31-Δ cells after a shift of 30 min to 37°C. We looked at TFIIS occupancy using the same chromatin preparations as for the Pol II ChIP experiments of Fig. 4B. Fig. 4D showed that no defect in TFIIS recruitment could be seen in the dst1-R200A mutant compared with wild-type. We concluded that TFIIS was recruited independently of Pol II on these genes.
TFIIS Cleavage Activity Is Not Required for Rescuing dst1-Δ Colethality with an Rpb1 CTD Deletion.
TFIIS inactivation exerts a synthetic effect with mutations in numerous genes, a number of which are implicated in transcription. Thus, we wanted to know whether the domain II and linker were sufficient to rescue the colethality of dst1-Δ with mutants other than med31-Δ. We investigated the well established colethality of DST1 deletion with the Rpb1 CTD truncation. In yeast, Rpb1 CTD is composed of 26 or 27 repeats of a heptapeptide. Reducing the number of these repeats to 10–12, as in the rpb1-Δ104 strain, is colethal with dst1-Δ (14). We modified the rpb1-Δ104 strain (15) by fully deleting DST1 and complementing it with a wild-type copy on a URA3 vector. This strain was transformed with vectors carrying the DST1 truncations used above, and cells containing the DST1 wild-type copy were counterselected on a 5FOA-containing medium. We confirmed the colethality of rpb1-Δ104 mutation with dst1-Δ (SI Fig. 7, last row). However, the colethality was fully complemented by a TFIIS fragment composed of domain II and linker alone (SI Fig. 7, row 4).
Discussion
In this work, we show that the TFIIS elongation factor has a function required for the optimal transcription of selected genes. Importantly, this function of TFIIS is independent of its cleavage activity but requires its Pol II-binding domain. We show that TFIIS is required in conjunction with the Med31 Mediator subunit for full Pol II occupancy in vivo and thus has a dual and independent role in transcription initiation and elongation at a subset of genes. This conclusion is also supported by the independent finding by Ranish and coworkers (11) that TFIIS is important for preinitiation complex formation in vitro.
Domain II binds strongly to RNA Pol II in vitro through residues of a basic patch (7, 8). We mutated the R200A basic patch residue and found that the mutation was conditionally colethal with med31-Δ but did not affect MPA sensitivity, strongly suggesting a lack of effect on transcription elongation. Indeed, Struhl and coworkers showed that in a TFIIS mutant sensitivity to 6-azauracil, which, like MPA, is a nucleotide-depleting drug, correlated with decreased transcription processivity, and stimulated transcription through an arrest site in vivo (16, 17). The dst1-R200A mutation in the med31-Δ background affected the transcription of 80–90 genes and the recruitment of Pol II on the three genes (ADH1, MET17, and VTC3) that were selected among those that were transcription-impaired. However, eliminating RNA cleavage stimulatory activity had no effect on Pol II occupancy. TFIIS was recruited to the promoters of these genes, which is not the general situation (10). Contrary to Pol II, TFIIS itself was still recruited to the impaired genes, implicating an independent recruitment mechanism for TFIIS and Pol II. TBP association with promoters was also not altered, indicating that the dst1-R200A mutation did not affect the early steps of preinitiation complex formation in vivo.
We propose the following model. TFIIS would be recruited by activators or coactivators independently of Pol II to selected gene promoters. Then, TFIIS, in conjunction with coactivators such as the Mediator complex, would stimulate Pol II recruitment or stabilization on promoters via a direct interaction with the enzyme through its Pol II-binding site.
In vitro, the TFIIS basic patch is important for TFIIS binding to RNA Pol II (7, 8). Whereas the TFIIS-R200A mutation reduced Pol II occupancy at promoters of selected genes, it did not affect sensitivity to MPA or occupation of TFIIS on ORFs, suggesting that in vivo, this mutation renders TFIIS Pol II binding limiting for preinitiation complex formation but not for transcription elongation.
