Abstract
α-Actinin-4 is a widely expressed protein that employs an actin-binding site with two calponin homology domains to crosslink actin filaments (F-actin) in a Ca2+-sensitive manner in vitro. An inherited, late-onset form of kidney failure is caused by point mutations in the α-actinin-4 actin-binding domain. Here we show that α-actinin-4/F-actin aggregates, observed in vivo in podocytes of humans and mice with disease, likely form as a direct result of the increased actin-binding affinity of the protein. We document that exposure of a buried actin-binding site 1 in mutant α-actinin-4 causes an increase in its actin-binding affinity, abolishes its Ca2+ regulation in vitro, and diverts its normal localization from actin stress fibers and focal adhesions in vivo. Inactivation of this buried actin-binding site returns the affinity of the mutant to that of the WT protein and abolishes aggregate formation in cells. In vitro, actin filaments crosslinked by the mutant α-actinin-4 exhibit profound changes of structural and biomechanical properties compared with WT α-actinin-4. On a molecular level, our findings elucidate the physiological importance of a dynamic interaction of α-actinin with F-actin in podocytes in vivo. We propose that a conformational change with full exposure of actin-binding site 1 could function as a switch mechanism to regulate the actin-binding affinity of α-actinin and possibly other calponin homology domain proteins under physiological conditions.
Keywords: cytoskeleton, kidney, podocyte, glomerulosclerosis
Discovered more than 40 years ago as a component of skeletal muscle, α-actinin was the first-recognized example of a nonmotor muscle molecule with a predominant actin filament (F-actin)-crosslinking function in vitro (1). Subsequently identified as an abundant protein in nonmuscle cells as well (2), extensive studies have defined the molecular structure of α-actinin and its in vitro F-actin-crosslinking activity. Each of the four highly homologous α-actinin genes encode a 100-kDa peptide with two N-terminal calponin homology (CH) domains, forming an F-actin-binding site [designated the head or the actin-binding domain (ABD)], four spectrin repeats, and a C-terminal calmodulin-like Ca2+-binding domain. α-Actinins form dumbbell-shaped antiparallel homodimers of ≈40 nm in length with an ABD on each end. Most studies have shown that these short-range crosslinking molecules promote alignment of F-actin as loose bundles (3), although α-actinin–F-actin networks are more isotropic under certain preparation conditions. In part, these differences reflect the dynamic properties of the binding of α-actinin to F-actin (4).
Despite this extensive research, however, the functions of α-actinins in general and the role of their F-actin binding in particular in vivo have been elusive. The notion that they represent key structural components for large-scale actin filament cohesion in cells required for cell shape and motility is called into question by the finding that Dictyostelium lacking α-actinin protein has few detectable phenotypic abnormalities (5). Further complicating the picture have been the recognition of two muscle-specific and two nonmuscle α-actinin isoforms encoded by different genes in mammals and a growing catalog of binding partners other than F-actin involved in cell signaling cascades (6). It is known that nonmuscle α-actinins are involved in a variety of cellular processes, among them cell motility, endocytosis, and adhesion (7–9). Cultured podocytes deficient in one of the nonmuscle isotypes, α-actinin-4, exhibit defective substrate adhesion (10), which is consistent with the well documented localization of α-actinin at focal adhesion sites (11), and is therefore probably responsible for the impaired translational locomotion of the α-actinin-4-deficient cells (7).
Five distinct point mutations in the α-actinin-4 head domain causing focal segmental glomerulosclerosis in affected humans all mediate increased actin binding (12, 13). However, this effect has not yet been studied in detail, and the mechanism of disease has so far remained elusive. Here we report in vitro and in vivo studies that take advantage of one mutant α-actinin-4 harboring a human disease-causing K255E amino acid exchange that leads to a similar disorder in a knockin mouse model (14). Podocytes of mice homozygous for this mutation develop large cytoplasmic aggregates, which are associated with the actin cytoskeleton (unpublished data). We show that the K255E mutant α-actinin has a markedly increased binding affinity for F-actin. As a result, the localization of this mutant α-actinin-4 in cells is abnormal and the actin cytoskeleton is perturbed. Our studies illuminate the molecular changes in the ABD of α-actinin-4 that mediate this increased actin-binding affinity and shed light on the structural basis of the binding of α-actinin to F-actin, resolving some controversy regarding this interaction. Furthermore, we link our in vitro data to the disease by studying the cellular distribution of WT and K255E mutant α-actinin-4. Our results provide evidence for an independent mechanism for regulating the actin-binding affinity of a CH domain protein under physiological conditions.
Results
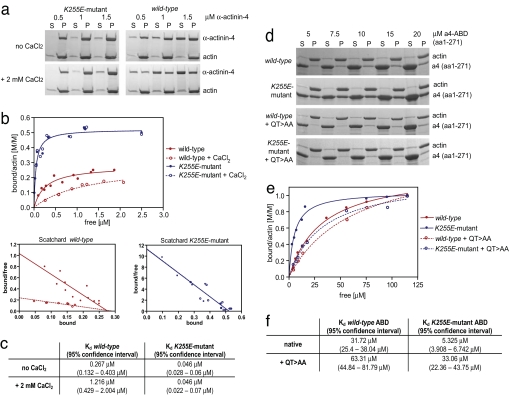
We initiated our studies by investigating the interaction of α-actinin-4 with actin in vitro by using purified recombinant K255E mutant and WT α-actinin-4 proteins. After confirming that K255E mutant α-actinin-4 alone does not aggregate in vitro [supporting information (SI) Fig. 7], we performed a detailed actin-binding analysis by using a standard actin cosedimentation assay. Representative Coomassie-stained polyacrylamide gels are shown in Fig. 1a. As shown in Fig. 1 b and c, the dissociation constant (Kd) of K255E α-actinin-4 was 0.046 μM, almost 6-fold lower than that of WT α-actinin-4 (0.267 μM). The K255E α-actinin-4 dimers saturate F-actin with a 1:2 stoichiometry, twice the ratio of WT dimers (1:4).
Fig. 1.
K255E mutant α-actinin-4 shows higher F-actin-binding affinity as the WT protein and is resistant to Ca2+ regulation. (a) Representative images of Coomassie-blue-stained polyacrylamide gels at three molar ratios. Increasing amounts of K255E mutant and WT α-actinin-4 without (upper gels) or with (lower gels) the addition of 2 mM CaCl2 to reaction buffer were polymerized with 5 μM G-actin and centrifuged at 100,000 × g. Equal amounts of supernatants and pellets were subjected to SDS/PAGE. α-Actinin-4 alone did not pellet at this speed (data not shown). (b) K255E mutant α-actinin-4 shows increased binding affinity to F-actin, a higher binding stoichiometry, and is insensitive to Ca2+. Saturation binding curves and Scatchard plots were generated by nonlinear regression from quantified results of Coomassie-blue-stained polyacrylamide gels. (c) The table shows respective Kd values and 95% confidence intervals. Inactivation of ABS1 by introducing the QT>AA mutation into the K255E mutant α-actinin-4 ABD (a4-ABD) returns its F-actin-binding affinity to that of the WT protein. (d) Representative images taken from Coomassie-blue-stained polyacrylamide gels. Increasing amounts (5–120 μM) of purified α-actinin-4 ABDs were polymerized with 5 μM G-actin and centrifuged at 100,000 × g, and equal amounts of supernatants and pellets were subjected to SDS/PAGE. (e) K255E mutant α-actinin-4 ABD shows higher binding affinity to F-actin, whereas K255E mutant α-actinin-4 ABD containing the ABS1 mutation QT>AA shows similar F-actin-binding affinity as the WT protein. The F-actin-binding affinity of WT α-actinin-4 ABD containing the QT>AA mutation was only minimally affected. (f) The table shows respective Kd values and 95% confidence intervals.
The antiparallel assembly of α-actinin-4 as dimers allows interaction of the N-terminal CH domain of one monomer with the C-terminal calmodulin-like domain of the other. It has been postulated that binding of Ca2+ to the calmodulin-like domain leads to a conformational change, possibly by repositioning of the calmodulin-like domain between actin and the α-actinin ABD, causing a decrease in actin-binding affinity (15). The addition of CaCl2 to the reaction buffer decreased the binding affinity of the WT protein to a Kd of 1.216 μM at 1.5 mM free Ca2+ in the otherwise Ca2+-free reaction buffer (Fig. 1 b and c). Even lower Ca2+ concentrations (e.g., 0.1 mM free Ca2+) led to a clear decrease in the affinity of WT α-actinin-4 (data not shown). By contrast, the affinity of K255E α-actinin-4 was unaffected even at high Ca2+ concentrations (Fig. 1 b and c), indicating that this mutant form adopts a conformation that is insensitive to Ca2+ regulation. Hence, the K255E mutation in α-actinin-4 results in a high-affinity actin-binding protein resistant to regulation by Ca2+.
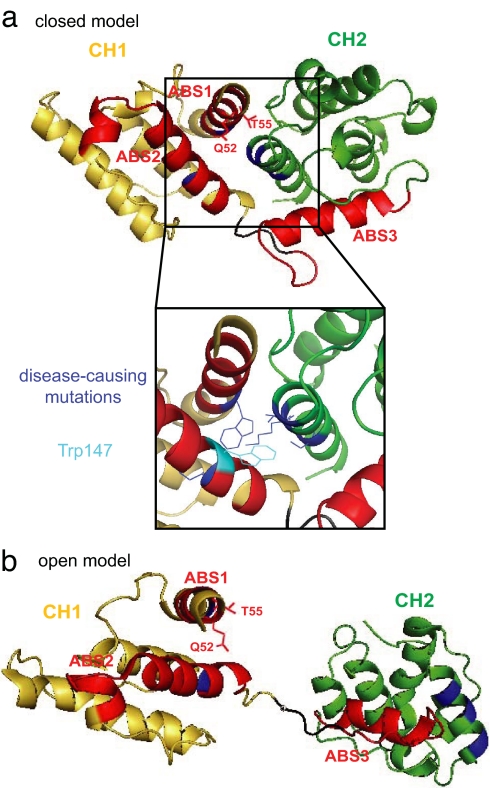
α-Actinin-4, like all α-actinins, contains three actin-binding sites (ABSs), designated ABS1, ABS2, and ABS3. In one model defined as closed, ABS2 and -3 form a continuous actin-binding surface spanning both CH domains (Fig. 2a). ABS1 is located in the CH1 domain and remains largely buried (16). Recently, the atomic structure of the α-actinin-1 ABD was published (17). α-Actinin-1 and -4 share >90% homology in the crystallized region; thus, the structural findings reported are likely true for α-actinin-4 as well. Based on conservation scores and structural data, the authors identified several amino acids within the interface of CH1 and CH2 that are likely to form atom-to-atom contacts between the two CH domains. Each one of these interactions likely contributes to keeping the two CH domains in a closed state; thus, the structure provides evidence for the possibility that limited molecular changes may lead to transition from a closed to an open conformation. Studies on plectin, another CH domain protein, revealed ABS1 is exposed in the open conformation and extends the actin-binding surface (18) (Fig. 2b). Close examination of the crystal structure of α-actinin-1 revealed that all of the disease-associated mutations in α-actinin-4 map to this highly conserved CH1-CH2 interface (Fig. 2). In fact, the lysine residue at respective position 255 in CH2 of α-actinin-4, mutated to a glutamate in the K255E mutant, is potentially a key interactor by forming a hinge-like connection to Trp-147 of CH1, presumably through the hydrophobic part of the lysine side chain. No local negative charges could be found in the vicinity of the mutation; thus, we believe the change of the charge balance from positive to negative introduced by the mutation most likely does not have any major effect.