The TFIIS minimal domain required for full complementation of dst1-Δ med31-Δ colethality and full occupancy of Pol II on promoters contained the linker domain in addition to domain II. We were able to construct a viable but very slow-growing dst1-Δ med31-Δ strain complemented by the first three α-helices of TFIIS domain II alone (data not shown). The presence of the linker sequence could protect domain II from degradation in its free form in vivo or might stabilize domain II association with Pol II.
TFIIS domain II and linker were sufficient to complement the colethality of the Rpb1 CTD truncation with dst1-Δ. Rpb1 CTD is bound by the Mediator complex and may be required for Pol II recruitment to activated promoters (18). Colethality of rpb1-Δ104 with dst1-Δ may also be due to a synergistic defect in Pol II recruitment to promoters. This possibility is reinforced by the colethality of rpb1-Δ104 with med31-Δ (19). Thus, TFIIS Pol II-binding activity may be required when the transcription machinery is defective in Pol II recruitment on activated promoters.
In our experiments with ADH1, MET17, and VTC3, we did not use mutants of the TFIIS domain I, because dst1-(133–265) lacking this domain was not colethal with med31-Δ. The Pol II ChIP results obtained using such a mutated strain were very variable, despite our best efforts (data not shown). This situation may be due to an unknown role of this domain. Indeed, it has been shown that domain I interacts with the Med13 (Srb9) Mediator and Spt8 SAGA subunits (20). Moreover, TFIIS domain I is required when the Rpb4 subunit of Pol II is absent. It is thus possible that TFIIS domain I, as the Pol II-binding domain, might have a role in transcription initiation at some promoters when subunits of the transcription machinery are lacking, as suggested by the data in the accompanying paper by Kim et al. (11), indicating that domain I contributes to PIC assembly in vitro.
Med31 is the most conserved subunit of Mediator in terms of protein sequence conservation and is present in most, if not all, eukaryotic organisms (21). It has been proposed previously that the most conserved subunits of Mediator form a core complex oriented toward Pol II (22). These subunits belong to the Mediator middle module known to contact Pol II (23), and they are essential in yeast. Med31 interacts with subunits of the middle module (24, 25), but is essential neither in S. cerevisiae nor in Schizosaccharomyces pombe (26) despite its strong conservation. This situation could be explained if the role of Med31 and TFIIS in conjunction during transcription initiation also holds in eukaryote species other than S. cerevisiae, a very likely possibility in view of the strong conservation of TFIIS throughout the eukaryotic kingdom.
In conclusion, we provide insights into the in vivo function of the TFIIS transcription factor independent of its well characterized stimulatory activity on Pol II RNA cleavage and requiring its Pol II-binding domain. TFIIS acts in transcription initiation in conjunction with the Med31 Mediator subunit and contributes to the optimal Pol II occupancy at a subset of gene promoters in vivo.
Experimental Procedures
Oligonucleotides.
The sequence of the oligonucleotides used in this study can be found in SI Text.
Plasmid Construction and Cloning.
All cloning was done by using the Gateway Invitrogen cloning method following the standard protocol (Invitrogen, Carlsbad, CA). The DST1 cloned sequences were transferred into pVV204 (CEN TRP1 pTetO7) vector (27) by the LR reaction (Gateway Technology; Invitrogen, Carlsbad, CA). R200A point mutation in DST1 was obtained by PCR overlap extension mutagenesis (28) and cloned by using the Gateway standard method and transferred into pVV204 as well.
MED31 was cloned the same way as DST1 and transferred into pVV208 (CEN URA3 pTetO7) vector (27).
Yeast Strains.
MED31 was deleted in YPH500 background (MATa ura3–52 his3–200 ade2–101 trp1–63 lys2–801 leu2–1) and replaced by a kan marker using the standard one-step method (29).