Fig. 2.
Structural models of the ABD of α-actinin based on two previously published atomic structures (15, 17). (a) This model is consistent with a closed conformation of the CH domains. CH1 is shown in yellow, CH2 in green. ABS1 to -3 are shown in red; side chains of Q52 and T55, mutated to alanines in the QT>AA mutant, are shown to demonstrate their inward-facing position within the model. The enlarged view of the CH1–CH2 interface demonstrates that amino acids changed by the five disease-associated mutations (W59R, I149del, K255E, T259I, and S262P), depicted in blue, all localize to the CH1–CH2 interface region. In addition, Lys-255, mutated to Glu in the K255E mutant, interacts with Trp-147 (shown in cyan) of CH1 to form a hinge-like connection that supposedly functions as a key connection that keeps the CH domains in a closed conformation. (b) This model shows an open conformation of the CH domains. Here, the two CH domains are widely separated, thereby fully exposing ABS1, including Q52 and T55. The interface regions, bearing the disease-associated mutations, are pointing in opposite directions, breaking apart all atom-to-atom connections between CH1 and CH2.
We therefore tested whether the observed increase in actin-binding affinity seen in the K255E α-actinin-4 could be the result of a higher propensity for adopting an open conformation of its ABD in solution, thereby leading to full exposure of ABS1. We used site-directed mutagenesis to inactivate ABS1, generating four different ABS1 point mutants of WT and K255E α-actinin-4 ABD as GST fusion proteins. By using these mutated ABDs in an actin cosedimentation assay, we identified one double mutation, 52Q>A + 55T>A (termed QT>AA) that significantly decreased actin binding of the K255E ABD but not of the WT ABD in a screen. This double mutation changed two amino acids with side chains buried in the closed conformation (see Fig. 2). Representative Coomassie-stained polyacrylamide gels (Fig. 1d) showed an in vitro actin-binding analysis with purified WT, K255E, WT+QT>AA, and K255E+QT>AA ABDs. It revealed that the ABS1 mutation in K255E ABD (Kd = 33.06 μM; Fig. 1 e and f) returned the affinity of the K255E mutant (Kd = 5.325 μM; Fig. 1 e and f) to that of the WT ABD (Kd = 31.72 μM; Fig. 1 e and f), indicating that ABS1 is required for the increased affinity of the mutant protein. Introducing the QT>AA mutation into the WT protein had only a slight effect on the actin-binding affinity (Kd = 63.31 μM; Fig. 1 e and f), indicating that ABS1 contributes minimally to F-actin binding in this conformation. Furthermore, the stoichiometric difference observed between full-length K255E mutant and WT dimers could not be observed by using the ABD alone.
Our finding is consistent with the fact that each α-actinin-4 ABD requires contact with two actin monomers for binding. Based on a previously published model (19), the obtained stoichiometric ratio of the full K255E dimer indicates that it fully saturates all available α-actinin-binding sites on the crosslinked actin filaments. Our results also imply that WT α-actinin-4 in vitro can bind to only every other α-actinin-4-binding site on the actin filament. The α-actinin rod exhibits a torsional twist to accommodate its parallel binding to actin filaments, possibly resulting in a sterically unfavorable constraint that prevents binding of an α-actinin dimer to the adjacent binding site (20). Thus, we propose that the higher stoichiometry of the K255E mutant dimer is most likely due to a different conformation of the mutant ABD, which avoids such hindrance by providing the K255E mutant with even higher flexibility than the WT protein regarding the orientation of the ABD with respect to the rod domain. Alternatively, the open conformation results in greater distance between the two CH domains, enabling the ABD to bind to sites that are not accessible to the WT dimer in the closed conformation and making it easier for both ends to bind actin under bundling conditions.
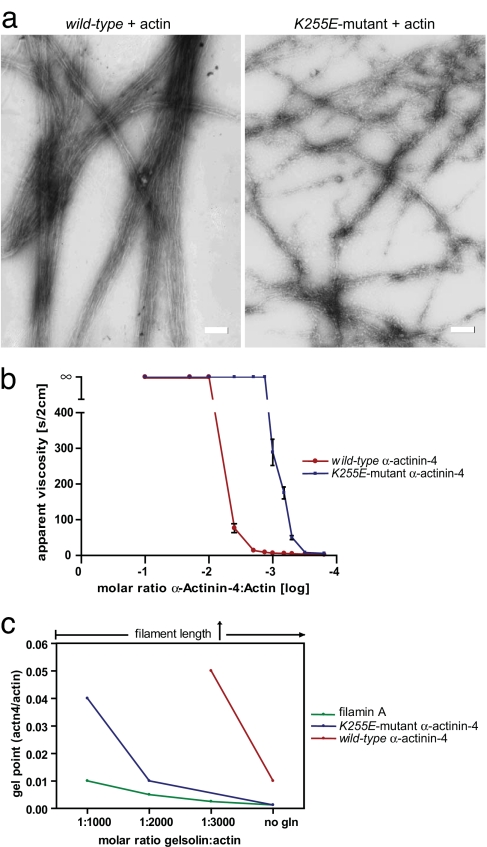
The rate of dissociation of an F-actin crosslinker can influence whether filaments are sufficiently mobile to align into bundles (21), leading to the prediction that the relatively low-affinity WT α-actinin-4 would allow for such bundling, whereas the high-affinity K255E α-actinin-4 mutant would not. We tested these predictions by observing negatively stained F-actin networks in the electron microscope (Fig. 3a) and by analyzing the effects of WT and K255E α-actinin-4 on F-actin mechanics by using miniature falling ball viscometry (Fig. 3b). Actin polymerized with WT α-actinin-4 appeared as thick parallel bundles (mean thickness, 310 nm; SD, 129 nm). Polymerization of actin with the K255E α-actinin-4 mutant, however, resulted in a disordered and entangled network of thin F-actin bundles (mean thickness, 83 nm; SD, 38 nm). As reported previously (22, 23), the ratio of α-actinin-4 to actin monomers assembled in its presence at which an abrupt increase in viscosity, or gel point, occurs, was 1:100 (Fig. 3b). In contrast, a molar ratio of 1:850 K255E mutant α-actinin-4 monomer per actin monomers was sufficient to induce a similar transition, a value approaching the gel point defined for the potent F-actin crosslinker filamin A (24). The F-actin-crosslinking efficiency of filamin A, however, depends on its ability to organize rod-like F-actin into an orthogonal network rather than on high-affinity binding to F-actin (25). α-Actinin-4, even with greatly increased affinity, would not be predicted to have this capability, suggesting that its ability to gel actin efficiently would occur only if actin filaments were sufficiently long to form entanglements that resisted parallel alignment. As expected, actin polymerized in the presence of increasing plasma gelsolin concentrations which proportionally shorten F-actin lengths (26), resulted in much more K255E α-actinin-4 being required to induce actin gelation than filamin A (Fig. 3c). This result shows that unlike filamin A, mutant α-actinin-4 does not directly oppose parallel alignment despite its enhanced affinity to actin. The ability of the K255E mutant protein to crosslink actin filaments at highly variable angles also indicates a greater geometric flexibility of the ABD toward the bound actin filament.
Fig. 3.
K255E mutant α-actinin-4 crosslinks F-actin into architecturally distinct structures. (a) Transmission electron microscopy of actin filaments alone and crosslinked by WT or K255E mutant α-actinin-4 in vitro. WT α-actinin-4 crosslinks actin filaments into thick parallel bundles with defined spacing. Mutant α-actinin-4 induces the formation of a disordered and entangled network of thin filament bundles. (Scale bars, 200 nm.) (b) Viscometry assay of actin in the presence of WT or K255E mutant α-actinin-4. Decreasing α-actinin-4/actin molar ratios were used for gel point assays and are expressed as a log scale. Solid-to-gel transition is shown by the ∞ sign. The gel point concentration of WT α-actinin-4 was determined at 1:100, whereas the gel point of the K255E mutant was found at 1:850. (c) Determination of actin gel points in the presence of filamin A and WT and K255E mutant α-actinin-4 as different actin filament lengths. The gel point concentrations are shown as a molar ratio of crosslinker to actin and at four different gelsolin concentrations. WT α-actinin-4 can only induce actin gelation with long filaments. With decreasing filament length, a higher concentration of K255E mutant α-actinin-4 than filamin A is required to induce gelation.
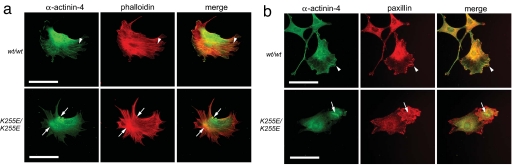
Fibroblasts isolated from a knockin Actn4K255E mouse model (14) served as a tool to study the effects of endogenously expressed WT and K255E mutant α-actinin-4 on the actin cytoskeleton (Fig. 4). We stained fibroblasts derived from Actn4WT/WT and Actn4K255E/K255E mice with a specific antibody against α-actinin-4. Immunoreactive α-actinin-4 associated with phalloidin-labeled F-actin stress fibers (Fig. 4a) and with focal adhesions identified by paxillin immunostaining (Fig. 4b) in WT/WT cells. In K255E/K255E fibroblasts, however, α-actinin-4 showed no association with focal adhesions, and, in most cells, was found associated with dense actin-containing aggregates. This actin aggregation behavior resembles the properties of mutant α-actinin-4 in the in vitro assays. We observed similar aggregates in K255E/K255E podocytes derived from the same mouse model. Coimmunostainings with antibodies against other actin-binding proteins expressed in podocytes, including cortactin and synaptopodin, showed that these proteins were also at least partially mislocalized into the α-actinin-4 aggregates (SI Fig. 8).