The ESH1 (20) strain deleted for DST1 was transformed with the CEN URA3 pTetO7:MED31 plasmid and then deleted for MED31 with a kan marker. The collection of cloned DST1 mutants was transformed into this strain. The rpb1-Δ104 strain (15) was transformed with a CEN URA3 pTetO7:DST1, then the chromosomal copy of DST1 was deleted by a HIS3MX6 marker (29). The resulting strain was transformed with the collection of cloned DST1 mutants.
5FOA and MPA Assays.
The collection of strains bearing the truncation mutants of DST1 was spotted on complete medium with 25 μg/ml MPA to test for elongation defect or streaked on synthetic dextrose (SD) medium supplemented with uracil and then spotted on 5FOA medium minus uracil to counterselect the wild-type MED31-bearing plasmid.
ChIP.
ChIP were done essentially as described (30, 31). The HA-tagged proteins were immunoprecipitated with 12CA5 antibody bound to IgG magnetic beads (Dynabead; Dynal Biotech ASA, Oslo, Norway). Pol II was immunoprecipitated with 8WG16 anti-CTD antibody (Covance). Rabbit antibody to recombinant yeast TFIIS was a gift from C. Kane. Anti-TBP antibody was a gift from A. Weil.
Immunoprecipitated DNA was analyzed by quantitative real-time PCR on an ABI Prism 7000 machine (Applied Biosystems, Foster City, CA). Relative quantification using a standard curve method was performed, and the occupancy level for a specific fragment was defined as the ratio of immunoprecipitated over total DNA. The respective locations of the amplified PCR products are indicated in Fig. 3.
When cells were shifted from 30°C to 37°C, one-half of an exponentially growing culture was collected by centrifugation, immediately mixed with medium heated at and incubated further at 37°C for 30 min under agitation. Three independent experiments were averaged. The corresponding standard deviations are indicated in Figs. 1–4. When cells were cultivated in YPD medium, the GAL1 ORF region was used as a nontranscribed control.
Quantitative RT-PCR.
mRNA levels were determined by quantitative RT-PCR. RNA was extracted as described by Schmitt et al. (32). Reverse transcription of 5-μg RNA samples was performed in the presence of SuperScript II and random primers (Invitrogen) for 1–2 h at 42°C in the appropriate buffer. 25S rRNA was used as internal control for normalization.
Microarray Analysis.
Gene expression was monitored with DNA microarrays manufactured by Service de Génomique Fonctionnelle (Commissariat à l'Energie Atomique/Evry, France), as described (13), except that an indirect cDNA-labeling protocol of the targets was used (35). The microarrays were scanned with a GENEPIX 4000B scanner. Spot intensities and fluorescence ratios were measured by using the GENEPIX 4.0 software (Molecular Devices, Sunnyvale, CA). Temperature shift of the culture was done as described for ChIP experiments. For each growth condition (30°C or 37°C), seven hybridizations were performed with three batches of RNA extracted from med31-Δ DST1 and med31-Δ dst1-(R200A) strains, and labels were exchanged in one-half of the hybridizations. The data were analyzed with the GeneSpring software (Agilent Technologies, Santa Clara, CA).
Supplementary Material
Acknowledgments
We thank L. Kuras for initiating us into the ChIP method; C. Kane for the anti-TFIIS antibody; A. Weil for the anti-TBP antibody; P. Thuriaux, M. Wery, and E. Shematorova (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia) for discussion and yeast strains; J. Ranish for sharing results before publication; C. Mann for improving the manuscript; and F. Amiot and X. Gidrol (Commissariat à l'Energie Atomique, Evry, France) for providing DNA microarrays. B.G. was supported by grants from the Ministère de l'Enseignement et de la Recherche and from the Association pour la Recherche sur le Cancer.
Abbreviations
- Pol II
polymerase II
- MPA
mycophenolic acid
- 5FOA
5-fluorooroatic acid
- YPD
yeast extract/peptone/dextrose
- CTD
C-terminal domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704534104/DC1.