Fig. 4.
Endogenous K255E mutant α-actinin-4 induces cytoplasmic F-actin aggregates in immortalized lung fibroblasts and is absent from peripheral stress fibers and focal adhesions. (a) Representative images of fibroblasts isolated from Actn4WT/WT and Actn4K255E/K255E mice. Cells were immunostained with anti-α-actinin-4 (green) and rhodamine-phalloidin (red). Images were merged; areas of colocalization appear yellow. Arrowheads point to expression of α-actinin-4 along peripheral actin stress fibers in WT/WT cells. In K255E/K255E cells, peripheral expression of α-actinin-4 is notably diminished, and arrows point to F-actin aggregates. (b) Representative images of fibroblasts isolated from Actn4WT/WT and Actn4K255E/K255E mice. Cells were immunostained with anti-α-actinin-4 (green) and anti-paxillin (red), and images were merged. Arrowheads point to colocalization of α-actinin-4 with paxillin at focal adhesions in WT/WT cells. In K255E/K255E cells, α-actinin-4 is absent from focal adhesions and the cell periphery and forms cytoplasmic aggregates (arrows). (Scale bars, 25 μm.)
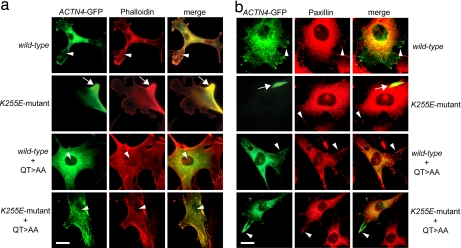
We investigated the effect of an inactivation of ABS1 on the cellular localization of K255E mutant and WT α-actinin-4. We introduced the QT>AA mutation into WT and K255E mutant ACTN4–GFP constructs and examined the localization of α-actinin-4 after transfection into Actn4-deficient murine fibroblasts (Fig. 5). Coimmunostainings with phalloidin (Fig. 4a) and paxillin (Fig. 5b) showed that WT α-actinin-4 localizes with focal adhesions and along actin stress fibers, whereas K255E mutant protein accumulates in large aggregates, as previously published (13), and is completely absent from focal adhesions and the distal portions of stress fibers. However, both K255E mutant and WT α-actinin-4-GFP containing the ABS1 mutation showed no aggregate formation and exhibited a distribution similar to that of the WT construct.
Fig. 5.
Actn4-deficient fibroblasts expressing WT or K255E mutant ACTN4–GFP containing the QT>AA mutation exhibit normal distribution of α-actinin-4–GFP. (a) Actn4-deficient fibroblasts were transfected with WT or K255E mutant ACTN4–GFP (green) without and with the QT>AA mutation and were stained with rhodamine–phalloidin (red). Images were merged; areas of colocalization appear yellow. Arrowheads point to normal distribution of α-actinin-4–GFP along actin stress fibers in cells expressing WT, WT+QT>AA, and K255E mutant+QT>AA ACTN4 constructs. In contrast, K255E mutant-expressing cells show α-actinin-4 staining associated exclusively with a large peripheral aggregate that also exhibits strong phalloidin staining, whereas F-actin stress fibers are absent. (b) Actn4-deficient fibroblasts were transfected with the various ACTN4–GFP constructs (green) and coimmunostained with an antibody against paxillin, a focal adhesion marker (red). All transfected cells exhibit paxillin staining at focal adhesion sites (arrowheads). Cells expressing WT, WT+QT>AA, or K255E mutant+QT>AA ACTN4–GFP showed normal peripheral expression of α-actinin-4 at focal adhesions that colocalized with paxillin staining. In contrast, K255E mutant-expressing cells show α-actinin-4 staining exclusively in a large aggregate (arrows), and α-actinin-4–GFP is absent from focal adhesions in these cells. (Scale bars, 25 μm.)
Discussion
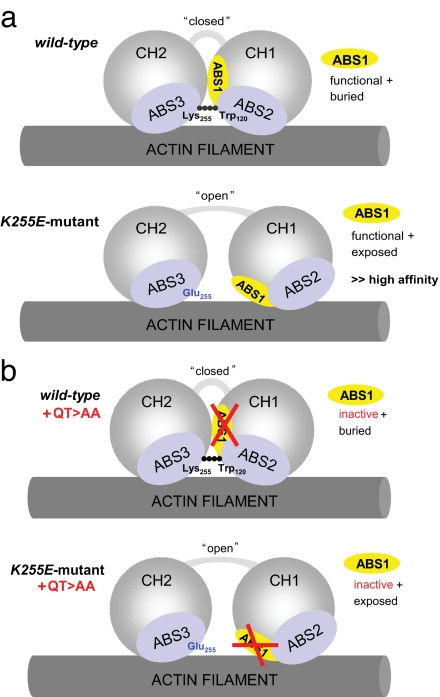
Mutations in α-actinin-4, including a K255E substitution, cause an autosomal dominant form of kidney failure. Our experiments provide evidence that the aggregation of this K255E mutant α-actinin-4 with F-actin is a direct result of its increased actin-binding affinity. We demonstrate that the conformational change also leads to an increase of the actin-binding stoichiometry of the protein as well as a resistance to regulation by Ca2+. The lack of sensitivity to Ca2+ might be mediated either by an inability to bind Ca2+ or by a different conformation that does not accommodate the structural changes necessary for the Ca2+-mediated effect. All of these effects result from a single point mutation in the ABD of the protein and lead to altered structure and biomechanical behavior of F-actin networks in vitro, likely accounting for the changes in α-actinin-4/F-actin distribution observed in cells. Given the structural data available at this point, the results suggest that the K255E mutation, and, given the similar disease phenotype, the other four disease-causing point mutations located in the CH1–CH2 interface region (see Fig. 2), all result in a structural alteration with higher propensity to an open conformation of the ABD, leading to full exposure of ABS1 (Fig. 6). Whether such a conformational change requires an interaction with F-actin remains to be determined.
Fig. 6.
Schematic illustration of the binding of the CH domains of WT and K255E mutant α-actinin-4 in open or closed conformation to an actin filament involving ABS1 to -3. (a) In the WT protein, the CH domains can adopt a closed conformation in which ABS1 (yellow) is buried between the CH domains. ABS2 and -3 (light blue) mainly account for F-actin binding in this conformation. The Lys-255–Trp-147 hinge-like connection contributes to keep the CH domains in a closed conformation. In the mutant protein, because of a Lys>Glu amino acid exchange at amino acid 255 (mutation shown in blue), the connection with Trp-147 is lost and the two CH domains adopt an “open” conformation. ABS1 is exposed and extends the actin-binding surface together with ABS2 and -3. (b) Inactivation of ABS1 (illustrated by the red crossed bars) by the QT>AA mutation does not affect actin binding of the WT protein because ABS1 does not contribute to actin binding in the closed conformation. In the K255E mutant, however, in which ABS1 is exposed because of the open conformation of the CH domains, ABS1 inactivation returns the actin-binding affinity back to that of the WT protein.
The relatively stable association of K255E mutant α-actinin-4 with F-actin and the effect of the mutant on the structure of the actin cytoskeleton are likely to adversely affect cell behavior (e.g., by interfering with actin dynamics and formation of actin-based contractile structures, which potentially results in significant adhesion and/or motility defects). Of note, we identified other actin-binding proteins in the aggregates, many of them known interactors of α-actinin-4 and of particular importance in podocytes. It is possible that a mislocalization of these proteins contributes to the phenotype, e.g., by disrupting signaling cascades important for actin reorganization or assembly (27).
Our findings give rise to interesting speculations about a possible physiological role of a transient exposure of ABS1 in the modulation of the actin-binding affinity of α-actinin. It is well established that Ca2+ and phosphoinositides regulate the affinity of α-actinin to F-actin. A switch between an open and closed conformation of the two CH domains, especially upon interaction with F-actin, could function as an additional, independent mechanism for affinity regulation. It will be interesting to identify factors leading to the conformational switch, not only in α-actinins but possibly also in other CH domain proteins. Illuminating such a pathway might not only help us understand the mechanism of disease but might also lead to the discovery of compounds that could be widely used experimentally for studying the regulation of actin-based processes in the cell.
In light of the large differences in biochemical behavior between WT and K255E mutant α-actinin-4 in vitro, the kidney-limited pathology is striking, although further research may reveal additional, nonrenal phenotypic effects. α-Actinin-4 is highly abundant in these specialized and terminally differentiated glomerular epithelial cells that are part of the glomerular filtration barrier (12). A highly organized actin cytoskeleton enables them to respond dynamically to forces generated by the hydrostatic pressure of the glomerular capillaries necessary for maintaining glomerular filtration (28). We believe that the dynamic actin-binding protein α-actinin-4 is a key regulator of actin filament reorganization in podocytes. Hence, this particular cell type may be especially vulnerable to an alteration of the actin-binding properties of α-actinin-4. One early pathologic finding in patients with ACTN4 mutations is podocyte foot process effacement at the basement membrane (12), a process known to be accompanied by disruption and redistribution of the actin cytoskeleton (29). In addition, mice homozygous for the K255E mutation in α-actinin-4 show a characteristic glomerular lesion characterized by capillary collapse (unpublished data), a phenotype that we also observe in kidneys of α-actinin-4-deficient mice (10). Thus, we believe that the phenotype observed in K255E/K255E mice, similar to that of Actn4−/− mice, might be a result of decreased podocyte adhesion because of the absence of α-actinin-4 at focal adhesions. K255E/K255E mice exhibit large F-actin/α-actinin-4 aggregates in their podocytes that are likely a result of the adynamic interaction of the mutant protein with F-actin. Although we see similar aggregates in podocytes of human individuals with disease, all heterozygous for one of the mutations, our studies do not yet prove that this is the definitive and sole disease-causing mechanism in these patients. However, there is a strong possibility that the presence of high-affinity mutant α-actinin-4 leads to entrapment of α-actinin-4 in adynamic aggregates with actin, thereby directly perturbing the intricate structural organization of the podocyte actin cytoskeleton, resulting in a gain-of-function phenotype that is not apparent until later in life. Mechanisms by which sequestration of mutant α-actinin-4 into cytoplasmic aggregates might lead to podocyte injury and to a loss-of-function phenotype in animals homozygous for the mutation (i.e., by resulting in a deficiency of α-actinin-4 at focal adhesions or by keeping it or its binding partners form entering the nucleus and affecting transcriptional activity) are certainly possible.
Materials and Methods
Generation of Recombinant Proteins.