References
- 1.Hampsey M. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koh SS, Ansari AZ, Ptashne M, Young RA. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 3.Lee YC, Park JM, Min S, Han SJ, Kim YJ. Mol Cell Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaman Z, Ansari AZ, Gaudreau L, Nevado J, Ptashne M. Cold Spring Harbor Symp Quant Biol. 1998;63:167–171. doi: 10.1101/sqb.1998.63.167. [DOI] [PubMed] [Google Scholar]
- 5.Wind M, Reines D. BioEssays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olmsted VK, Awrey DE, Koth C, Shan X, Morin PE, Kazanis S, Edwards AM, Arrowsmith CH. J Biol Chem. 1998;273:22589–22594. doi: 10.1074/jbc.273.35.22589. [DOI] [PubMed] [Google Scholar]
- 7.Awrey DE, Shimasaki N, Koth C, Weilbaecher R, Olmsted V, Kazanis S, Shan X, Arellano J, Arrowsmith CH, Kane CM, Edwards AM. J Biol Chem. 1998;273:22595–22605. doi: 10.1074/jbc.273.35.22595. [DOI] [PubMed] [Google Scholar]
- 8.Kettenberger H, Armache KJ, Cramer P. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 9.Malagon F, Tong AH, Shafer BK, Strathern JN. Genetics. 2004;166:1215–1227. doi: 10.1534/genetics.166.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prather DM, Larschan E, Winston F. Mol Cell Biol. 2005;25:2650–2659. doi: 10.1128/MCB.25.7.2650-2659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim B, Nesvizhskii AI, Geetha Rani P, Hahn S, Aebersold R, Ranish JA. Proc Natl Acad Sci USA. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ubukata T, Shimizu T, Adachi N, Sekimizu K, Nakanishi T. J Biol Chem. 2003;278:8580–8585. doi: 10.1074/jbc.M211384200. [DOI] [PubMed] [Google Scholar]
- 13.Fauchon M, Lagniel G, Aude J-C, Lombardia LJ, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. Mol Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- 14.Lindstrom DL, Hartzog GA. Genetics. 2001;159:487–497. doi: 10.1093/genetics/159.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nonet ML, Young RA. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason PB, Struhl K. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Kulish D, Struhl K. Mol Cell Biol. 2001;21:4162–4168. doi: 10.1128/MCB.21.13.4162-4168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson CM, Koleske AJ, Chao DM, Young RA. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 19.Fan HY, Cheng KK, Klein HL. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wery M, Shematorova E, Van Driessche B, Vandenhaute J, Thuriaux P, Van Mullem V. EMBO J. 2004;23:4232–4242. doi: 10.1038/sj.emboj.7600326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linder T, Gustafsson CM. J Biol Chem. 2004;279:49455–49459. doi: 10.1074/jbc.M409046200. [DOI] [PubMed] [Google Scholar]
- 22.Bjorklund S, Gustafsson CM. Adv Protein Chem. 2004;67:43–65. doi: 10.1016/S0065-3233(04)67002-1. [DOI] [PubMed] [Google Scholar]
- 23.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Mol Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grallert A, Grallert B, Zilahi E, Szilagyi Z, Sipiczki M. Yeast. 1999;15:669–686. doi: 10.1002/(SICI)1097-0061(19990615)15:8<669::AID-YEA411>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 27.Van Mullem V, Wery M, De Bolle X, Vandenhaute J. Yeast. 2003;20:739–746. doi: 10.1002/yea.999. [DOI] [PubMed] [Google Scholar]
- 28.Higuchi R, Krummel B, Saiki RK. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philipsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Kuras L, Struhl K. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 31.Kuras L, Borggrefe T, Kornberg RD. Proc Natl Acad Sci USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt ME, Brown TA, Trumpower BL. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Dietrich FS. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basehoar AD, Zanton SJ, Pugh BF. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 35.Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, Werner M. Mol Cell Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.