Baculovirus-expressed recombinant K255E mutant and WT α-actinin-4 proteins were produced by ProteinOne (College Park, MD). In brief, the full-length human ACTN4 cDNA sequence was subcloned into FastBac (Invitrogen, Carlsbad, CA), transfected into SF21 insect cells, and titrated and amplified to an appropriate concentration; this was followed by miniscale infection, purification, and analysis by Western blot. The selected virus was amplified, and a cell extract of the cultured virus was prepared. The proteins were affinity purified. Molecular weight and immunoreactivity were confirmed by SDS/PAGE, Coomassie blue staining, and immunoblotting.
Electron Microscopy of Recombinant Proteins.
The morphology of recombinant α-actinin-4 and the structures formed by mixtures of 1 μM actin with α-actinin-4 at 1:10 were studied with a JEOL 1010 transmission electron microscope (JEOL, Tokyo, Japan) by negative staining and low-angle rotary shadowing according to established protocols (30). The thickness of the actin bundles was measured in a blinded fashion on 10 randomly selected locations on four images each for WT and mutant α-actinin-4.
Generation of GST Fusion Proteins.
Fragments encoding the ABD of α-actinin-4 (amino acids 1–271) were generated by PCR from WT and K255E mutant α-actinin-4 cDNAs, subcloned into pGEX-4T1-HTa and sequence verified. GST purification and protein cleavage were carried out according as described previously (31). Site-directed mutagenesis of ABS1 was performed by using the QuikChange kit (Stratagene, La Jolla, CA) according to the instructions of the manufacturer.
Actin Filament Sedimentation Assays.
Recombinant ABDs were mixed with 5 μM G-actin in a solution containing 20 mM Tris·HCl, pH 7.4, 0.5 mM ATP, 2 mM MgCl2, 120 mM NaCl, and 0.5 mM EGTA for 1 h at room temperature. For the Ca2+-sensitivity assay, CaCl2 was added to a final concentration of 2 mM. For determining the dissociation constant (Kd), 5 μM G-actin was incubated with increasing concentrations of crosslinker. F-Actin was sedimented at 100,000 × g for 30 min at 22°C. Proteins in supernatants and pellets were solubilized in equal amounts of SDS sample buffer, boiled, and subjected to SDS/PAGE. Protein bands were visualized by Coomassie blue staining, and band intensities were quantified by using ImageJ software (National Institutes of Health, Bethesda, MD) (n = 2). Nonlinear regression analysis for one-site binding (hyperbola), Scatchard analysis, calculation of Kd, and 95% confidence intervals were performed by using Prism version 4.0 (GraphPad, San Diego, CA).
Measurement of Apparent Viscosity and Gelation.
Apparent viscosity and gelation were determined by using a miniature falling ball viscometer as described previously (31). G-Actin (24 μM) was mixed with increasing concentrations (1:20 to 1:2,000) of α-actinin-4 without gelsolin and in the presence of increasing gelsolin concentrations. Nonlinear regression (curve fit) (n = 2, error bars = SEM) was performed by using Prism version 4.0 (GraphPad).
Cell Culture, Transfection, and Immunocytochemistry.
Immortalized lung fibroblasts from Actn4WT/WT, Actn4K255E/K255E, and Actn4−/− mice were obtained as described by Yao et al. (14). Cells were grown on collagen I-coated coverslips (BD Biosciences, Palo Alto, CA). Full-length human ACTN4 cDNA was subcloned into pEGFP-N1, and the ABS1 mutation was introduced by using the QuikChange Kit (Stratagene). Constructs were sequence verified. Transfection with pEGFP constructs was carried out by using Fugene6 transfection reagent (Roche, Indianapolis, IN). After 48 h, cells were fixed with 4% PFA, permeabilized in 0.3% Triton, and stained with 1° antibodies [polyclonal anti-α-actinin-4 (12)], mouse monoclonal anti-paxillin (Transduction Laboratories, Franklin Lakes, NJ), mouse monoclonal anti-synaptopodin G1 (ProGen, Heidelberg, Germany), mouse monoclonal anti-cortactin (Upstate, Lake Placid, NY), and Cy2- or Cy3-conjugated 2° antibodies (The Jackson Laboratory, Bar Harbor, ME) or rhodamine-phalloidin (Sigma, St. Louis, MO). Stainings were analyzed with an Eclipse 80i fluorescence microscope (Nikon, Tokyo, Japan) by using a ×100/1.30 oil immersion objective. Images were obtained with an RT Monochrome Spot cooled CCD camera (Diagnostics Instruments, Sterling Heights, MI). Postacquisition processing was performed by using Photoshop software (Adobe, San Jose, CA).
Structural Models.
Structural models of the α-actinin ABD that are shown in this report are based on published crystal structures (17, 32). Structural images were generated by using the PyMol software (DeLano Scientific, Palo Alto, CA).
Supplementary Material
Acknowledgments
We thank Roberto Dominguez and Jagesh Shah for helpful discussions. This work was supported by a National Kidney Foundation Research Fellowship Award (A.W.); National Institutes of Health (NIH) Training Grants T32-DK007527-20 and F32-DK074308-01 (to J.S.S.); NIH Grants DK59588 (to M.R.P.) and HL19429 (to T.P.S.); and the American Cancer Society (T.P.S.). M.R.P. is an Established Investigator of the American Heart Association.
Abbreviations
- ABD
actin-binding domain
- ABS
actin-binding site
- CH
calponin homology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702451104/DC1.
References
- 1.Maruyama K, Ebashi S. J Biochem (Tokyo) 1965;58:13–19. doi: 10.1093/oxfordjournals.jbchem.a128158. [DOI] [PubMed] [Google Scholar]
- 2.Mimura N, Asano A. Nature. 1978;272:273–276. doi: 10.1038/272273a0. [DOI] [PubMed] [Google Scholar]
- 3.Pelletier O, Pokidysheva E, Hirst LS, Bouxsein N, Li Y, Safinya CR. Phys Rev Lett. 2003;91:148102. doi: 10.1103/PhysRevLett.91.148102. [DOI] [PubMed] [Google Scholar]
- 4.Wachsstock DH, Schwarz WH, Pollard TD. Biophys J. 1994;66:801–809. doi: 10.1016/s0006-3495(94)80856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witke W, Schleicher M, Noegel AA. Cell. 1992;68:53–62. doi: 10.1016/0092-8674(92)90205-q. [DOI] [PubMed] [Google Scholar]
- 6.Otey CA, Carpen O. Cell Motil Cytoskeleton. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 7.Honda K, Yamada T, Hayashida Y, Idogawa M, Sato S, Hasegawa F, Ino Y, Ono M, Hirohashi S. Gastroenterology. 2005;128:51–62. doi: 10.1053/j.gastro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Yan Q, Sun W, Kujala P, Lotfi Y, Vida TA, Bean AJ. Mol Biol Cell. 2005;16:2470–2482. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajfur Z, Roy P, Otey C, Romer L, Jacobson K. Nat Cell Biol. 2002;4:286–293. doi: 10.1038/ncb772. [DOI] [PubMed] [Google Scholar]
- 10.Dandapani SV, Sugimoto H, Matthews BD, Kolb RJ, Sinha S, Gerszten RE, Zhou J, Ingber DE, Kalluri R, Pollak MR. J Biol Chem. 2007;282:467–477. doi: 10.1074/jbc.M605024200. [DOI] [PubMed] [Google Scholar]
- 11.Geiger B, Bershadsky A, Pankov R, Yamada KM. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 12.Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodriguez-Perez JC, Allen PG, Beggs AH, et al. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 13.Weins A, Kenlan P, Herbert S, Le TC, Villegas I, Kaplan BS, Appel GB, Pollak MR. J Am Soc Nephrol. 2005;16:3694–3701. doi: 10.1681/ASN.2005070706. [DOI] [PubMed] [Google Scholar]
- 14.Yao J, Le TC, Kos CH, Henderson JM, Allen PG, Denker BM, Pollak MR. PLoS Biol. 2004;2:e167. doi: 10.1371/journal.pbio.0020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J, Taylor DW, Taylor KA. J Mol Biol. 2001;310:845–858. doi: 10.1006/jmbi.2001.4789. [DOI] [PubMed] [Google Scholar]
- 16.Franzot G, Sjoblom B, Gautel M, Djinovic Carugo K. J Mol Biol. 2005;348:151–165. doi: 10.1016/j.jmb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Borrego-Diaz E, Kerff F, Lee SH, Ferron F, Li Y, Dominguez R. J Struct Biol. 2006;155:230–238. doi: 10.1016/j.jsb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Alvarez B, Bobkov A, Sonnenberg A, de Pereda JM. Structure (London) 2003;11:615–625. doi: 10.1016/s0969-2126(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 19.McGough A, Way M, DeRosier D. J Cell Biol. 1994;126:433–443. doi: 10.1083/jcb.126.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Djinovic-Carugo K, Young P, Gautel M, Saraste M. Cell. 1999;98:537–546. doi: 10.1016/s0092-8674(00)81981-9. [DOI] [PubMed] [Google Scholar]
- 21.Wachsstock DH, Schwartz WH, Pollard TD. Biophys J. 1993;65:205–214. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araki N, Hatae T, Yamada T, Hirohashi S. J Cell Sci. 2000;113(Pt 18):3329–3340. doi: 10.1242/jcs.113.18.3329. [DOI] [PubMed] [Google Scholar]
- 23.Bennett JP, Zaner KS, Stossel TP. Biochemistry. 1984;23:5081–5086. doi: 10.1021/bi00316a039. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura F, Osborn E, Janmey PA, Stossel TP. J Biol Chem. 2002;277:9148–9154. doi: 10.1074/jbc.M111297200. [DOI] [PubMed] [Google Scholar]
- 25.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 26.Yin HL, Hartwig JH, Maruyama K, Stossel TP. J Biol Chem. 1981;256:9693–9697. [PubMed] [Google Scholar]
- 27.Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, Mundel P. Nat Cell Biol. 2006;8:485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- 28.Pavenstadt H, Kriz W, Kretzler M. Physiol Rev. 2003;83:253–307. doi: 10.1152/physrev.00020.2002. [DOI] [PubMed] [Google Scholar]
- 29.Oh J, Reiser J, Mundel P. Pediatr Nephrol. 2004;19:130–137. doi: 10.1007/s00467-003-1367-y. [DOI] [PubMed] [Google Scholar]
- 30.Hartwig JH, Stossel TP. J Mol Biol. 1981;145:563–581. doi: 10.1016/0022-2836(81)90545-3. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura F, Hartwig JH, Stossel TP, Szymanski PT. J Biol Chem. 2005;280:32426–32433. doi: 10.1074/jbc.M502203200. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Taylor DW, Taylor KA. J Mol Biol. 2004;338:115–125. doi: 10.1016/j.jmb.2004.02.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.